Abstract
In this study we report on the development and application of a novel method for efficiently extracting and detecting single Cryptosporidium oocysts from archived glass slides. Laser capture microscopy was used to extract low numbers of oocysts from archived glass slides. Highly sensitive real-time PCR methods were then developed to enable the rapid detection and identification of Cryptosporidium oocysts from these samples. The method was applied to fecal smears stained with a variety of standard oocyst stains and water samples. This application, with samples derived from both public health and water service laboratories, highlighted the strong potential of this method to be used as a rapid high-throughput screening tool for the routine monitoring of Cryptosporidium and other medically important pathogens from clinical, veterinary, and environmental water samples. Importantly, the application of our protocol could be used to type Cryptosporidium and other pathogens from stored archived glass slides in public health and water service laboratories, providing vital epidemiological updates and helping to identify and trace pathogens and their routes of infection and ultimately improve their control.
Cryptosporidium spp. are obligate intracellular protozoan parasites that infect a wide range of vertebrates and undergo endogenous development, culminating in the production of an encysted stage (oocyst), which is discharged in the feces of their host (10, 14). Globally, Cryptosporidium is responsible for the majority of gastrointestinal parasitic infections, representing a significant cause of morbidity and mortality in its hosts (35, 53). There are currently 16 recognized species of Cryptosporidium (54-57), and cryptosporidiosis in humans and livestock is mainly caused by the zoonotic pathogen Cryptosporidium parvum and the anthroponotic pathogen C. hominis (21). Cryptosporidiosis in humans usually results in self-limited watery diarrhea in immunocompetent subjects but has far more devastating effects on immunocompromised patients, e.g., AIDS patients, and can be life-threatening due to dehydration caused by chronic diarrhea (10, 11). While no effective therapy for cryptosporidiosis is currently known (8), a rapid effective diagnosis of cryptosporidiosis is essential in immunocompromised patients because the results could influence therapeutic procedures (49, 50).
Surveys in the United Kingdom and the United States have shown that between 50 and 80% of standing water is contaminated with Cryptosporidium oocysts (26, 55). Cryptosporidium oocysts are highly resistant to chlorination (36) and can survive for several months in standing water (38, 52), rendering them a serious problem for large-scale water suppliers and users of private water supplies. The recognition of Cryptosporidium as an important pathogen and the global increase in immunocompromised populations have resulted in a strong demand for sensitive and reliable detection and typing systems for both clinical and environmental water samples (21, 32, 51). This demand is further highlighted because different species of Cryptosporidium oocysts are morphologically indistinguishable from each other (15). In developed countries, regulations exist which require that drinking water suppliers carry out monitoring and risk assessments to establish whether there is a significant risk from Cryptosporidium oocysts in water supplied from their treatment works for human consumption (4, 5, 6). During these activities, samples of water are fixed onto glass slides for Cryptosporidium analysis (4, 5, 6). Where it is established that there is such a risk, suppliers must use a process for treating the water to ensure that the average number of Cryptosporidium oocysts per 10 liters of water is less than 1 (4, 6). Breach of this drinking water quality standard in the United Kingdom is a criminal offense, and to facilitate investigations all slides must be stored in a refrigerator at +2°C to +8°C for a period of at least three calendar months following completion of analysis (4, 5, 6). After this time, the slides become accessible for further analysis; thus a major resource becomes available for further examination. The application of molecular techniques for species identification and genotyping from archived glass slides has the potential to provide detailed epidemiological information on the cryptosporidia in circulation in each water catchment. However, until recently, few procedures were sensitive and accurate enough to recover low numbers of oocysts fixed to glass slides (42, 44).
Samples from livestock, immunocompromised patients, water, and foodstuffs are routinely fixed onto glass slides for identification of Cryptosporidium. Here we report a novel and efficient laser capture microscopy method for extracting single Cryptosporidium oocysts from slides, facilitating rapid and accurate detection of this organism and allowing strain discrimination using real-time PCR and DNA sequencing.
MATERIALS AND METHODS
Cryptosporidium strains.
Clinical fecal samples containing C. parvum and C. hominis were provided by the Northern Ireland Public Health Laboratories, Belfast City Hospital, Belfast, United Kingdom. C. parvum (reference strain) was obtained from Moredun Scientific Ltd., Penicuik, Scotland, United Kingdom. C. parvum oocysts, genotype C (isolate ISSC6), and C. hominis oocysts, Peru (human) isolate, were also used in this study. All Cryptosporidium oocysts were stored at 4°C.
DNA extraction from stained oocysts.
An initial study was carried out to ensure that PCR inhibitors and a range of commonly used laboratory stains for detecting Cryptosporidium oocysts were effectively removed during the DNA extraction protocol (34). Purified C. parvum oocysts were enumerated using a hemocytometer, and pellets containing approximately 500 oocysts each were washed twice in double-distilled H20 (ddH20) by centrifugation at 5,000 × g for 2 min. The oocysts were then resuspended in 100 μl of each of the following staining solutions and/or, where appropriate, combinations of staining solutions: Ziehl-Neelsen (ZN), carbol fuchsin (CF) (GCC Diagnostics, Flintshire, United Kingdom), malachite green (GCC Diagnostics), immunofluorescence assay (IFA) fluorescein (Microgen Bioproducts Limited, Surrey, United Kingdom) (17), DAPI (4′,6′-diamidino-2-phenyindole; Roche Diagnostics Ltd., Sussex, United Kingdom) (43), and propidium iodide (Sigma, Dorset, United Kingdom). In each case, where appropriate, the time periods and temperatures of incubation were in accordance with each manufacturer's instructions.
