Abstract
Human African trypanosomiasis (HAT) or sleeping sickness is a neglected disease that affects poor rural populations across sub-Saharan Africa. Confirmation of diagnosis is based on detection of parasites in either blood or lymph by microscopy. Here we present the development and the first-phase evaluation of a simple and rapid test (HAT-PCR-OC [human African trypanosomiasis-PCR-oligochromatography]) for detection of amplified Trypanosoma brucei DNA. PCR products are visualized on a dipstick through hybridization with a gold-conjugated probe (oligochromatography). Visualization is straightforward and takes only 5 min. Controls both for the PCR and for DNA migration are incorporated into the assay. The lower detection limit of the test is 5 fg of pure T. brucei DNA. One parasite in 180 μl of blood is still detectable. Sensitivity and specificity for T. brucei were calculated at 100% when tested on blood samples from 26 confirmed sleeping sickness patients, 18 negative controls (nonendemic region), and 50 negative control blood samples from an endemic region. HAT-PCR-OC is a promising new tool for diagnosis of sleeping sickness in laboratory settings, and the diagnostic format described here may have wider application for other infectious diseases.
Human African trypanosomiasis (HAT) is a complex of protozoan infections, fatal if untreated, which can be caused by infection with either Trypanosoma brucei gambiense (chronic HAT) or Trypanosoma brucei rhodesiense (acute HAT). Both human infective subspecies are cyclically transmitted by tsetse flies (genus Glossina). HAT is a severely neglected disease, typically affecting rural populations of poor people across sub-Saharan Africa (23). More than 60 million people are at risk of contracting the disease, with an estimated 50,000 to 70,000 persons newly infected annually (2). In the absence of treatment, HAT patients inevitably die after a more or less prolonged period of grave illness. Disease control for T. b. gambiense relies heavily on accurate diagnosis and effective treatment of patients and for T. b. rhodesiense on treatment of patients and of the animal reservoir (24). Classical diagnosis requires demonstration of parasites in blood or lymph, which for T. b. gambiense infection is problematic due to extremely low parasitemias in infected people. It is estimated that 20 to 30% of the patients remain undiagnosed by standard parasitological techniques (20). New molecular techniques based on the PCR have been developed as a surrogate for parasite detection. Applications of PCR for detecting T. brucei and its subspecies have been reported which can be highly effective for detection of T. b. rhodesiense and for distinguishing T. b. rhodesiense from T. b. gambiense (7, 16, 17, 18, 22, 25). The use of these highly specific and sensitive PCR methods is, however, suitable only for screening for disease prevalence and for research purposes, and they are not commonly used to inform diagnosis of HAT (5). This is partly due to the cumbersome methods of PCR product detection. Amplicons are normally identified using UV transillumination after being electrophoresed in the presence of ethidium bromide (a carcinogen). Alternative methods for PCR product detection, such as real-time PCR, PCR-enzyme-linked immunosorbent assay, or mass spectrometry, have been developed (3, 10, 21) but are complex, expensive, and equipment, recourse, and personnel hungry. There is a demand for a simplified method of amplification and product detection which would make these tools usable in the facilities available in regional sleeping sickness diagnostic laboratories.
Oligochromatography (OC) provides a simple and rapid dipstick format for detection of amplified PCR products (Coris BioConcept, Gembloux, Belgium; patent no, WO 2004/099438A1) (12, 13, 15, 19). PCR products are visualized by hybridization with a gold-conjugated probe. This PCR product detection format takes only 5 min, and no equipment other than a dry heating block and a pipette are needed. An internal control (IC) for the PCR and a control for the chromatographic migration are incorporated in the assay.
We present here the development and phase I evaluation of a T. brucei-specific PCR-oligochromatography test, called HAT-PCR-OC (human African trypanosomiasis-PCR-oligochromatography). We have chosen the 18S rRNA gene as the target for the HAT-PCR-OC, since it is a multicopy gene that contains sequences conserved within trypanosomatids and species-specific sequences (11).
MATERIALS AND METHODS
Parasite DNA.
