Abstract
We describe a case of Solobacterium moorei bacteremia in a 43-year-old woman presenting with acute proctitis complicating radiotherapy for cervical carcinoma. Phenotypic tests failed to identify the bacterium, which was subsequently identified by 16S rRNA gene sequencing. 16S rRNA gene sequencing could help better define the pathogenicity of S. moorei.
CASE REPORT
A 43-year-old Philippine woman was admitted for radiotherapy of stage IIIB carcinoma of the cervix. Following teleradiotherapy, she received four cycles of intracavitary brachytherapy. Five days after the first cycle of intracavitary brachytherapy, she complained of fever, chills, and rigor associated with vomiting, lower abdominal and anal pain, and watery diarrhea. Her oral temperature was 39°C. Blood was present on rectal examination. Her total leukocyte count was 5.9× 109/liter, her hemoglobin level was 11.2 g/dl, and her platelet count was 521 × 109/liter. Her serum urea was 3.6 mmol/liter, her creatinine was 131 μmol/liter, her albumin was 40 g/liter, her total globulin was 42 g/liter, her bilirubin was 6 μmol/liter, her alkaline phosphatase was 427 IU/liter, her aspartate aminotransferase was 233 IU/liter, and her alanine aminotransferase was 180 IU/liter. Two sets of blood cultures were performed. Empirical intravenous cefuroxime was commenced. Both sets of anaerobic blood cultures were positive for a short, gram-positive bacillus. Piperacillin-tazobactam was subsequently given for 2 weeks. Her fever and diarrhea responded, and brachytherapy was continued.
Anaerobic gram-positive bacilli are a heterogeneous group of bacteria that are difficult to identify in clinical microbiology laboratories. As a result, the clinical significance of many species is poorly understood. Since the recognition of the 16S rRNA gene as the standard for classification and identification of bacteria (11, 12), these bacteria can be accurately identified and revisions in classification and introductions of new genera and species can be made (1a, 10, 14, 16). Recently, we have reported the use of this technique for identifying and defining the clinical significance of these bacteria and the discovery of three novel species (6-8, 17-20).
The genus Solobacterium was first described in 2000, when three anaerobic, gram-positive, non-spore-forming bacillus strains were characterized in Japan (4). These three Eubacterium-like strains were isolated from human feces. On the basis of 16S rRNA gene sequence analysis, they were classified under a new genus, Solobacterium, as Solobacterium moorei, the only species in the genus. S. moorei was later found in infected root canals of patients with endodontic infections by cloning of 16S rRNA genes (13). During a subsequent study using a similar technique, it was also identified from tongue dorsum scrapings and subgingival plaque samples and, together with several other bacteria, found to be associated with halitosis (5). However, the clinical significance of this rarely encountered bacterium remains largely unknown and there have been no reports of Solobacterium bacteremia or invasive infections in humans in the literature.
Microbiological data.
The BACTEC 9240 blood culture system (Becton Dickinson) was used. All isolates were identified by standard conventional biochemical methods (3, 9), the Vitek System (ANI; bioMérieux Vitek), and the API 20A system (bioMérieux Vitek, Hazelwood, Mo.). Antimicrobial susceptibility was tested by the E-test (AB Biodisk, Solna, Sweden) according to the CLSI criteria for anaerobic bacteria (1). On day 2 postincubation, both sets of anaerobic blood culture bottles turned positive for a short, gram-positive bacillus. It grew slowly on anaerobic blood agar as grayish white, nonhemolytic colonies that were 0.5 mm in diameter after 72 h of incubation at 37°C in an anaerobic environment. It did not grow in a 5% CO2 aerobic or microaerophilic environment. It was nonmotile, and spores were not found. It was positive for gelatin hydrolysis, α-galactosidase, α-glucosidase, leucine arylamidase, and proline arylamidase but negative for catalase and indole hydrolysis. The Vitek system (ANI) indicated that it was unidentified. The API 20A system showed that it was 53.2% likely to be a Clostridium sp., 32.6% likely to be Clostridium histolyticum, and 7.9% likely to be a Peptostreptococcus sp. The MIC of vancomycin was 0.5 μg/ml, that of penicillin was 0.012 μg/ml, that of metronidazole was 0.25 μg/ml, and that of cefotaxime was 0.032 μg/ml.