Source and preparation of water sample slides.
Water sample slides were prepared for this study by the Northern Ireland Department of Environment, Water Executive, Cryptosporidium Reference Laboratory. Briefly, serial dilutions of C. parvum oocysts (Moredun strain), ranging from 1 to 1,000 oocysts, were fixed and stained on glass slides in accordance with the Drinking Water Inspectorate standard operating procedure (4, 5) for monitoring Cryptosporidium oocysts in United Kingdom treated water supplies. Each dilution was repeated in triplicate, and oocysts were enumerated from each slide surface using standard IFA microscopic-examination methods (5, 17). The fluorescence microscope (Nikon, Kawasaki, Kanagawa, Japan) was fitted with a fluorescein isothiocyanate filter system. The maximum excitation wavelength was 490 nm, and the emission wavelength was 530 nm. Magnifications were ×40 and ×100. Oocyst counts were recorded, and the slides were stored at 4°C in darkness for a minimum period of 3 months, after which they were made available for molecular analysis under blind trial conditions.
Fecal-slide preparation.
Between January 2003 and December 2003, in the greater Belfast area, fecal specimens were obtained from both community patients with a recent history of gastrointestinal infection who were attending their family practitioners and hospitalized patients in this area. Fresh fecal specimens were homogenized in a 1.1:1.0 (vol/vol) ratio with sterile saline (Sigma; 0.9%, wt/vol), and slides were prepared in triplicate for microscopic examination following ZN-CF acid-fast staining, as previously described (7). The slides were then blot dried and examined under a phase-contrast light microscope using an oil immersion lens at ×63 magnification. Diarrheal feces, in which no Cryptosporidium oocysts were observed under microscopic examination, were also retained for nested-PCR and real-time PCR analyses. All clinical slides were stored for a period of up to 3 months at 4°C, without preservative, prior to extraction of cryptosporidial DNA from slide surfaces.
Stain inhibition detection studies.
Clinical fecal samples which previously tested positive for Cryptosporidium species using a simple acid-fast staining technique were genotyped using a previously described nested-PCR-restriction fragment length polymorphism (RFLP) technique targeting a polymorphic region of the 18S rRNA genes of Cryptosporidium species (54). Cryptosporidium genotyping was carried out by RFLP analysis following digestion of the secondary product with VspI and SspI endonucleases (Promega Ltd., Southampton, United Kingdom) in accordance with the manufacturer's instructions (54). The digested products were fractionated on a 1.5% agarose gel and visualized by ethidium bromide staining.
Laser capture microscopy (LCM).
Glass slides archived for up to 3 years were examined for the presence of oocysts with the aid of a fluorescence microscope fitted with a fluorescein isothiocyanate filter system. Oocyst counts were measured from each slide, and, when low numbers were observed (i.e., 1 to 20 oocysts) (56, 57), a map of the slide surface was drawn on grid paper so that the oocysts/clumps could be accurately targeted during the removal process (18). The PALM (Microlaser, Bernried, Germany) laser microdissection system was used to detect and dissect single oocysts or groups of oocysts from normal archived or polyethylene foil slides (9). Briefly, the PALM Microlaser uses a UV laser to circumcise selected areas of interest (Fig. 1A and B). A laser pulse then catapults (laser pressure catapulting [LPC]) the dissected material into an appropriate container, which in this case was a 0.5-ml microcentrifuge tube, above the sample slides (39). For sensitivity tests, triplicate samples were obtained by individually catapulting 1 to 50 oocysts from a slide into 0.5-ml microcentrifuge tubes containing 10 μl sterile ddH2O using the LPC function of the microscope. Isolated oocysts were then stored at −80°C until further analysis was carried out.
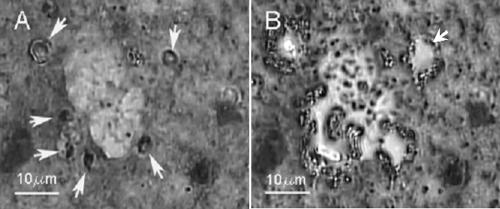
FIG. 1.
(A and B) Application of LCM for the recovery of Cryptosporidium parvum oocysts from archived fecal smears. (A) Fecal smears on glass slides were acid fast stained using the ZN stain method. This acid-fast staining resulted in clearly visible oocysts, which are indicated by the arrows. (B) The oocysts were then recovered from slides by using the LPC function of the LCM, which catapulted the oocysts into a microcentrifuge tube. The arrow indicates an area of the glass slide from which a ZN-stained oocyst was recovered using LCM.
Because of the high purity of samples, the DNA of microdissected oocysts could be extracted using a freeze-thaw method followed by an ethanol precipitation step (20, 41). Briefly, collected oocysts were suspended in 10 μl ddH2O. The oocyst suspensions were frozen in liquid nitrogen for 5 min, followed by thawing at 65°C for 5 min. Samples were then centrifuged at 12,000 × g for 10 min, and this procedure was repeated five times. DNA was then purified using an ethanol precipitation step (20, 41).
Real-time PCR detection.