DNA was obtained from T. b. gambiense (LiTat 1.3) and T. b. rhodesiense (STIB 382) parasites amplified in HsdCpb:WU rats (Harlan, The Netherlands). Trypanosomes were separated from the blood using DEAE chromatography (9), followed by repeated centrifugation (20 min, 2,000 × g) and sediment washes with phosphate-buffered saline glucose (38 mM Na2HPO4 · 2H20, 2 mM NaHPO4, 80 mM glucose, 29 mM NaCl, pH 8.0). Trypanosome pellets were stored at −80°C. Twenty microliters of trypanosome pellet (approximately 2 × 107 cells) was resuspended in 200 μl of phosphate-buffered saline (8.1 mM Na2HPO4 · 2H2O, 1.4 mM NaHPO4, 140 mM NaCl, pH 7.4), and the DNA was extracted with the QIAamp DNA mini kit (QIAGEN, Hilden, Germany), resulting in pure DNA in 200 μl Tris-EDTA elution buffer. The typical yield of DNA extracted from a 20-μl pellet was 150 ng/μl or 30 μg of total DNA. The extracts obtained were diluted 200 times in water and divided into aliquots of 2 ml in microcentrifuge tubes for storage at −20°C. Purified DNA from other pathogens, i.e., Leishmania donovani, Trypanosoma cruzi, Mycobacterium tuberculosis, Plasmodium falciparum, and Schistosoma mansoni, was obtained from other research groups.
Blood samples.
Informed consent was obtained from patients or their parents or guardians and from nondiseased persons. The human and animal experimentation guidelines of the Institute of Tropical Medicine (Antwerp, Belgium) were followed.
(i) Nonendemic controls.
Venous blood on EDTA was obtained from 28 healthy human Belgian blood donors who had never visited a HAT-endemic country (6).
(ii) Endemic controls.
Venous blood on EDTA was obtained from 50 healthy human volunteers in Kinshasa, Democratic Republic of the Congo.
(iii) Sleeping sickness patients.
Venous blood on heparin was obtained from 26 T. b. gambiense patients enrolled in a clinical study performed in Democratic Republic of the Congo and with confirmed presence of parasites in the blood.
(iv) Spiked blood.
(iv) Blood on EDTA spiked with T. b. gambiense (LiTat 1.3) parasites was used throughout the development of the assay and for the estimation of its lower detection limit. Bloodstream form trypanosomes were grown in rats. At day 3 postinfection, tail blood was taken and diluted in phosphate-buffered saline glucose and the number of parasites per ml was counted in a KOVA cell counting chamber (Hycor Biomedical, Inc., California). A 10-fold dilution series of parasites was made in freshly taken naive human blood, ranging from 10,000 parasites/180 μl to 1 parasite/180 μl blood. Nonspiked blood was used as a negative control.
DNA extraction from blood samples.
As recommended by QIAGEN, 180 μl of blood was mixed with an equal volume of AS1 buffer (QIAGEN, Hilden, Germany) and stored at ambient temperature until DNA extraction. Blood can be stored in this stabilizing buffer for up to 3 months in the dark without loss of DNA quality. DNA was extracted with the QIAamp DNA mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions and stored at −20°C. Briefly, DNA is adsorbed onto the silica gel membrane in the QIAamp spin column, washed by two centrifugation steps, and eluted in 50 μl elution buffer. A 180-μl sample of whole blood will yield an estimated 3 to 12 μg of DNA. The internal control line in the oligochromatography (see below) showed the presence of inhibitory factors in the extracted DNA in some samples (1). In such cases, DNA was further purified as follows. A 1/10 volume of 3 M sodium acetate, pH 5.2, and 2 volumes of ice-cold 100% ethanol were added, followed by overnight DNA precipitation at −20°C. The suspension was centrifuged at 16,100 × g for 15 min and the DNA resuspended in 500 μl of 70% ethanol. The suspension was again centrifuged for 10 min at 16,100 × g, and the ethanol was removed. DNA was dried in a vacuum chamber for at least 30 min and finally resuspended in 50 μl of water and stored at −20°C.
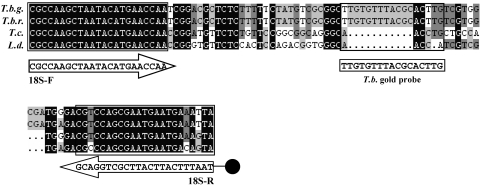
Primers and probes (Fig. 1).
FIG. 1.