16S rRNA gene sequencing and phylogenetic characterization.
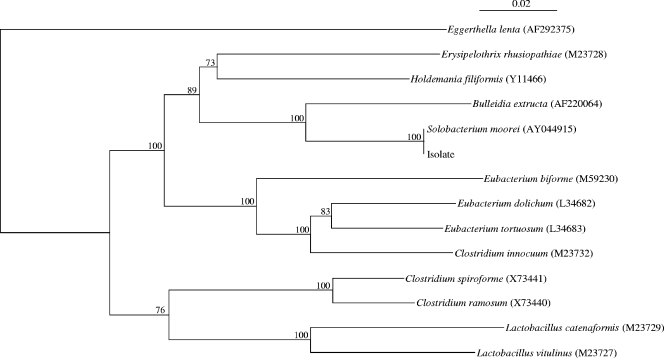
PCR amplification and DNA sequencing of the 16S rRNA gene of the isolate were performed as described previously (8, 21). LPW57 (5′-AGTTTGATCCTGGCTCAG-3′) and LPW205 (5′-CTTGTTACGACTTCACCC-3′) (Gibco BRL, Rockville, MD) were used as the PCR and sequencing primers. The sequences of the PCR products were compared with known 16S rRNA gene sequences in the GenBank database by BLAST search and multiple sequence alignment with the closest matches performed with the ClustalW program (15). Phylogenetic tree construction was performed with ClustalX version 1.81 (2) and by the neighbor-joining method with GrowTree (Genetics Computer Group, Inc., San Diego, CA.). A total of 1,303 nucleotide positions were included in the analysis. PCR of the 16S rRNA gene of the isolate showed a band at about 1,430 bp. The 16S rRNA gene sequence of the isolate had no nucleotide difference from that of S. moorei (GenBank accession no. AY044915); 8% nucleotide difference from that of Bulleidia extructa (GenBank accession no. AF220064), and 12% nucleotide difference from that of Holdemania filiformis (GenBank accession no. Y11466), indicating that the isolate was a strain of S. moorei (Fig. 1).
FIG. 1.
Phylogenetic tree showing the relationships of the blood culture isolate to an S. moorei isolate and members of other, related genera. The tree was constructed by using the neighbor-joining method and bootstrap values calculated from 1,000 trees. The scale bar indicates the estimated number of substitutions per 100 bases using the Jukes-Cantor correction. The accession numbers shown are those in the GenBank database.
We describe a case of S. moorei bacteremia associated with acute proctitis complicating radiotherapy for carcinoma of the cervix. The clinical significance of the isolate is evident by its isolation from blood in association with the development of fever, lower abdominal pain, and diarrhea and the prompt response to antibiotic therapy. The source was likely the gastrointestinal tract, as the patient had symptoms of acute proctitis during the bacteremic episode and S. moorei has been found in human feces (4). We speculate that the bacterium entered the bloodstream by translocation through the inflamed intestinal mucosa. However, it cannot be determined if the bacterium contributed to the proctitis. Nevertheless, the isolation of S. moorei from blood suggests that the bacterium can be of clinical significance instead of being normal gut flora only.
The variations in biochemical profiles among different strains of S. moorei make identification of the bacterium, and hence understanding of its epidemiology and clinical disease association, difficult. Both the present and previously described strains are obligately anaerobic, non-spore-forming, Eubacterium-like, gram-positive bacilli and are indole negative. They grow slowly and produce few positive biochemical reactions in commercially available identification kits. Moreover, the phenotypic characteristics of the present isolate exhibited considerable differences from those described for S. moorei (Table 1), which are likely due to interstrain variations. Isolates of S. moorei reported in the literature, including the present one, were all identified by 16S rRNA gene sequencing. This tech- nique is more reliable for identification of S. moorei and would help us better understand its epidemiology and disease spectrum.