Unless stated otherwise, all real-time PCR equipment and reagents were obtained from Roche Diagnostics GmbH, Mannheim, Germany, and Roche Diagnostics, Hertfordshire, United Kingdom. Real-time PCR was performed with a LightCycler using the LightCycler FastStart DNA Master SYBR Green I “Hot Start” kit (45), which was used in accordance with the manufacturer's instructions.
Using 18S rRNA as a target, the detection sensitivities of real-time PCR and nested-PCR systems were compared. Nested PCR of 18S rRNA was carried out as previously described (54) for stain inhibition studies. Real-time PCR was carried out using an internal primer pair (54), under the following conditions: initial denaturation of 10 min at 95°C was followed by 45 cycles of 15 s of denaturation at 95°C, 30 s of annealing at 58°C, and 60 s of extension at 72°C. To assess the sensitivity of real-time PCR detection of oocysts recovered from archived slides by laser capture microscopy, three different genes were targeted: the genes encoding Cryptosporidium-specific thrombospondin-related adhesive proteins (TRAP-C2; primers A and B) (37) and lactate dehydrogenase 1 (LDH1) (Table 1) and the 18S rRNA gene (54). Using identical PCR mixtures, primers C and D (Table 1) were designed to target a 241-bp region of the C. parvum LDH1 gene and primers E and F (Table 1) were designed to target a 232-bp region of 18S RNA in C. parvum. For PCR of the LDH1 gene, initial denaturation was carried out for 10 min at 95°C, followed by 50 cycles of 10 s of denaturation at 95°C, 15 s of annealing at 55°C, and 20 s of extension at 72°C. For the detection of 18S rRNA, initial denaturation was carried out for 5 min at 95°C, followed by 45 cycles of 30 s of denaturation at 95°C, 15 s of annealing at 55°C, and 15 s extension at 72°C. Apart from 4 mM MgCl2, the stock PCR mixtures were identical to those from previous reactions.
TABLE 1.
Oligonucleotide primers used to target Cryptosporidium genes in this study
| Primer | 5′-3′ nucleotide sequence | Comments | Gene copy no. | 5′-3′ gene coordinates from corresponding accession no. | Reference or source |
|---|---|---|---|---|---|
| A | CAT ATT CCC TGT CCC TTG AGT TGT | Forward primer for Cryptosporidium TRAP-C2 gene | 1 | 812-831 from X77586 | 34 |
| B | TGG ACA ACC CAA ATG CAG AC | Reverse primer for Cryptosporidium TRAP-C2 gene | 1 | 1180-1171 from X77586 | 34 |
| C | AGA ACA TTC ATT GCA CAA CA | Forward primer for Cryptosporidium LDH1 gene | 1 | 463-482 from AF274310 | This study |
| D | CAA AGT AGG CAG TTC CTG TC | Reverse primer for Cryptosporidium LDH1 gene | 1 | 703-684 from AF274310 | This study |
| E | AGA AAC GGC TAC CAC ATC TA | Forward primer for real-time PCR of 18S rRNA | 5 | 380-399 from L16996 | This study |
| F | ACG AGC TTT TTA ACT GCA AC | Reverse primer for real-time PCR of 18S rRNA | 5 | 1134-1115 from L16996 | This study |
After optimization of these protocols, the sensitivity of the real-time assay was determined for direct detection of Cryptosporidium oocysts from fecal smears fixed on glass slides, as previously described for clinical slide preparation. Finally, the feasibility of the assay for use in routine clinical diagnosis of cryptosporidiosis was investigated using Cryptosporidium-positive fecal smears from five patients. Cryptosporidium-positive slides up to 2 years were obtained from the Department of Bacteriology, Belfast City Hospital, Belfast, United Kingdom, and were analyzed in triplicate.
Discrimination of C. parvum isolates.
Samples of purified oocysts of two different isolates (Moredun and genotype C [Rome isolate ISSC6]) were counted using a hemocytometer and mixed at a 1:1 ratio. The mixture was serially diluted and applied to slides to give approximately 30 oocysts per slide. Using LCM and 18S rRNA real-time PCR with primers E and F (Table 1) as previously described, individual oocysts were catapulted into separate microcentrifuge tubes and 232-bp PCR amplicons were acquired and sequenced for typing. This assay was performed in triplicate.
Gel electrophoresis and sequence analysis.
In order to confirm PCR primer specificity and demonstrate that PCR products were the correct size, all products were recovered from the LightCycler glass capillaries at the end of each experimental run and analyzed. PCR products were separated on 1.5% agarose gels (1 h at 100 V) and visualized using ethidium bromide staining. Bands of the desired sizes of PCR products were extracted from agarose gels using the Wizard SV gel and PCR clean-up system (Promega). Samples were then prepared for sequencing (with their corresponding primers (Table 1) using a BigDye Terminator V3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) with ethanol precipitation as described in the kit protocol and an ABI/Hitachi (Arcade, NY) 3100 genetic analyzer capillary action sequencer. DNA sequences were then analyzed using Chromas version 2.3 (Technelysium, Tewantin Qld, Australia) and NCBI BLAST.
Nucleotide sequence accession numbers.
DNA sequences were deposited on the GenBank sequence database system under DQ336672 for the TRAP-C2 gene; DQ336673 for the LDH1 gene; and DQ431245, DQ656104, and DQ656105 for 18S rRNA from real-time PCRs.