Alignment of the HAT-PCR-OC target sequence within the 18S rRNA gene of the Trypanosomatidae parasites Trypanosoma brucei gambiense (T.b.g.) (GenBank accession number AJ009141), Trypanosoma brucei rhodesiense (T.b.r.) (GenBank accession number AJ009142), Trypanosoma cruzi (T.c.) (GenBank accession number AF303660), and Leishmania donovani (L.d.) (GenBank accession number X07773). Situation of the forward primer, 18S-F, the biotin-labeled reverse primer, 18S-R, and the T. brucei (T.b.) gold probe. Gaps in the sequence are represented by dots. Black, dark-gray, and light-gray shading indicates consensus among all four parasites, three parasites, or two parasites, respectively. The target regions of the 18S-F and 18S-R primers and the T. brucei gold probe are boxed.
Primers, biotinylated primers, internal control DNA, and probes were synthesized by Sigma (Bornem, Belgium).
(i) Primers.
Sequences of the 18S rRNA gene of the Trypanosomatidae parasites T. brucei gambiense (GenBank accession number AJ009141), T. brucei rhodesiense (GenBank accession number AJ009142), T. cruzi (GenBank accession number AF303660) and L. donovani (GenBank accession number X07773) were aligned, and primers (18S-F and 18S-R) were designed to amplify a sequence of the Trypanosomatidae 18S rRNA gene using the “DNAman” software (Lynnon Corporation, Quebec, Canada). Primers are situated in Trypanosomatidae conserved sequences that surround a T. brucei conserved sequence. The reverse primer, 18S-R, was biotinylated at the 5′ end.
(ii) T. brucei gold probe.
The alignments described above were used to design a 17-bp DNA probe that can hybridize with a T. brucei-specific sequence situated between the Trypanosomatidae conserved primer sequences. The absence of putative secondary structures within the expected PCR amplicon was checked via mfold version 3.1 (26). The probe was conjugated with gold particles using the procedure described in patent WO 2004/099438A1 (19).
(iii) Internal control DNA.
The IC DNA was constructed to be of the same length as the T. brucei sequence (106 bp). Its sequence is identical to the T. brucei sequence except for the 17-bp central part. This central part was designed to have the same GC content as the T. brucei 17-bp sequence (patent WO 2004/099438A1) (19).
(iv) IC gold probe.
A DNA probe was designed that can hybridize with the 17-bp IC-specific sequence within the IC DNA. This IC probe was conjugated with gold particles as described above.
(v) Migration control probe.
The migration control probe is identical to the 17-bp central sequence of the IC DNA.
PCR amplification.
An asymmetric PCR was designed using fourfold more biotinylated reverse primer than forward primer. The 50-μl reaction mixture contained 10 mM Tris-HCl (pH 8.3 at 25°C), 50 mM KCl (PCR buffer, Sigma, Bornem, Belgium), 200 μM (each) deoxynucleotide triphosphate (Roche, Mannheim, Germany), 2.5 mM MgCl2 (Sigma, Bornem, Belgium), 0.2 μM forward primer 18S-F, 0.8 μM biotinylated reverse primer 18S-R, 0.1 mg/ml acetylated bovine serum albumin (Promega, Madison, Wis.), 3.2 aM IC DNA, 1 U of HotStar Taq polymerase (QIAGEN, Hilden, Germany) and 5 μl of sample DNA. An initial denaturation step of 94°C for 15 min to activate the HotStar Taq polymerase was followed by 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s with a final extension at 72°C for 5 min. Amplification was conducted in 200-μl thin-wall PCR tubes (Abgene, Epsom, United Kingdom) in a T3 thermocycler 48 (Biometra, Göttingen, Germany).
Oligochromatography. (i) Preparation.
