Abstract
We identified genotypes of Enterocytozoon bieneusi from 33 stool samples of Thai human immunodeficiency virus (HIV)-infected adult patients. Genotype D was identified at the highest frequency (36.4%), while genotype E was the second most common (15.1%). Genotypes O and PigEBITS 7, previously found only in pigs, were observed in Thai HIV-infected patients. Phylogenetic analysis supported a zoonotic nature for E. bieneusi.
Human infections from microsporidia have been reported globally, with the majority of cases involving human immunodeficiency virus (HIV)-infected patients. The most widely distributed among humans is Enterocytozoon bieneusi, causing diarrhea and weight loss in immunocompromised patients (6). In Thailand, E. bieneusi genotype A was identified in HIV-negative and HIV-positive children in an orphanage (9). Information on genotypes of E. bieneusi infection in HIV-positive adult patients has never been studied. In this study, we used the 243-bp internal transcribed spacer (ITS) region of the rRNA genes for genotypic characterization of E. bieneusi in stool specimens of Thai HIV-infected adults.
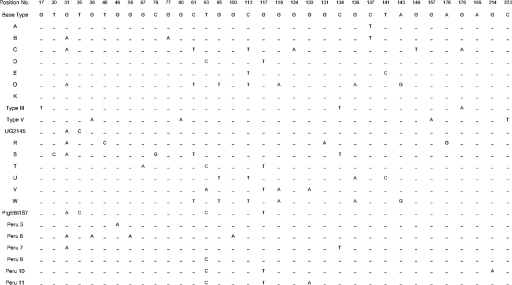
Thirty-three positive E. bieneusi stool specimens collected during 1999 to 2002 at Siriraj Hospital, Bangkok, Thailand, were analyzed. All specimens were examined for microsporidia with Gram-chromotrope staining as previously described (13) and screened for E. bieneusi using electron microscopy (26). DNA extraction was performed using FTA filter paper (Whatman Bioscience) (19). The E. bieneusi rRNA genes were amplified using MSP3 and MSP4B primers, which amplify a 508-bp fragment containing 122 bp of the 3′ end of the small-subunit rRNA gene, 243 bp of the ITS, and 143 bp of the 5′ region of the large-subunit rRNA gene (8). The purified PCR product was cloned using the TOPO TA Cloning kit (Invitrogen), following the manufacturer's instructions. Direct DNA sequencing of the PCR products as well as cloning PCR products prior to sequencing validated the accuracy of nucleotide sequences. The genotype of E. bieneusi from each specimen was classified by the homology of the published sequences of alignments performed by Clustal X 1.83 for Windows (24). Figure 1 shows 33 polymorphic sites in the E. bieneusi ITS rRNA genes from 23 representative sequences of E. bieneusi isolated from humans, compared with nucleotides of the consensus sequence. Among the 33 stool samples, 24 exhibited 5 previously identified genotypes (D, E, O, Peru 11, and PigEBITS7). Six new genotypes (R, S, T, U, V, and W) were observed. Genotype D was the most common with 36.4% (12 of 33), followed by genotype E with 15.2% (5 of 33). Genotypes PigEBITS 7 and S were each identified for four patients (12.1%), while genotype Peru-11 was identified for two patients (6.1%). Additionally, genotypes O, R, T, U, V, and W were each found in one patient (3%). No mixed infections of E. bieneusi genotypes were found in this study based on the results from sequences of clone PCR products.
FIG. 1.
Summary of the polymorphic sites of the ITS region of the rRNA genes of 23 genotypes of Enterocytozoon bieneusi reported for humans (2, 11, 16, 22, 25; this study).
