Abstract
Eighty Vibrio cholerae O1 strains selected to represent the 1998-to-1999 history of the largest cholera epidemic in Kenya were characterized by ribotyping, antimicrobial susceptibility, and random amplified polymorphic DNA patterns. Except for 19 strains from 4 local outbreaks in North Eastern Province along the Somalia border, the other 61 strains from 25 outbreaks occurring in districts scattered around the country were all ribotype B27 and resistant to chloramphenicol, spectinomycin, streptomycin, sulfamethoxazole, and trimethoprim. The 61 strains showed similar and specific amplified DNA patterns. These findings indicate that the predominant strains that caused the Kenyan epidemic had a clonal origin and suggest that ribotype B27 strains, which first appeared in West Africa in 1994, have had a rapid spread to eastern Africa.
Cholera is a widespread, severe diarrheic disease which continues to be a global threat (12). Seven pandemics of cholera have been recorded since 1817. The ongoing seventh pandemic was caused by Vibrio cholerae O1 biotype El Tor and started in Indonesia in 1961 (5). V. cholerae O1 El Tor spread rapidly through Asia and the Middle East, affecting Africa in 1970. In the following two decades, cholera reached virtually all African countries, accounting for more than 20% of the world cholera cases notified to the WHO in the early 1990s. This percentage increased to 75% in 1996 and, on average, remained higher than 85% from 1997 to 2004.
Since 1971, Kenya has suffered several waves of cholera recrudescence. Its largest epidemic started in 1997 (14), with 17,200 cases notified to the WHO. In 1998 and 1999, the epidemic progressed with more than 33,400 notified cases. The final official figures of the Kenyan epidemic were 10% of all cholera cases reported from the African continent in the same 3 years (14, 17, 18).
In this paper, we present phenotypic and genotypic features of 80 V. cholerae O1 strains representative of the 1998-to-1999 period of the epidemic occurring in Kenya. Strains were characterized by PCR detection of phage CTXΦ genes and pathogenicity genes, BglI ribotyping, antimicrobial susceptibility testing, and a random amplified polymorphic DNA (RAPD) assay.
Collection and identification of isolates.
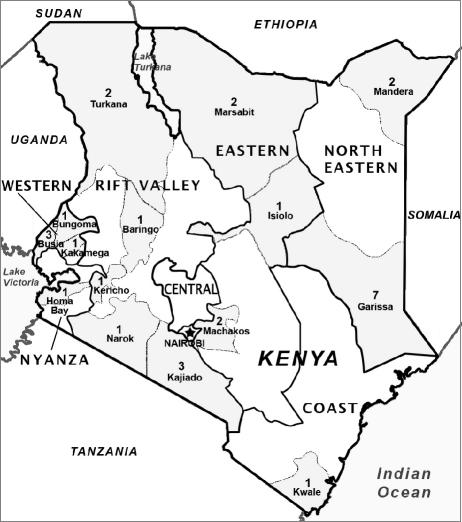
Eighty strains were selected among V. cholerae O1 isolates collected from 29 outbreaks in different provinces of Kenya in 1998 and 1999 (Table 1; Fig. 1). The strains were isolated from rectal swabs in Cary-Blair transport medium as described previously (8). All strains were biotype El Tor, 57 were serotype Ogawa, and 23 were serotype Inaba. Original stock cultures of isolates were kept in 20% glycerol Luria-Bertani broth at −70°C.
TABLE 1.
Geographical distribution, ribotypes, resistance patterns, and RAPD cluster types of 80 V. cholerae O1 strains isolated in Kenya in 1998 and 1999
| Province | No. of strains | District(s) of isolation (no. of strains)a | Ribotype(s) (no. of strains) | Resistance pattern (no. of strains) | RAPD cluster type (no. of strains) |
|---|---|---|---|---|---|
| Western | 10 | Bungoma (1), Kakamega (2) | B27 (10) | CHL, SPT, STR, SMX, TMP (7) | IV (10) |
| Busia (7) | CHL, DOX, SPT, STR, SMX, TET, TMP (3)c | ||||
| Nyanza | 3 | Homa Bay (3) | B27 (3) | CHL, DOX, SPT, STR, SMX, TET, TMP (3)c | IV (3) |
| Rift Valley | 25 | Turkana (10), Baringo (3), Kericho (1), Narok (4), Kajiado (7) | B27 (21), B27kb (4) | CHL, SPT, STR, SMX, TMP (25) | IV (25) |
| Eastern | 10 | Marsabit (6), Isiolo (1), Machakos (3) | B27 (10) | CHL, SPT, STR, SMX, TMP (10) | IV (10) |
| Coast | 5 | Kwale (5) | B27 (5) | CHL, SPT, STR, SMX, TMP (5) | IV (5) |
| North Eastern | 27 | Mandera (12) | B5a (10), B8a (1), B21a (1) | SPT (12) | VII (12) |
| Garissa (15) | B8a (7) | AMP, CHL, SPT, STR, SMX, TMP (7) | VIII (7) | ||
| B27 (8) | CHL, SPT, STR, SMX, TMP (8) | IV (8) |
The distribution of isolation districts by province is shown in Fig. 1.