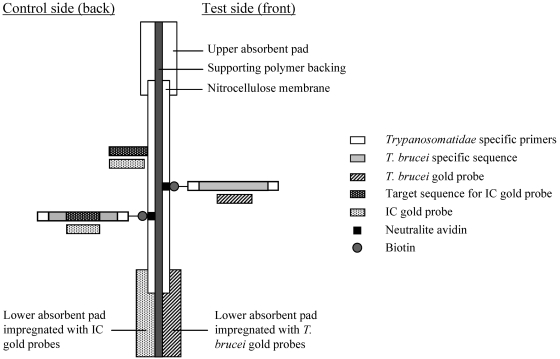
The OC dipstick is constructed with polymer backing faced on each side with a lower absorbent pad which overlaps a nitrocellulose membrane (in the middle) and an upper absorbent pad (Fig. 2). The facing side represents the test for T. brucei, and the back is the control. The lower absorbent pad at the test side is impregnated with the T. brucei gold probe, and the lower absorbent pad at the control side is impregnated with the IC gold probe. On the membrane at the test side, a line of Neutralite avidin (Belovo SA, Bastogne, Belgium) is coated. On the membrane at the control side, two control lines are coated. A first line consists of Neutralite avidin (control for PCR) and a second line of a probe that is complementary to the IC gold probe (migration control). The OC conditions for the phase I evaluation were as follows. The PCR product was denaturated at 94°C for 30 s and kept on ice. Forty microliters was mixed with an equal volume of migration buffer preheated at 55°C, followed by dipping the OC dipstick into the solution.
FIG. 2.
HAT-PCR-OC test principle. Test side. In case of a positive sample, during migration, the T. brucei gold probes hybridize with the T. brucei amplicons that will accumulate on the Neutralite avidin line on the test side of the stick. Control side. During migration the IC gold probes hybridize with IC amplicons that will accumulate on the Neutralite avidin line on the control side of the stick (control for PCR). The unbound IC gold probes hybridize with the complementary migration control probes (control for migration).
(ii) Principle.
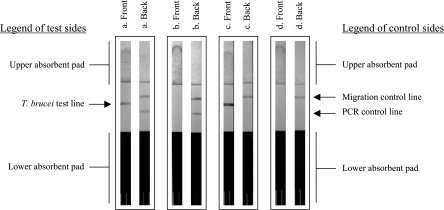
After amplification of a sample containing T. brucei DNA, the PCR product solution contains both IC amplicons and T. brucei amplicons. During migration at 55°C through the lower absorbent pad, the solution takes up the impregnated gold probes. The T. brucei gold probe hybridizes with the biotinylated strands of the T. brucei amplicons during migration. These complexes accumulate on the Neutralite avidin line at the test side, resulting in a visible red line. On the back side, the IC gold probe hybridizes with the biotinylated strands of the IC amplicons, and these complexes accumulate on the Neutralite avidin line. Unbound IC gold probe migrates further and is trapped by the complementary migration control probe line. Both control lines determine whether the HAT-PCR-OC is valid or invalid (Fig. 3). The migration control line should always be visible to validate the test. A test is invalid when both the IC amplicon control line and the T. brucei test line are invisible (Fig. 3d), indicating that the PCR failed, possibly due to inhibitory factors in the extracted DNA. When a sample contains very high concentrations of Trypanosomatidae DNA, competition between this target DNA and the IC template DNA for the primers can result in an invisible IC amplicon control line combined with a strongly visible T. brucei test line (Fig. 3c). In the latter case, the test is considered valid.
FIG. 3.
Possible HAT-PCR-OC test result. a. Valid test, positive result. b. Valid test, negative result. c. Valid test, positive result of a sample with high T. brucei DNA content. d. Invalid test.
RESULTS
Analytical sensitivity.
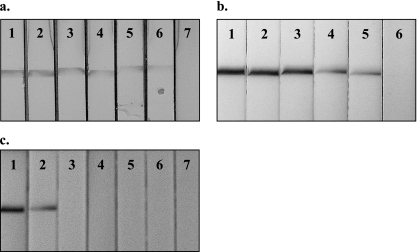
The detection limit of the HAT-PCR-OC was evaluated on a serial dilution of T. b. gambiense DNA in water. The lower detection limit was 5 fg of parasite DNA, which is about 1/40 of the DNA content of one parasite (Fig. 4a). Moreover, the test was able to detect 10 times less DNA than agarose gel electrophoresis (data not shown). The lower detection limit was also evaluated with DNA extracted from a blood sample series spiked with decreasing numbers of living T. b. gambiense parasites. HAT-PCR-OC was consistently able to detect a single parasite in 180 μl of blood (Fig. 4b). The assay remained negative when nonspiked control blood samples were tested.
FIG. 4.