Information compiled in previous studies indicated that the genotypes of E. bieneusi may differ in their geographical distributions (1, 4, 10, 11, 14, 17). Genotype B was the most common genotype reported for HIV-positive patients in France (10, 11), Germany (14, 4), Switzerland (1), and also the United Kingdom (17), while the less-common genotypes were A, C, D, and K, respectively. In South America, the most-common genotypes were Peru-1 (or genotype A) and Peru-2 (or genotype K) for HIV-positive patients in Peru (20). In contrast, genotype K was the most prevalent reported for HIV-negative children in Uganda (25). The observation of predominant genotypes found in different geographical areas suggests differences in the epidemiology of E. bieneusi infection. Further studies utilizing greater numbers of infected patients will be needed to confirm this hypothesis.
All four E. bieneusi genotypes E, D, O, and PigEBITS7 were not host specific and could infect both humans and pigs. It has been shown that genotype D was not only reported for HIV positive patients but also identified and named differently for varieties of animals, i.e., pigs (PigEBITS9: AF348477) (2), macaques (WL13: AY237221) (3, 7), beavers, foxes, muskrats, and raccoons (WL8: AY237216) (21). It was found that 30 to 35% of pigs were infected with previously reported E. bieneusi genotypes D and F and nine new genotypes, EBITS1 to EBITS9 (2). Genotype E was reported for HIV-positive patients in Germany (4) and in Switzerland (1). However, the sequence of genotype E was identical to those previously described for pigs (EbpC: AF076042) (1) and those found in beavers, muskrats, raccoons, otters, and foxes (WL13: AY237221) (21). Our study describes the first observation of genotype O and PigEBITS7 for Thai HIV-positive patients. Both genotypes O and PigEBITS7 were previously found in pigs in Germany (4) and in the United States (2), respectively.
The 243-nucleotide positions of the ITS sequences from E. bieneusi of human genotypes and some animal genotypes were aligned using Clustal X 1.83 for Windows (10, 11, 17, 20, 25, 1, 2, 4, 14, 16, 21, 22). The inferred phylogenetic tree in Fig. 2 was constructed by the neighbor-joining analysis (18) of PAUP 4.0 beta 10 (23) to examine the evolutionary relationship between those E. bieneusi isolates. The inferred phylogenetic tree was demonstrated by the TREEVIEW program provided by the Institute of Biomedical and Life Sciences, University of Glasgow, Scotland, United Kingdom. The reliability of the tree topology was determined by bootstrap analysis using 1,000 replicates for the neighbor-joining analysis. The Enterocytozoon sequence from a dog was used as an outgroup taxon (AF059610) (12). The scale bar of phylogenetic tree branch length (3.5 cm) represents 0.1 nucleotide substitutions per site. The inferred NJ tree showed the relatedness of the isolates grouped by sequence identity (18). The isolates included in the same genotype clustered with each other with good bootstrap support. Genotype A of E. bieneusi from humans in Thailand (9) and genotype Peru-1 in Peru (20) were nested with each other. The inferred phylogenetic tree that showed the genetic relatedness between humans and some animal genotypes also shows the likelihood of a zoonotic potential from these isolates. For example, genotype E of E. bieneusi from this report and genotype Peru-4 were clustered together with genotype WL13 of isolates from beavers, muskrats, raccoons, otters, and foxes in the United States (21) and genotype E of isolates from pigs in Germany and Switzerland (5, 1). Genotype O was grouped by E. bieneusi isolates from an HIV-positive patient in this study and E. bieneusi from pigs in Germany (4). The genotype D clade was nested by E. bieneusi isolates from humans in Thailand, Germany (15), and Peru (20) together with genotype WL8 of E. bieneusi from beavers, muskrats, raccoons, and foxes in the United States (21) and the genotype PigEBITS9 of E. bieneusi from pigs in the United States (2).
FIG. 2.
A bootstrap consensus phylogram inferred by the neighbor-joining analysis of the 243 bp of the ITS region of E. bieneusi human genotypes and genotypes in animals showing zoonotic potential. Numbers in bold letters on branches are bootstrapping values greater than 50% using 1,000 replicates. Bold letters represent 11 genotypes identified from HIV-infected patients in Thailand in this study. Names of the isolates and accession numbers in GenBank are shown in parentheses.