TABLE 1.
Phenotypic characteristics of the blood culture isolate and three S. moorei isolates reported in the literature
| Phenotypic characteristic | S. mooreia | Blood culture isolate |
|---|---|---|
| Catalase | − | |
| Gelatin hydrolysis | − | + |
| Arginine dehydrogenase | − | |
| Alkaline phosphatase | − | |
| Indole production | − | − |
| Phosphate choline | − | |
| Urease | − | |
| Reduction of nitrate | − | |
| Reduction of triphenyltetrazolium | − | |
| H2S production | − | |
| Oxidation or fermentation of: | ||
| Arabinose | − | − |
| Cellobiose | − | − |
| Fructose | + | |
| Galactose | + | − |
| Glucose | + | − |
| Glycogen | − | − |
| Inositol | − | |
| Lactose | − | − |
| Maltose | + | − |
| Mannitol | − | − |
| Mannose | − | − |
| Melezitose | − | − |
| Raffinose | − | − |
| Rhamnose | − | − |
| Ribose | + | − |
| Salicin | − | − |
| Sorbitol | − | − |
| Sucrose | − | − |
| Trehalose | − | − |
| Xylose | − | − |
| Starch | − | |
| α-Arabinosidase | − | |
| α-Fucosidase | − | |
| β-Fucosidase | − | |
| α-Galactosidase | + | |
| β-Galactosidase | − | |
| α-Glucosidase | + | |
| β-Glucosidase | − | |
| β-Glucuronidase | − | |
| α-Mannosidase | − | |
| β-Lactosidase | − | |
| β-Xylosidase | − | |
| N-Acetyl-glucosaminidase | − | |
| Alanine arylamidase | − | |
| Glutamyl glutamic acid arylamidase | − | |
| Leucine arylamidase | + | |
| Lysine arylamidase | − | |
| Proline arylamidase | + |
The data shown are from reference 4.
Acknowledgments
This work was partly supported by the University Development Fund, a University Research Grant Council grant (HKU 7236/02 M), and a Committee of Research and Conference grant from The University of Hong Kong.
REFERENCES
- 1.Clinical and Laboratory Standards Institute. 2003. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard M11-A5, 5th ed. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 1a.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 2.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with ClustalX. Trends Biochem. Sci. 10:403-405. [DOI] [PubMed] [Google Scholar]
- 3.Jousimies-Somer, H. R., P. Summanen, D. M. Citron, E. J. Baron, H. M. Wexler, and S. M. Finegold. 2002. Wadsworth anaerobic bacteriology manual, 6th ed. Star Publishing, Belmont, Calif.