Analytical sensitivity and species specificity of the HAT-PCR-OC test. a. Serial dilution of T. b. gambiense DNA in water. Dipsticks 1 to 7: 500 pg, 50 pg, 5 pg, 500 fg, 50 fg, 5 fg, or 0.5 fg per PCR. b. Serial dilution of living T. b. gambiense bloodstream form parasites in naive human blood. Dipsticks 1 to 6: 10,000, 1,000, 100, 10, 1, or 0 parasites in 180 μl of blood. c. HAT-PCR-OC results obtained with DNA from T. b. gambiense (dipstick 1), T. b. rhodesiense (dipstick 2), Leishmania donovani (dipstick 3), Trypanosoma cruzi (dipstick 4), Mycobacterium tuberculosis (dipstick 5), Plasmodium falciparum (dipstick 6), or Schistosoma mansoni (dipstick 7).
Analytical specificity.
The species specificity of the HAT-PCR-OC was assessed with purified DNA from T. b. gambiense, T. b. rhodesiense, Leishmania donovani, Trypanosoma cruzi, Mycobacterium tuberculosis, Plasmodium falciparum, and Schistosoma mansoni. Positive results were obtained only with the two T. brucei subspecies (Fig. 4c).
Diagnostic sensitivity and specificity (Fig. 5).
FIG. 5.
Diagnostic sensitivity and specificity of the HAT-PCR-OC. a. HAT-PCR-OC results for 26 blood samples from confirmed T. b. gambiense sleeping sickness patients (dipsticks 1 to 26) and one negative control (−). b. HAT-PCR-OC results for one positive control (+) and for five blood samples from area-of-endemicity negative controls (dipsticks 1 to 5).
HAT-PCR-OC was positive with the blood samples from all 26 T. b. gambiense sleeping sickness patients and negative with blood samples from all 28 healthy Belgian blood donors. With the 50 endemic negative-control blood samples, 47 tests were valid and negative by HAT-PCR-OC. Three test results were invalid, since the IC for PCR was negative, indicating the presence of PCR-inhibitory factors in the extracted DNA. Therefore, DNA of these samples was ethanol precipitated and resuspended in water. With retesting, valid negative results were obtained. Thus, HAT-PCR-OC results corresponded fully with the infection status of the sampled persons, indicating 100% sensitivity and specificity.
DISCUSSION
A major constraint for effective sleeping sickness control is imperfect diagnosis. Reliable and fast diagnostic tools are desperately needed. The HAT-PCR-OC test described here has been developed as a simplified molecular test for rapid and sensitive detection of T. brucei parasites in blood samples. HAT-PCR-OC combines both the sensitivity and specificity of PCR with the simplicity and speed of membrane chromatography (visible after 5 min by the naked eye). OC, in contrast to conventional amplicon detection techniques, doesn't require postamplification preparation, neither does it require sophisticated equipment. The data presented here show that the assay is able to detect a single parasite in a 180-μl volume of blood. When evaluated using both trypanosome-positive patient samples and trypanosome-negative control samples, HAT-PCR-OC results corresponded 100% with the infection status of the tested persons.
HAT-PCR-OC shows clear potential for implementation as a reference diagnostic test in midlevel-equipped laboratory facilities (clean laboratories with power and cold storage) and will be of particular value for evaluation of cure assessment during clinical trials of new drugs or drug combinations. The simple detection methodology described here may facilitate PCR application in HAT-endemic countries. Further refinement of this assay will widen its application. New simple nucleic acid extraction techniques together with novel isothermal nucleic acid amplification methods, such as loop-mediated isothermal amplification (8, 14) and nucleic acid based amplification (4), will mean that the constraint for field PCR (namely, the need for thermocycling reactions) could be eliminated.
A further application of the HAT-PCR-OC test format would be for diagnosis of all Trypanozoon infections, since the detection probe is specific for all Trypanozoon taxa, including T. b. brucei, Trypanosoma evansi, and Trypanosoma equiperdum, that affect cattle, buffaloes, small ruminants, camels, and horses and that are responsible for severe losses in the agricultural sector. This is of particular value for T. b. rhodesiense HAT, in which case the disease is maintained in the domestic animal reservoir (24).
A common drawback of PCR is the risk of sample contamination with PCR products leading to false-positive results. When implementing HAT-PCR-OC, one should take measures to avoid this risk. The oligochromatography procedure should be performed in closed test tubes, and pre- and post-PCR manipulations should be physically separated, preferably in separate lab spaces. Finally, used dipsticks should be thrown away after the result is read or should be archived in a postamplification room.