Our data supported a zoonotic nature of genotypes E, D, O, and PigEBITS7 of E. bieneusi, in which pigs could be a significant source of transmission to humans, especially for immunocompromised patients. More studies of prevalence surveys of specific genotypes of E. bieneusi in pigs in Thailand should be performed.
Nucleotide sequence accession numbers.
The GenBank accession numbers assigned to the sequences determined in this study are as follows: genotype R, AY945808; genotype S, AY945809; genotype T, AY945810; genotype U, AY945811; genotype V, AY945812; and genotype W, AY945813.
Acknowledgments
This study was financially supported by Thailand-Tropical Disease Research Programme (T-2) (grant no. ID 02-2-ARI-24-007).
REFERENCES
- 1.Breitenmoser, A. C., A. Mathis, E. Burgi, R. Weber, and P. Deplazes. 1999. High prevalence of Enterocytozoon bieneusi in swine with four genotypes that differ from those identified in humans. Parasitology 118:447-453. [DOI] [PubMed] [Google Scholar]
- 2.Buckholt, M. A., J. H. Lee, and S. Tzipori. 2002. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 68:2595-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalifoux, L. V., A. Carville, D. Pauley, B. Thompson, A. A. Lackner, and K. G. Mansfield. 2000. Enterocytozoon bieneusi as a cause of proliferative serositis in simian immunodeficiency virus-infected immunodeficient macaque (Macaca mulatta). Arch. Pathol. Lab. Med. 124:1480-1484. [DOI] [PubMed] [Google Scholar]
- 4.Dengjel, B., M. Zahler, W. Hermanns, K. Heinritzi, T. Spillmann, A. Thomschke, T. Loscher, R. Gothe, and H. Rinder. 2001. Zoonotic potential of Enterocytozoon bieneusi. J. Clin. Microbiol. 39:4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deplazes, P., A. Mathis, C. Muller, and R. Weber. 1996. Molecular epidemiology of Encephalitozoon cuniculi and first detection of Enterocytozoon bieneusi in faecal samples of pigs. J. Eukaryot. Microbiol. 43:93S. [DOI] [PubMed] [Google Scholar]
- 6.Didier, E. S. 2005. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop. 94:61-67. [DOI] [PubMed] [Google Scholar]
- 7.Green, L. C., P. J. Didier, L. C. Bowers, and E. S. Didier. 2004. Natural and experimental infection of immunocompromised rhesus macaques (Macaca mulatta) with the microsporidian Enterocytozoon bieneusi genotype D. Microb. Infect. 6:996-1002. [DOI] [PubMed] [Google Scholar]
- 8.Katzwinkel-Wladarsch, S., M. Lieb, W. Helse, T. Löscher, and H. Rinder. 1996. Direct amplification and species determination of microsporidian DNA from stool specimen. Trop. Med. Int. Health 1:373-378. [DOI] [PubMed] [Google Scholar]
- 9.Leelayoova, S., I. Subrungruang, R. Rangsin, P. Chavalitshewinkoon-Petmitr, J. Worapong, T. Naaglor, and M. Mungthin. 2005. Transmission of Enterocytozoon bieneusi genotype A in a Thai orphanage. Am. J. Trop. Med. Hyg. 73:104-107. [PubMed] [Google Scholar]
- 10.Liguory, O., F. David, C. Sarfati, F. Derouin, and J. M. Molina. 1998. Determination of types of Enterocytozoon bieneusi strains isolated from patients with intestinal microsporidiosis. J. Clin. Microbiol. 36:1882-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liguory, O., C. Sarfati, F. Derouin, and J. M. Molina. 2001. Evidence of different Enterocytozoon bieneusi genotypes in patients with and without human immunodeficiency virus infection. J. Clin. Microbiol. 39:2672-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathis, A., A. C. Breitenmoser, and P. Deplazes. 1999. Detection of new Enterocytozoon genotypes in faecal samples of farm dogs and a cat. Parasite 6:189-193. [DOI] [PubMed] [Google Scholar]