Subtype of ribotype B27.
Strains from areas on the shore of Lake Victoria.
FIG. 1.
Province distribution of isolation districts of 80 V. cholerae O1 El Tor strains from 29 outbreaks during the large epidemic in Kenya in the late 1990s. The number of local outbreaks investigated is indicated above each district name. The provinces shown are Western, Nyanza, Rift Valley, Eastern, Central, Nairobi, Coast, and North Eastern.
Detection of phage CTXΦ genes and pathogenicity genes.
Detection of genes encoding cholera toxin subunit A (ctxA), zonula occludens toxin (zot), accessory cholera enterotoxin (ace), and toxin-coregulated pilus for intestinal colonization (tcpA) was performed by PCR as described previously (10, 16). A negative control (reaction mixture without template) and a toxin-positive control (V. cholerae O1 strain O395) were also included. Of 80 strains, 77 were PCR positive for genes ctxA, zot, ace, and tcpA, 2 strains were negative for gene tcpA, and 1 was negative for gene zot.
Ribotyping.
Molecular characterization by BglI restriction patterns of 16S and 23S rRNA genes was performed as described previously (4, 6, 7, 11). On the basis of the restriction patterns observed, the 80 Kenyan strains of V. cholerae O1 could be divided into two groups. The first group was composed of 61 strains showing the standard reference pattern of ribotype B27 (57 strains) or of its subtypes (4 strains from Kajiado District) (Table 1). The second group (North Eastern Province) contained 19 strains belonging to ribotypes B5a (10 strains), B8a (8 strains), and B21a (1 strain) (Table 1).
The 61 strains of ribotype B27 represented 25 local outbreaks that had occurred in different districts scattered around the entire country (Fig. 1), whereas the 19 strains of ribotypes B5a, B8a, and B21a were from 4 outbreaks all localized in North Eastern Province, which lies along the Somalia border. Moreover, in the 1990s, ribotypes B5a, B8a, and B21a were systematically prevalent in focuses in Somalia where V. cholerae is endemic and epidemic (unpublished results).
These data indicated that ribotype pattern B27 was a distinguishing molecular character of the most widely distributed V. cholerae O1 strains responsible for the epidemic in Kenya.
Antimicrobial susceptibility.
Antimicrobial susceptibility was determined by the disk diffusion method as described previously (2, 13). The antimicrobial disks were used at the following concentrations: 10 μg ampicillin (AMP), 30 μg chloramphenicol (CHL), 30 μg doxycycline (DOX), 30 μg kanamycin, 10 μg spectinomycin (SPT), 10 μg streptomycin (STR), 25 μg sulfamethoxazole (SMX), 30 μg tetracycline (TET), and 5 μg trimethoprim (TMP).
Escherichia coli ATCC 25922 was used as a quality control strain. Genetic techniques for conjugation and incompatibility testing were the same as those used previously (2).
The 61 strains of ribotype B27 were resistant to CHL, SPT, STR, SMX, and TMP; 6 of them, isolated from areas on the shore of Lake Victoria (Table 1; Fig. 1), were also resistant to DOX and TET. These additional resistances were due to the presence of a conjugative plasmid belonging to incompatibility class C and encoding resistance to DOX, SMX, and TET. Acquisition of this plasmid by a ribotype B27 strain resistant to CHL, SPT, STR, SMX, and TMP was the most probable and simple event for explaining the origin of this Lake Victoria group of TET-resistant strains.
The 19 strains of ribotypes different from B27 exhibited two resistance patterns. The 12 strains of ribotypes B5a, B8a, and B21a from Mandera District (Fig. 1) were resistant to SPT and susceptible to all the other antimicrobials tested independent of ribotype. The seven strains of ribotype B8a from Garissa District were resistant to AMP, CHL, SPT, STR, SMX, and TMP. These resistance patterns corresponded to the two prevalent patterns identified in V. cholerae O1 strains in Somalia in the 1990s (unpublished results).
RAPD assay.