To our knowledge this is the first nucleic acid-based diagnostic test in dipstick format for vector-borne diseases. The proof of principle for HAT-PCR-OC presented in this study opens perspectives for its use in laboratories within countries where sleeping sickness is endemic. The same format can be applied for other infectious diseases, such as malaria, tuberculosis, leishmaniasis, Chagas' disease, schistosomiasis, etc., for which simpler and more applicable molecular diagnostic tools are needed.
Acknowledgments
This study received financial support from the International Atomic Energy Agency (IAEA, Vienna, Austria, contract number 12851/RBF) and from the International Livestock Research Institute (ILRI, Nairobi, Kenya).
We gratefully acknowledge C. Grevelding (Institüt für Genetik, Heinrich-Heine-Universität, Düsseldorf, Germany) for providing us with DNA from Schistosoma mansoni. We are grateful to S. De Doncker, F. Portaels, and U. D'Alessandro (Institute of Tropical Medicine, Antwerp) for providing DNA from Leishmania donovani, Trypanosoma cruzi, Mycobacterium tuberculosis, and Plasmodium falciparum. Special thanks to S. C. Welburn for critical reading of the manuscript.
REFERENCES
- 1.Al-Soud, W. A., L. J. Jönsson, and P. Radström. 2000. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J. Clin. Microbiol. 38:345-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 2006. Human African trypanosomiasis (sleeping sickness): epidemiological update. Wkly. Epidemiol. Rec. 81:69-80. [PubMed] [Google Scholar]
- 3.Becker, S., J. R. Franco, P. P. Simarro, A. Stich, P. M. Abel, and D. Steverding. 2004. Real-time PCR for detection of Trypanosoma brucei in human blood samples. Diagn. Microbiol. Infect. Dis. 50:193-199. [DOI] [PubMed] [Google Scholar]
- 4.Chang, A. B., and J. D. Fox. 1999. NASBA and other transcription based amplification methods for research and diagnostic microbiology. Rev. Med. Microbiol. 10:185-196. [Google Scholar]
- 5.Chappuis, F., L. Loutan, P. Simarro, V. Lejon, and P. Büscher. 2005. Options for the field diagnosis of human African trypanosomiasis. Clin. Microbiol. Rev. 18:133-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Doncker, S., V. Hutse, S. Abdellati, S. Rijal, B. M. Singh Karki, S. Decuypere, D. Jacquet, D. Le Ray, M. Boelaert, S. Koirala, and J. C. Dujardin. 2005. A new PCR-ELISA for diagnosis of visceral leishmaniasis in blood of HIV-negative subjects. Trans. R. Soc. Trop. Med. Hyg. 99:25-31. [DOI] [PubMed] [Google Scholar]
- 7.Kabiri, M., J. R. Franco, P. P. Simarro, J. A. Ruiz, M. Sarsa, and D. Steverding. 1999. Detection of Trypanosoma brucei gambiense in sleeping sickness suspects by PCR amplification of expression-site-associated genes 6 and 7. Trop. Med. Int. Health 4:658-661. [DOI] [PubMed] [Google Scholar]
- 8.Kuboki, N., N. Inoue, T. Sakurai, F. Di Cello, D. J. Grab, H. Suzuki, C. Sugimoto, and T. Igarashi. 2003. Loop-mediated isothermal amplification for detection of African trypanosomes. J. Clin. Microbiol. 41:5517-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanham, S. M., and D. G. Godfrey. 1970. Isolation of salivarian trypanosomes from man and other mammals using DEAE-cellulose. Exp. Parasitol. 28:521-534. [DOI] [PubMed] [Google Scholar]
- 10.Martin-Sanchez, J., J. A. Pineda, M. Andreu-Lopez, J. Delgado, J. Macias, R. De La Rosa, and F. Morillas-Marquez. 2002. The high sensitivity of a PCR-ELISA in the diagnosis of cutaneous and visceral leishmaniasis caused by Leishmania infantum. Ann. Trop. Med. Parasitol. 96:669-677. [DOI] [PubMed] [Google Scholar]
- 11.Maslov, D. A., J. Lukes, M. Jirku, and L. Simpson. 1996. Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs: implications for the evolution of parasitism in the trypanosomatid protozoa. Mol. Biochem. Parasitol. 75:197-205. [DOI] [PubMed] [Google Scholar]
- 12.Mertens, P., I. Renuart, R. Boreux, M.-P. Hayette, and T. Leclipteux. 2003. Detection of Toxoplasma gondii by oligochromatography of nested-PCR amplicons. Presented at 103rd General Meeting of the American Society of Medicine, Washington, D.C.