- 4.Kageyama, A., and Y. Benno. 2000. Phylogenic and phenotypic characterization of some Eubacterium-like isolates from human feces: description of Solobacterium moorei gen. nov., sp. nov. Microbiol. Immunol. 44:223-227. [DOI] [PubMed] [Google Scholar]
- 5.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41:558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau, S. K. P., K. H. L. Ng, P. C. Y. Woo, K. T. Yip, A. M. Y. Fung, G. K. S. Woo, K. M. Chan, T. L. Que, and K. Y. Yuen. 2006. Usefulness of the MicroSeq 500 16S rDNA bacterial identification system for identification of anaerobic gram-positive bacilli isolated from blood cultures. J. Clin. Pathol. 59:219-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau, S. K. P., P. C. Y. Woo, A. M. Y. Fung, K. M. Chan, G. K. S. Woo, and K. Y. Yuen. 2004. Anaerobic, non-sporulating, gram-positive bacilli bacteremia characterized by16S ribosomal RNA gene sequencing. J. Med. Microbiol. 53:1247-1253. [DOI] [PubMed] [Google Scholar]
- 8.Lau, S. K. P., P. C. Y. Woo, G. K. S. Woo, A. M. Y. Fung, K. M. Wong, K. M. Chan, S. F. Tang, and K. Y. Yuen. 2004. Eggerthella hongkongensis sp. nov. and Eggerthella sinensis sp. nov., two novel Eggerthella species, account for half of the cases of Eggerthella bacteremia. Diagn. Microbiol. Infect. Dis. 49:255-263. [DOI] [PubMed] [Google Scholar]
- 9.Murray, P. R., E. J. Baro, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken. 2003. Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 10.Nakazawa, F., M. Sato, S. E. Poco, T. Hashimura, T. Ikeda, S. Kalfas, G. Sundqvist, and E. Hoshino. 2000. Description of Mogibacterium pumilum gen. nov., sp. nov. and Mogibacterium vescum gen. nov., sp. nov., and reclassification of Eubacterium timidum (Holdeman et al. 1980) as Mogibacterium timidum gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 50:679-688. [DOI] [PubMed] [Google Scholar]
- 11.Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tompkins. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N. Engl. J. Med. 323:1573-1580. [DOI] [PubMed] [Google Scholar]
- 12.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 13.Rolph, H. J., A. Lennon, M. P. Riggio, W. P. Saunders, D. MacKenzie, L. Coldero, and J. Bagg. 2001. Molecular identification of microorganisms from endodontic infections. J. Clin. Microbiol. 39:3282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taras, D., R. Simmering, M. D. Collins, P. A. Lawson, and M. Blaut. 2002. Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 52:423-428. [DOI] [PubMed] [Google Scholar]
- 15.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wade, W. G., J. Downes, D. Dymock, S. J. Hiom, A. J. Weightman, F. E. Dewhirst, B. J. Paster, N. Tzellas, and B. Coleman. 1999. The family Coriobacteriaceae: reclassification of Eubacterium exiguum (Poco et al. 1996) and Peptostreptococcus heliotrinreducens (Lanigan 1976) as Slackia exigua gen. nov., comb. nov. and Slackia heliotrinreducens gen. nov., comb. nov., and Eubacterium lentum (Prevot 1938) as Eggerthella lenta gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:595-600. [DOI] [PubMed] [Google Scholar]
- 17.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, G. Y. Wong, and K. Y. Yuen. 2002. Diagnosis of pelvic actinomycosis by 16S ribosomal RNA gene sequencing and its clinical significance. Diagn. Microbiol. Infect. Dis. 43:113-118. [DOI] [PubMed] [Google Scholar]
- 18.Woo, P. C. Y., A. M. Y. Fung, S. K. P. Lau, J. L. L. Teng, B. H. L. Wong, M. K. M. Wong, E. Hon, G. W. K. Tang, and K. Y. Yuen. 2003. Actinomyces hongkongensis sp. nov. A novel Actinomyces species isolated from a patient with pelvic actinomycosis. Syst. Appl. Microbiol. 26:518-522. [DOI] [PubMed] [Google Scholar]
- 19.Woo, P. C. Y., S. K. P. Lau, G. K. S. Woo, A. M. Y. Fung, V. P. Y. Yiu, and K. Y. Yuen. 2004. Clostridium hathewayi bacteremia in a patient with acute appendicitis. J. Clin. Microbiol. 42:5947-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woo, P. C. Y., S. K. P. Lau, K. M. Chan, A. M. Y. Fung, B. S. F. Tang, and K. Y. Yuen. 2005. Clostridium bacteremia characterized by 16S ribosomal RNA gene sequencing. J. Clin. Pathol. 58:301-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuen, K. Y., P. C. Y. Woo, J. L. L. Teng, K. W. Leung, M. K. M. Wong, and S. K. P. Lau. 2001. Laribacter hongkongensis gen. nov., sp. nov., a novel gram-negative bacterium isolated from a cirrhotic patient with bacteremia and empyema. J. Clin. Microbiol. 39:4227-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]