RESULTS
No inhibitory effect of laboratory stains towards Cryptosporidium PCR detection.
There was no observed inhibitory effect on the ability of 18S rRNA nested PCR to detect DNA which had been extracted from Cryptosporidium oocysts prestained with ZN, CF, malachite green, IFA fluorescein, DAPI-propidium iodide, or a combination of these stains (data not shown). No significant difference in the detection sensitivity of unstained versus stained oocysts was found, and similar results were found when the assay was tested for use with prestained aliquots of fecal samples (Table 2 and data not shown).
TABLE 2.
Comparisons of the sensitivities of detection of 18S rRNA from Cryptosporidium oocysts using nested PCR and real-time LightCycler PCR detectiona
| No. of oocysts or type of test | Sensitivityb of:
|
|
|---|---|---|
| Nested PCR | LightCycler real-time PCR | |
| 1,000, 1,000, 997 | +, +, + | +, +, + |
| 500, 497, 518 | +, +, + | +, +, + |
| 100, 108, 106 | +, +, + | +, +, + |
| 78, 74, 77 | +, +, + | +, +, + |
| 58, 53, 62 | +, +, + | +, +, + |
| 13, 10, 11 | +, +, + | +, +, + |
| 5, 3, 3 | −, −, − | +, +, + |
| 1, 1, 1 | −, −, − | +, +, + |
| Negative control (blind trial) | −, −, − | −, −, − |
High sensitivity of real-time PCR detection method.
A 10-fold increase in sensitivity was observed when using real-time PCR technology, when compared with a conventional nested-PCR approach to detect cryptosporidial 18S rRNA from immunomagnetic separation (IMS)-recovered oocysts (Table 2). A sensitivity analysis of the assay determined that it was routinely capable of detecting three oocysts, from the method which utilized IMS for oocyst recovery (data not shown). LightCycler protocols were optimized to enable the successful detection of the DNA from low numbers of Cryptosporidium oocysts recovered from surfaces of archived glass slides by LCM. In order to determine the sensitivity of the combined LCM-LightCycler method, extractions were performed on oocysts removed from water slides using LCM. Both the Cryptosporidium-specific TRAP-C2 gene and the LDH1 gene were successfully targeted using the LightCycler as the amplification system. Routinely, we were able to detect three or seven oocysts per slide when the LDH1 and TRAP-C2 genes, respectively, were targeted (data not shown). Using 18S RNA as a target, we were routinely able to detect a single C. parvum oocyst from clinical slides (Fig. 2A and B).
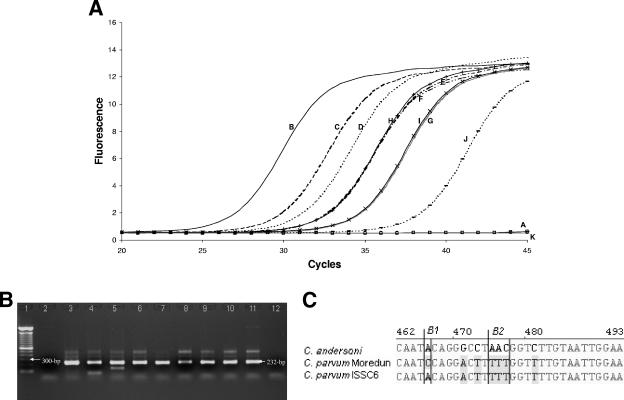
FIG. 2.
(A through C) Detection and identification of Cryptosporidium parvum from archived clinical glass slides using real-time LightCycler PCR detection of 18S rRNA, which produced 232-bp PCR products. (A) Sensitivity of PCR detection of C. parvum, where a single oocyst was routinely detected. (B) Agarose gel (1.5%) of 232-bp PCR products from 18S rRNA. Lane 1, 100-bp DNA ladder; lane 2: negative control; lane 3, positive control, C. parvum (Moredun strain); lane 4, 50 oocysts; lane 5, 20 oocysts; Lane 6, 10 oocysts; lane 7, 7 oocysts; lane 8, 5 oocysts; lane 9, 3 oocysts; lane 10, 2 oocysts; lane 11, 1 oocyst; lane 12, no oocysts. (C) Clustal alignment of C. parvum isolate Moredun (accession no. DQ431245), C. parvum isolate ISSC6 (accesion no. DQ656104), and C. andersoni (accesion no. AY954885) sequences show the ability to genotype and differentiate between C. parvum isolates after sequencing LightCycler PCR products. Shaded gray areas indicate sequence variations between isolates, and examples of sequence variations are indicated within boxes B1 and B2.
Detection and typing of Cryptosporidium from clinical slides.
To determine the adaptability of our new technique, developed with water samples, we applied it to clinical fecal samples that had been previously tested for the presence of Cryptosporidium using acid-fast stains. All clinical slides had been stored for a period of up to 3 years at 4°C. Based on sensitivity tests, 10 oocysts were randomly recovered by individually catapulting them from a Cryptosporidium-positive fecal smear stained with ZN acid-fast stain. Storage of recovered, extracted oocysts for more than 3 days had a negative effect on DNA extraction and prevented amplification of the target (data not shown). Therefore, DNA extractions were carried out immediately after oocysts were recovered by LCM and were subjected to real-time PCR analysis targeting the TRAP-C2 gene (37). With the appropriate real-time PCR, all analyzed positive samples gave identical average melting temperatures and real-time PCR curves. This indicated that only one genotype of Cryptosporidium was detected from each sample and was confirmed by gel electrophoresis (Fig. 3) and sequencing. All sequences showed at least 97% homology to the C. parvum TRAP-C2 gene.