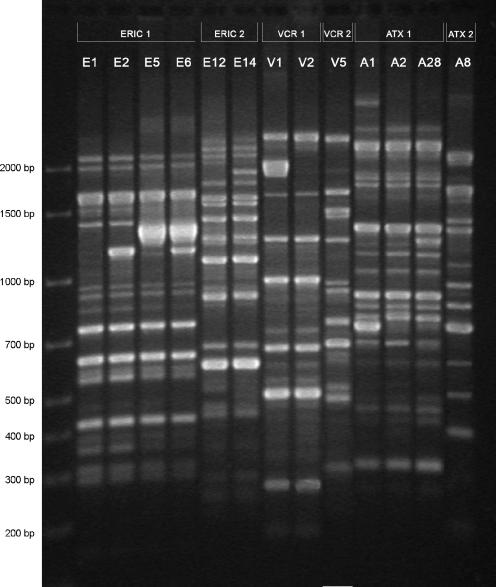
Genomic DNA extraction and PCRs were performed as described previously (9). Each strain was characterized by the combination (cluster type) of six RAPD patterns (Fig. 2) generated by six single primers selected from enterobacterial repetitive intergenic consensus sequences (ERIC1 and ERIC2), from V. cholerae repetitive sequences (VCR1 and VCR2), and from phage CTXΦ sequences (ATX1 and ATX2). Combinations with six identical amplified DNA patterns or with one different amplicon in only one pattern out of the six were classified in the same specific RAPD cluster type.
FIG. 2.
RAPD patterns of genomic DNA of V. cholerae O1 strains generated by six primers. All representative variants of single RAPD patterns are included. Each specific pattern is designated by a letter with a serial number (E1, V1, or A1), and the corresponding primer is reported at the top of the gel (ERIC1, VCR1, or ATX1). Molecular size markers are given on the left.
Three major RAPD cluster types were identified. As reported in Table 1, their designations were IV, VII, and VIII.
The 61 strains of ribotype B27 were all classified as RAPD cluster type IV whose reference combination was E5/E12/V2/V5/A1/A8 (Fig. 2). Fifty-five strains produced the exact reference combination, and six strains, all of the Lake Victoria group (Table 1), produced cluster E6/E12/V2/V5/A1/A8 (three strains) and cluster E6/E12/V1/V5/A1/A8 (three strains). The difference between patterns E5 and E6 consisted in one band at 1.2 kb (Fig. 2) associated with the IncC DOX-SMX-TET resistance plasmid. Lake Victoria strains cured of the plasmid shifted from pattern E6 to pattern E5. In comparison with pattern V2, pattern V1 contained one more band at 1.9 kb. The genetic nature of that variation remains unexplained.
The 12 strains of ribotypes B5a, B8a, and B21a from Mandera District and the 7 strains of ribotype B8a from Garissa District had clusters E1/E12/V2/V5/A28/A8 and E2/E14/V2/V5/A2/A8, respectively. These two combinations were distinct (Fig. 2) from those of RAPD cluster type IV and belonged to two different cluster types, VII and VIII.
Characterization of 80 strains of V. cholerae O1 isolated during the Kenyan epidemic in 1998 and 1999 demonstrated that 61 strains were ribotype B27, showed an identical and stable multiple resistance to CHL, SPT, STR, SMX, and TMP, and produced the same cluster type of six random amplified DNA patterns. This uniformity of properties and the fact that the 61 strains examined were representative of 25 outbreaks (out of 29 investigated) occurring in districts scattered over the entire area of the country (Fig. 1) provided strong genetic and epidemiological evidence that the predominant strains causing the epidemic had a clonal origin. Identification of 19 strains, with traits typical of V. cholerae O1 strains active in Somalia, from four outbreaks in North Eastern Province indicated that province as an epidemic zone where the Kenyan clone and Somali strains were overlapping and presumably competing.
Ribotype B27 was first identified in Calcutta in 1993 and introduced into the western African country of Guinea-Bissau in 1994 (3, 15). In 1995 and 1996, ribotype B27 was identified among V. cholerae O1 strains, causing cholera outbreaks in Senegal (1). Our findings suggest that this emerging ribotype has had a rapid spread into eastern Africa.
Acknowledgments
This study was supported by grant IC18-CT97-0231 (F.M.) from the European Commission under the INCO-DC program and, in part, by research funds of the Faculty of Sciences, University of Bari, and the Ph.D. course in Genetics and Molecular Evolution at the University of Bari.
We thank Nicola Pugliese for technical assistance.