- 13.Mertens, P., L. Vijgen, C. Olungu, S. Degallaix, I. Renuart, M. Van Ranst, and T. Leclipteux. 2004. Detection of SARS-CoV by oligochromatography of RT-PCR amplicons. Clin. Microbiol. Infect. 10(Suppl. 3):526-527. [Google Scholar]
- 14.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28:7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olungu, C., P. Mertens, S. Degallaix, I. Renuart, O. Legeay, R. Laube, and T. Leclipteux. 2004. Detection of HAV by oligochromatography of RT-PCR amplicons. Presented at 104th General Meeting of the American Society of Medicine. New Orleans, 23 to 27 May, 2004.
- 16.Picozzi, K., E. M. Fèvre, M. Odiit, M. C. Eisler, I. Maudlin, and S. C. Welburn. 2006. Sleeping sickness in Uganda: a thin line between two fatal diseases. BMJ 331:1238-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radwanska, M., M. Chamekh, L. Vanhamme, F. Claes, S. Magez, E. Magnus, P. De Baetselier, P. Büscher, and E. Pays. 2002. The serum resistance-associated gene as a diagnostic tool for the detection of Trypanosoma brucei rhodesiense. Am. J. Trop. Med. Hyg. 67:684-690. [DOI] [PubMed] [Google Scholar]
- 18.Radwanska, M., F. Claes, S. Magez, E. Magnus, D. Perez-Morga, E. Pays, and P. Büscher. 2002. Novel primer sequences for a polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am. J. Trop. Med. Hyg. 67:289-295. [DOI] [PubMed] [Google Scholar]
- 19.Renuart, I., P. Mertens, and T. Leclipteux. 18November2004. One step oligochromatographic device and method of use. EU patent WO2004/099438:A1.
- 20.Robays, J., M. M. C. Bilengue, P. Van der Stuyft, and M. Boelaert. 2004. The effectiveness of active population screening and treatment from sleeping sickness control in the Democratic republic of Congo. Trop. Med. Int. Health 9:542-550. [DOI] [PubMed] [Google Scholar]
- 21.Sampath, R., S. A. Hofstadler, L. B. Blyn, M. W. Eshoo, T. A. Hall, C. Massire, H. M. Levene, J. C. Hannis, P. M. Harrell, B. Neuman, M. J. Buchmeier, Y. Jiang, R. Ranken, J. J. Drader, V. Samant, R. H. Griffey, J. M. McNeil, S. T. Crooke, and D. J. Ecker. 2005. Rapid identification of emerging pathogens: coronavirus. Emerg. Infect. Dis. 11:373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schares, G., and D. Mehlitz. 1996. Sleeping sickness in Zaire: a nested polymerase chain reaction improves the identification of Trypanosoma (Trypanozoon) brucei gambiense by specific kinetoplast DNA probes. Trop. Med. Int. Health 1:59-70. [DOI] [PubMed] [Google Scholar]
- 23.Trouiller, P., P. Olliaro, E. Torreele, J. Orbinski, R. Laing, and N. Ford. 2002. Drug development for neglected diseases: a different market and a public-health policy failure. Lancet 359:2188-2194. [DOI] [PubMed] [Google Scholar]
- 24.Welburn, S. C., P. G. Coleman, I. Maudlin, E. M. Fèvre, M. Odiit, and M. C. Eisler. 2006. Crisis, what crisis? Control of Rhodesian sleeping sickness. Trends Parasitol. 22:123-128. [DOI] [PubMed] [Google Scholar]
- 25.Welburn, S. C., K. Picozzi, E. M. Fèvre, P. G. Coleman, M. Odiit, M. Carrington, and I. Maudlin. 2001. Identification of human-infective trypanosomes in animal reservoir of sleeping sickness in Uganda by means of serum-resistance-associated (SRA) gene. Lancet 358:2017-2019. [DOI] [PubMed] [Google Scholar]
- 26.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]