- 13.Moura, H., J. L. da Silva, F. C. Sodre, P. Brasil, D. Walmo, S. Wahlquist, G. P. Croppo, and G. S. Visvesvara. 1996. Gram-chromotrope: a new technique that enhances detection of microsporidial spores in clinical samples. J. Eukaryot. Microbiol. 43:94S-95S. [DOI] [PubMed] [Google Scholar]
- 14.Rinder, H., S. Katzwinkel-Wladarsch, and T. Loscher. 1997. Evidence for the existence of genetically distinct strains of Enterocytozoon bieneusi. Parasitol. Res. 83:670-672. [DOI] [PubMed] [Google Scholar]
- 15.Rinder, H., S. Katzwinkel-Wladarsch, A. Thomschke, and T. Loscher. 1998. Strain differentiation in microsporidia. Tokai J. Exp. Clin. Med. 23:433-437. [PubMed] [Google Scholar]
- 16.Rinder, H., A. Thomschke, B. Dengjel, R. Gothe, T. Luscher, and M. Zahler. 2000. Close genotypic relationship between Enterocytozoon bieneusi from humans and pigs and first detection in cattle. J. Parasitol. 86:185-188. [DOI] [PubMed] [Google Scholar]
- 17.Sadler, F., N. Peake, R. Borrow, P. L. Rowl, E. G. L. Wilkin, and A. Curry. 2002. Genotyping of Enterocytozoon bieneusi in AIDS patients from the North West of England. J. Infect. 44:39-42. [DOI] [PubMed] [Google Scholar]
- 18.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 19.Subrungruang, I., M. Mungthin, P. Chavalitshewinkoon-Petmitr, R. Rangsin, T. Naaglor, and S. Leelayoova. 2004. Evaluation of DNA extraction and PCR methods for detection of Enterocytozoon bieneusi in stool specimens. J. Clin. Microbiol. 42:3490-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulaiman, I. M., C. Bern, R. Gilman, V. Cama, V. Kawai, D. Vargas, E. Ticona, A. Vivar, and L. Xiao. 2003. A molecular biologic study of Enterocytozoon bieneusi in HIV-infected patients in Lima, Peru. J. Eukaryot. Microbiol. 50:S591-S596. [DOI] [PubMed] [Google Scholar]
- 21.Sulaiman, I. M., R. Fayer, A. A. Lal, J. M. Trout, F. W. Schaefer III, and L. Xiao. 2003. Molecular characterization of microsporidia indicates that wild mammals harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 69:4495-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulaiman, I. M., R. Fayer, C. Yang, M. Santin, O. Matos, and L. Xiao. 2004. Molecular characterization of Enterocytozoon bieneusi in cattle indicates that only some isolates have zoonotic potential. Parasitol. Res. 92:328-334. [DOI] [PubMed] [Google Scholar]
- 23.Swofford, D. L., and G. L. Olsen. 1990. Phylogeny reconstruction, p. 411-501. In D. M. Hillis and C. Moritz (ed.), Molecular systematics. Sinauer Associates, Sunderland, Mass.
- 24.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tumwine, J. K., A. Kekitiinwa, N. Nabukeera, D. E. Akiyoshi, M. A. Buckholt, and S. Tzipori. 2002. Enterocytozoon bieneusi among children with diarrhea attending Mulago Hospital in Uganda. Am. J. Trop. Med. Hyg. 67:299-303. [DOI] [PubMed] [Google Scholar]
- 26.Visvesvara, G. S., G. J. Leitch, H. Moura, S. Wallace, R. Weber, and R. T. Bryan. 1991. Culture, electron microscopy, and immunoblot studies on a microsporidian parasite isolated from the urine of a patient with AIDS. J. Protozool. 38:105S-111S. [PubMed] [Google Scholar]