FIG. 3.

Detection and genotyping of Cryptosporidium parvum oocysts from clinical samples (fecal glass smears) using LCM followed by real-time LightCycler PCR targeting the TRAP-C2 gene. PCR products were run on a 1.5% agarose gel, and positive results were indicated by the presence of a 369-bp PCR product. Lane l, 100-bp DNA ladder; lane 2, negative control; lane 3, patient sample Tn3 (slide number), stored for 3 years; lane 4, patient sample Tn23, stored for 2 years; lane 5, patient sample Tn24, stored for 2 years.
Discrimination of different C. parvum isolates from archived slides.
Oocysts of two different isolates (Moredun and genotype C [Rome isolate ISSC6]) were mixed together at a defined ratio to highlight the discriminatory power of the system. The isolates could not be distinguished morphologically. Individual oocysts were recovered, and real-time PCR amplicons of 232 bp were generated (data not shown). Based on the analysis of 18S rRNA amplicons (Fig. 2C), sequence analysis showed that these C. parvum isolates were clearly and consistently differentiated. From 21 PCR amplicons generated from single captured oocysts, 12 were typed as C. parvum Moredun isolates and 9 were identified as C. parvum Rome isolates.
DISCUSSION
High sensitivity and rapid detection and typing.
While microdissection has previously been used to excise C. andersoni oocysts from slides (23), our work is the first study to report the successful combination of LCM and real-time PCR for the rapid and sensitive detection of Cryptosporidium from glass slides. In our study, the combination of morphological analysis from stained slides and recovery of selected material makes LCM unique from any other extraction method for fixed material on slides (1-3). In addition, no adverse effect on the recovery of Cryptosporidium oocysts or subsequent extraction of DNA was observed for the different staining procedures (34). Gel electrophoresis and sequencing of the real-time PCR amplicons showed the successful amplification of target sequence and also importantly allowed us to genotype the recovered Cryptosporidium oocysts. LCM enabled us to recover selected oocysts without the use of detergents and reduced the amount of PCR inhibitors by catapulting only oocysts, and not surrounding fecal material, enabling the procurement of purer samples, limiting the effect of PCR inhibitors (31, 34). The results of our study also compare favorably with previously designed real-time systems where similar detection sensitivities were obtained (16, 22, 48), as we were routinely able to detect a single oocyst when targeting 18S rRNA. Nested PCR at the 18S rRNA locus is a robust and sensitive molecular technique capable of discriminating between all species of Cryptosporidium. However, real-time PCR allows for a quicker turnaround time in diagnosis, and importantly we demonstrated that LCM could be used not only for genotyping single Cryptosporidium oocysts but also for differentiating between different C. parvum isolates.
Cryptosporidium detection.
Molecular methods are essential for rapidly detecting Cryptosporidium in immunocompromised patients and for generating epidemiological data, which are of the utmost importance in the event of any outbreak of cryptosporidiosis (50, 56). In the majority of modern clinical laboratories, the most widely used staining methods for the detection of relatively low numbers of Cryptosporidium are IFA conjugates (46, 47). It is often necessary to use an oocyst concentration technique such as IMS to maintain an acceptable level of assay sensitivity (31, 34). The antibodies used in IMS bind selectively to the oocyst wall proteins enabling them to be concentrated in a suspension free from inhibitory debris. As useful as these laboratory techniques are, they are unable to discriminate between species of Cryptosporidium and do not lend themselves to the generation of any useful epidemiological data (31, 32, 34).
PCR methods have been shown to be more sensitive and specific than traditional microscopic techniques for detecting Cryptosporidium in both clinical and environmental samples (28, 29, 33). Recently, real-time PCR procedures for the detection and genotyping of oocysts of Cryptosporidium provide a reliable, specific, and rapid detection method alternative to nested PCR, with a baseline sensitivity of between 1 and 10 oocysts (16, 22, 27, 48), which our study agreed with. Previously, several studies have linked acute waterborne outbreak situations to the source using molecular detection methods (13, 19, 30). Postoutbreak, archived environmental slides and clinical slides produced from fecal smears are potential sources of genetic material for further epidemiological studies (2, 3). We have clearly demonstrated that the application of LCM followed by real-time PCR provides a valuable tool for unlocking archived Cryptosporidium genotype information, which is important epidemiological data that had previously been unobtainable.
Laser capture microscopy and PCR.
While LCM has an initially high capital price, e.g., £100,000, the technique has an extremely low unit cost (per sample), making application in diagnostic laboratories a competitive economic proposition. LCM has rapidly developed over the last few years and can be used as a tool to select and recover biological material from fixed/mounted slides (12, 24). Although this technology has predominantly been used as a tool for gene expression of fixed tissue section and in vitro-cultivated cell lines (12, 25), recent literature indicates the possibility of using it for the recovery of single life stages of parasites (40, 42, 44). Previous attempts to recover oocysts from archived slides were based on washing fixed oocyst/cryptosporidial DNA from slides (2, 3). These studies also did not find any inhibitory effect on PCR sensitivity following extraction (2, 3). However, the detection system employed in these studies was not as sensitive as our protocol; 30 μl of samples was fixed onto slides, equating to at least 30 oocysts for guaranteed detection (2, 3). Using our detection and identification system on glass slides we routinely tracked, extracted, detected, and typed single Cryptosporidium oocysts from archived glass slides.