REFERENCES
- 1.Aidara, A., S. Koblavi, C. S. Boye, G. Raphenon, A. Gassama, F. Grimont, and P. A. D. Grimont. 1998. Phenotypic and genotypic characterization of Vibrio cholerae isolates from a recent cholera outbreak in Senegal: comparison with isolates from Guinea-Bissau. Am. J. Trop. Med. Hyg. 58:163-167. [DOI] [PubMed] [Google Scholar]
- 2.Coppo, A., M. Colombo, C. Pazzani, R. Bruni, K. A. Mohamud, K. H. Omar, S. Mastrandrea, A. M. Salvia, G. Rotigliano, and F. Maimone. 1995. Vibrio cholerae in the Horn of Africa: epidemiology, plasmids, tetracycline resistance gene amplification, and comparison between O1 and non-O1 strains. Am. J. Trop. Med. Hyg. 53:351-359. [DOI] [PubMed] [Google Scholar]
- 3.Dalsgaard, A., H. F. Mortensen, F. Mølbak, F. Dias, O. Serichantalergs, and P. Echeverria. 1996. Molecular characterization of Vibrio cholerae O1 strains isolated during cholera outbreaks in Guinea-Bissau. J. Clin. Microbiol. 34:1189-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damian, M., S. Koblavi, I. Carle, N. Nacescu, F. Grimont, C. Ciufecu, and P. A. D. Grimont. 1998. Molecular characterization of Vibrio cholerae O1 strains isolated in Romania. Res. Microbiol. 149:745-755. [DOI] [PubMed] [Google Scholar]
- 5.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fields, P. I., T. Popovic, K. Wachsmuth, and Ø. Olsvik. 1992. Use of polymerase chain reaction for detection of toxigenic Vibrio cholerae O1 strains from the Latin American cholera epidemic. J. Clin. Microbiol. 30:2118-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machado, J., F. Grimont, and P. D. A. Grimont. 1998. Computer identification of Escherichia coli rRNA gene restriction patterns. Res. Microbiol. 149:119-135. [DOI] [PubMed] [Google Scholar]
- 8.Maimone, F., A. Coppo, C. Pazzani, S. O. Ismail, R. Guerra, P. Procacci, G. Rotigliano, and K. H. Omar. 1986. Clonal spread of multiply resistant strains of Vibrio cholerae O1 in Somalia. J. Infect. Dis. 153:802-803. [DOI] [PubMed] [Google Scholar]
- 9.Pazzani, C., M. Scrascia, A. M. Dionisi, F. Maimone, and I. Luzzi. 2006. Molecular epidemiology and origin of cholera reemergence in Italy and Albania in the 1990s. Res. Microbiol. 157:508-512. [DOI] [PubMed] [Google Scholar]
- 10.Pourshafie, M. R., F. Grimont, M. Saifi, and P. A. D. Grimont. 2000. Molecular epidemiology study of Vibrio cholerae isolates from infected patients in Teheran, Iran. J. Med. Microbiol. 49:1085-1090. [DOI] [PubMed] [Google Scholar]
- 11.Regnault, B., F. Grimont, and P. A. D. Grimont. 1997. Universal ribotyping method using a chemically labelled oligonucleotide probe mixture. Res. Microbiol. 148:649-665. [DOI] [PubMed] [Google Scholar]
- 12.Salim, A., R. Lan, and R. Reeves. 2005. Vibrio cholerae pathogenic clones. Emerg. Infect. Dis. 11:1758-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scrascia, M., M. Forcillo, F. Maimone, and C. Pazzani. 2003. Susceptibility to rifaximin of Vibrio cholerae strains from different geographical areas. J. Antimicrob. Chemother. 52:303-305. [DOI] [PubMed] [Google Scholar]
- 14.Shapiro, R. L., R. M. Otieno, P. M. Adcock, A. Penelope, P. Howard, W. A. Hawley, L. Kamur, P. Waiyaki, B. L. Nahlen, and L. Slutsker. 1999. Transmission of epidemic Vibrio cholerae O1 in rural western Kenya associated with drinking water from Lake Victoria: an environmental reservoir for cholera? Am. J. Trop. Med. Hyg. 60:271-276. [DOI] [PubMed] [Google Scholar]
- 15.Sharma, C., A. Ghosh, A. Dalsgaard, A. Forslund, R. K. Ghosh, S. K. Bhattacharya, and G. B. Nair. 1998. Molecular evidence that a distinct Vibrio cholerae O1 biotype El Tor strain in Calcutta may have spread to the African continent. J. Clin. Microbiol. 36:843-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamayo, M., S. Koblavi, F. Grimont, E. Castañeda, and P. D. A. Grimont. 1997. Molecular epidemiology of Vibrio cholerae O1 isolates from Columbia. J. Med. Microbiol. 46:611-616. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization. 1999. Cholera, 1998. Wkly. Epidemiol. Rec. 74:257-264. [PubMed] [Google Scholar]
- 18.World Health Organization. 2000. Cholera, 1999. Wkly. Epidemiol. Rec. 75:249-256. [PubMed] [Google Scholar]