In conclusion, we have shown that the combination of LCM and real-time detection provides a rapid, sensitive, and highly specific method to perform molecular analysis of recovered Cryptosporidium oocysts fixed onto glass slides. Our novel assay not only opens new routes to study archived material of previous outbreaks but also could be used to perform molecular analysis of a wide variety of environmental, clinical, and histological samples.
Acknowledgments
We thank Elise Cruthers and Kiernan McCorry from the Northern Ireland Public Health laboratories at Belfast City Hospital for providing clinical slides for analysis in this study. We also thank the Northern Ireland Cryptosporidium Reference Laboratory (Northern Ireland Department of Environment, Water Executive) for the provision of water-based slide samples for analysis. C. hominis oocysts, Peru (human) isolate, were kindly provided by the Centers for Disease Control and Prevention (CDC), Atlanta, GA. C. parvum oocysts, genotype C (isolate ISSC6), were kindly provided by Edoardo Pozio, Istituto Superiore di Sanita, Rome, Italy.
This work was partly funded through a LabLinks grant awarded by Safefood, the Food Safety Promotion Board. C.J.L., J.E.M., and J.S.G.D. were supported in part by funds from an EU Fifth Framework grant (PLK1-CT-1999-00775). Colm Lowery was also supported by a Sir Winston Churchill fellowship administered through the University of Ulster, Coleraine, and the CDC.
REFERENCES
- 1.Abe, N., I. Kimata, and M. Iseki. 2002. Comparative study of PCR-based Cryptosporidium discriminating techniques with a review of the literature. Kansenshogaku Zasshi. 76:869-881. [DOI] [PubMed] [Google Scholar]
- 2.Amar, C., S. Pedraza-Diaz, and J. McLauchlin. 2001. Extraction and genotyping of Cryptosporidium parvum DNA from faecal smears on glass slides stained conventionally for direct microscope examination. J. Clin. Microbiol. 39:401-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amar, C. F., R. M. Chalmers, K. Elwin, P. Tynan, and J. McLauchlin. 2002. Blinded evaluation of DNA extraction and genotyping of stained Cryptosporidium on glass slides. Lett. Appl. Microbiol. 35:486-488. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. 1999. Standard operating protocol for the monitoring of Cryptosporidium oocysts in treated water supplies to satisfy water supply (water quality) (amendment) regulations. SI no. 1524. Her Majesty's Stationery Office, London, United Kingdom.
- 5.Anonymous. 2000. The water supply (water quality) regulations 2000. Statutory instrument 2000, no. 3184. Queen’s Printer of Acts of Parliament, London, United Kingdom. [Online.] http://www.opsi.gov.uk/si/si2000/20003184.htm.
- 6.Anonymous. 2003. Water act, chapter 37, part 2. Queen’s Publisher of Acts of Parliament, London, United Kingdom.
- 7.Baron, E. J., L. R. Peterson, and S. M. Finegold. 1994. Bailey and Scott's diagnostic microbiology, p. 776-861. In B. A. Forbes, D. F. Sahm, and A. S. Weissfeld (ed.), Laboratory methods for the diagnosis of parasitic infections. Mosby, Inc., St. Louis, Mo.
- 8.Belkind-Valdovinos, U., J. Belkind-Gerson, D. Sanchez-Francia, M. M. Espinoza-Ruiz, and E. Lazcano-Ponce. 2004. Evaluacion de la nitazoxanida en dosis unica y por tres dias en parasitosis intestinal. Salud Publica Mex. 46:333-340. [DOI] [PubMed] [Google Scholar]
- 9.Burbach, G. J., D. Dehn, B. Nagel, D. Del Turco, and T. Deller. 2004. Laser microdissection of immunolabeled astrocytes allows quantification of astrocytic gene expression. J. Neurosci. Methods 138:141-148. [DOI] [PubMed] [Google Scholar]
- 10.Caccio, S. M. 2005. Molecular epidemiology of human cryptosporidiosis. Parassitologia 47:185-192. [PubMed] [Google Scholar]
- 11.Chen, X. M., S. P. O'Hara, B. Q. Huang, P. L. Splinter, J. B. Nelson, and N. F. LaRusso. 2005. Localized glucose and water influx facilitates Cryptosporidium parvum cellular invasion by means of modulation of host-cell membrane protrusion. Proc. Natl. Acad. Sci. USA 102:6338-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Martino, D., G. Giuffre, N. Staiti, A. Simone, P. Todaro, and L. Saravo. 2004. Laser microdissection and DNA typing of cells from single hair follicles. Forensic Sci. Int. 146:S155-S157. [DOI] [PubMed] [Google Scholar]
- 13.Elwin, K., R. M. Chalmers, R. Roberts, E. C. Guy, and D. P. Casemore. 2001. Modification of a rapid method for the identification of gene-specific polymorphisms in Cryptosporidium parvum and its application to clinical and epidemiological investigations. Appl. Environ. Microbiol. 67:5581-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection, and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 15.Fayer, R., M. Santin, and L. Xiao. 2005. Cryptosporidium bovis n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). J. Parasitol. 91:624-629. [DOI] [PubMed] [Google Scholar]
- 16.Fontaine, M., and E. Guillot. 2002. Development of a TaqMan quantitative PCR assay specific for Cryptosporidium parvum. FEMS Microbiol. Lett. 214:13-17. [DOI] [PubMed] [Google Scholar]
- 17.Giangaspero, A., U. Molini, R. Iorio, D. Traversa, B. Paoletti, and C. Giansante. 2005. Cryptosporidium parvum oocysts in seawater clams (Chameleagallina) in Italy. Vet. Med. 69:203-212. [DOI] [PubMed] [Google Scholar]
- 18.Giorgi, P. P., and M. L. Graydon. 1987. Retinal wholemounts: a simple method for precise mapping. J. Neurosci. Methods 22:119-124. [DOI] [PubMed] [Google Scholar]
- 19.Glaberman, S., J. E. Moore, C. J. Lowery, R. M. Chalmers, I. Sulaiman, K. Elwin, P. J. Rooney, B. C. Millar, J. S. Dooley, A. A. Lal, and L. Xiao. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guy, R. A., P. Payment, U. J. Krull, and P. A. Horgen. 2003. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl. Environ. Microbiol. 69:5178-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashim, A., G. Mulcahy, B. Bourke, and M. Clyne. 2006. Interaction of Cryptosporidium hominis and Cryptosporidium parvum with primary human and bovine intestinal cells. Infect. Immun. 74:99-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins, J. A., R. Fayer, J. M. Trout, L. Xiao, A. A. Lal, S. Kerby, and M. C. Jenkins. 2001. Real-time PCR for the detection of Cryptosporidium parvum. J. Microbiol. Methods 47:323-337. [DOI] [PubMed] [Google Scholar]
- 23.Hijjawi, N. S., B. P. Meloni, U. M. Ryan, M. E. Olson, and R. C. Thompson. 2002. Successful in vitro cultivation of Cryptosporidium andersoni: evidence for the existence of novel extracellular stages in the life cycle and implications for the classification of Cryptosporidium. Int. J. Parasitol. 32:1719-1726. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, P. T., M. N. Brown, B. C. Pulliam, D. H. Anderson, and L. V. Johnson. 2005. Synaptic pathology, altered gene expression, and degeneration in photoreceptors impacted by drusen. Investig. Ophthalmol. Vis. Sci. 46:4788-4795. [DOI] [PubMed] [Google Scholar]
- 25.Khanna, S., G. Cheng, B. Gong, M. J. Mustari, and J. D. Porter. 2004. Genome-wide transcriptional profiles are consistent with functional specialization of the extraocular muscle layers. Investig. Ophthalmol. Vis. Sci. 45:3055-3066. [DOI] [PubMed] [Google Scholar]
- 26.Le Chevallier, M. W., W. D. Norton, and R. G. Lee. 1991. Occurrence of Giardia and Cryptosporidium spp. in surface water supplies. Appl. Environ. Microbiol. 57:2610-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limor, J. R., A. A. Lal, and L. Xiao. 2002. Detection and differentiation of Cryptosporidium parasites that are pathogenic for humans by real-time PCR. J. Clin. Microbiol. 40:2335-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowery, C. J., J. E. Moore, B. C. Millar, D. P. Burke, K. A. McCorry, E. Crothers, and J. S. Dooley. 2000. Detection and speciation of Cryptosporidium spp. in environmental water samples by immunomagnetic separation, PCR and endonuclease restriction. J. Med. Microbiol. 49:779-785. [DOI] [PubMed] [Google Scholar]
- 29.Mayer, C. L., and C. J. Palmer. 1996. Evaluation of PCR, nested PCR, and fluorescent antibodies for detection of Giardia and Cryptosporidium species in wastewater. Appl. Environ. Microbiol. 62:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLauchlin, J., C. Amar, S. Pedraza-Diaz, and G. L. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, W. A., I. A. Gardner, E. R. Atwill, C. M. Leutenegger, M. A. Miller, R. P. Hedrick, A. C. Melli, N. M. Barnes, and P. A. Conrad. 2005. Evaluation of methods for improved detection of Cryptosporidium spp. in mussels (Mytilus californianus). J. Microbiol. Methods 65:367-379. [DOI] [PubMed] [Google Scholar]
- 32.Moore, J. E., and B. C. Millar. 2002. Need for improved molecular biology training for biomedical scientists in NHS microbiology laboratories. Br. J. Biomed. Sci. 59:180. [PubMed] [Google Scholar]
- 33.Morgan, U. M., L. Pallant, B. W. Dwyer, D. A. Forbes, G. Rich, and R. C. A. Thompson. 1998. Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens: clinical trial. J. Clin. Microbiol. 36:995-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nichols, R. A., J. E. Moore, and H. V. Smith. 2005. A rapid method for extracting oocyst DNA from Cryptosporidium-positive human faeces for outbreak investigations. J. Microbiol. Methods 65:512-524. [DOI] [PubMed] [Google Scholar]
- 35.Nydam, D. V., and H. O. Mohammed. 2005. Quantitative risk assessment of Cryptosporidium species infection in dairy calves. J. Dairy Sci. 88:3932-3943. [DOI] [PubMed] [Google Scholar]
- 36.Payment, P. 1999. Poor efficacy of residual chlorine disinfectant in drinking water to inactivate waterborne pathogens in distribution systems. Can. J. Microbiol. 45:709-715. [PubMed] [Google Scholar]
- 37.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. Mac Kenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson, L. J., A. T. Campbell, and H. V. Smith. 1992. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl. Environ. Microbiol. 58:3494-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubes, J., S. Kubickova, P. Musilova, M. E. Amaral, R. M. Brunner, and T. Goldammer. 2005. Assignment of chromosome rearrangements between X chromosomes of human and cattle by laser microdissection and Zoo-FISH. Chromosome Res. 13:569-574. [DOI] [PubMed] [Google Scholar]
- 40.Sacci, J. B., Jr., J. M. Ribeiro, F. Huang, U. Alam, J. A. Russell, P. L. Blair, A. Witney, D. J. Carucci, A. F. Azad, and J. C. Aguiar. 2005. Transcriptional analysis of in vivo Plasmodium yoelii liver stage gene expression. Mol. Biochem. Parasitol. 142:177-183. [DOI] [PubMed] [Google Scholar]
- 41.Schlaak, C., P. Hoffmann, K. May, and A. Weimann. 2005. Desalting minimal amounts of DNA for electroporation in E. coli: a comparison of different physical methods. Biotechnol. Lett. 27:1003-1005. [DOI] [PubMed] [Google Scholar]
- 42.Semblat, J. P., O. Silvie, J. F. Franetich, L. Hannoun, W. Eling, and D. Mazier. 2002. Laser capture microdissection of Plasmodium falciparum liver stages for mRNA analysis. Mol. Biochem. Parasitol. 121:179-183. [DOI] [PubMed] [Google Scholar]
- 43.Simmons, O. D., III, M. D. Sobsey, C. D. Heaney, F. W. Schaefer III, and D. S. Francy. 2001. Concentration and detection of Cryptosporidium oocysts in surface water samples by method 1622 using ultrafiltration and capsule filtration. Appl. Environ. Microbiol. 67:1123-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sokolova, Y. Y., L. R. McNally, J. R. Fuxa, and S. B. Vinson. 2004. Spore morphotypes of Thelohania solenopsae (microsporidia) described microscopically and confirmed by PCR of individual spores microdissected from smears by position ablative laser microbeam microscopy. Microbiology 150:1261-1270. [DOI] [PubMed] [Google Scholar]
- 45.Stephan, C., G. M. Yousef, A. Scorilas, K. Jung, M. Jung, G. Kristiansen, S. Hauptmann, T. Kishi, T. Nakamura, S. A. Loening, and E. P. Diamandis. 2004. Hepsin is highly over expressed in and a new candidate for a prognostic indicator in prostate cancer. J. Urol. 171:187-191. [DOI] [PubMed] [Google Scholar]
- 46.Sterling, C. R., and M. J. Arrowood. 1986. Detection of Cryptosporidium sp. infections using a direct immunofluorescent assay. Pediatr. Infect. Dis. 5:S139-S142. [DOI] [PubMed] [Google Scholar]
- 47.Stibbs, H. H., and J. E. Ongerth. 1986. Immunofluorescence detection of Cryptosporidium oocysts in fecal smears. J. Clin. Microbiol. 24:517-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanriverdi, S., A. Tanyeli, F. Baslamisli, F. Koksal, Y. Kilinc, X. Feng, G. Batzer, S. Tzipori, and G. Widmer. 2002. Detection and genotyping of oocysts of Cryptosporidium parvum by real-time PCR and melting curve analysis. J. Clin. Microbiol. 40:3237-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thielman, N. M., and R. L. Guerrant. 2004. Clinical practice. Acute infectious diarrhea. N. Engl. J. Med. 350:38-47. [DOI] [PubMed] [Google Scholar]
- 50.Tzipori, S. 1998. Cryptosporidiiosis: laboratory investigation and chemotherapy. Adv. Parasitol. 40:187-221. [DOI] [PubMed] [Google Scholar]
- 51.Umejiego, N. N., C. Li, T. Riera, L. Hedstrom, and B. Striepen. 2004. Cryptosporidium parvum IMP dehydrogenase: identification of functional, structural, and dynamic properties that can be exploited for drug design. J. Biol. Chem. 279:40320-40327. [DOI] [PubMed] [Google Scholar]
- 52.Unguen, C., P. Brasseur, A. Moreno-Sabater, L. Favennec, and J. J. Ballet. 1997. Evaluation of viability and infectivity of waterborne Cryptosporidium parvum oocysts. J. Eukaryot. Microbiol. 44:73S. [DOI] [PubMed] [Google Scholar]
- 53.Wetzel, D. M., J. Schmidt, M. S. Kuhlenschmidt, J. P. Dubey, and L. D. Sibley. 2005. Gliding motility leads to active cellular invasion by Cryptosporidium parvum sporozoites. Infect. Immun. 73:5379-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali,R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao, L., A. Singh, J. Limor, T. K. Graczyk, S. Gradus, and A. Lal. 2001. Molecular characterization of Cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 67:1097-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao, L., R. Fayer, U. Ryan, and S. J. Upton. 2004. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 17:72-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao L., A. A. Lal, and J. Jiang. 2004. Detection and differentiation of Cryptosporidium oocysts in water by PCR-RFLP. Methods Mol. Biol. 268:163-176. [DOI] [PubMed] [Google Scholar]