Abstract
GB virus C (GBV-C; also called hepatitis G virus) is a common cause of infection associated with prolonged survival among HIV-infected individuals. The prevalences of GBV-C viremia vary widely in different studies, and there has been poor agreement among different laboratories performing GBV-C RNA detection in quality control studies. To determine the optimal method of measuring GBV-C RNA in clinical samples, samples obtained from 939 HIV-infected subjects were studied using reverse transcription (RT)-PCR methods amplifying four separate regions of the GBV-C genome. Primers amplifying the E2 coding region were 100% specific; however, their sensitivity was only 76.6%. In contrast, primers amplifying three additional conserved regions of the GBV-C genome (the 5′ nontranslated region and the nonstructural protein-coding regions 3 and 5A) were more sensitive but produced higher rates of false-positive results. Using low-specificity primer sets influenced the significance of association between GBV-C viremia and response to antiretroviral therapy. Using a quantitative GBV-C RNA method, the GBV-C RNA concentration did not correlate with baseline or set point HIV RNA levels; however, a correlation between negative, low, and high GBV-C RNA levels and increasing reduction in HIV RNA following antiretroviral therapy was observed. Subjects with both GBV-C E2 antibody and viremia had significantly lower GBV-C RNA levels than did viremic subjects without E2 antibody. These studies demonstrate that accurate detection of GBV-C RNA by nested RT-PCR requires the use of primers representing multiple genome regions. Analyses based on testing with single primers do not lead to reliable conclusions about the association between GBV-C infection and clinical outcomes.
GB virus type C (GBV-C, also called hepatitis G virus) is classified within the family Flaviviridae and is the human virus most closely related to hepatitis C virus (17, 24). GBV-C contains a single-stranded, positive-sense RNA genome encoding a long polyprotein that is proteolytically cleaved into structural and nonstructural proteins (reviewed in reference 26). Epidemiologic studies have failed to identify any association between GBV-C and acute or chronic hepatitis or any other human disease (reviewed in references 2 and 19). Although GBV-C viremia may persist for decades in some infected humans, the majority of immune-competent individuals clear GBV-C RNA and thereafter have detectable antibody to the GBV-C surface envelope glycoprotein E2 (26). The presence of E2 antibody is associated with decreased risk of subsequent transfusion-related infection with GBV-C (29, 31), suggesting that E2 antibodies possess neutralizing activity.
Due to shared modes of transmission (7, 8, 10, 16, 22, 34), GBV-C infection is common in human immunodeficiency virus (HIV)-infected people (20, 27, 36), with active viremia or evidence of past infection (E2 antibody) present in as many as 86% (33). Active viremia with GBV-C has been detected by reverse transcription (RT)-PCR methods in 17% (9) to 43% (20) of HIV-positive individuals. In several, though not all, studies, HIV-infected people who were coinfected with GBV-C had decreased mortality (9, 14, 30, 33, 36, 37) and favorable clinical markers of HIV disease progression (30, 33, 37) compared to those without GBV-C viremia. A meta-analysis found a highly significant association with prolonged survival in HIV-infected individuals when GBV-C RNA was detected five or more years following HIV infection (39). In addition, several, though not all, studies found an association between GBV-C viremia and improved response to antiretroviral therapy (ART) (3, 6, 21, 25).
GBV-C viremia is measured by detecting viral RNA in serum or plasma using RT-PCR methods designed to amplify conserved sequences of the viral genome. Early studies of RT-PCR detection of GBV-C utilized primers that amplified the nonstructural-protein-coding regions 3 and 5A (NS3 and NS5A) (5, 12, 15); however, most subsequent studies have used primers that amplified the conserved 5′ nontranslated region (5′ NTR) of the genome (6, 9, 14, 21, 36, 37). We previously designed primers to amplify two regions of the 5′ NTR, the 3′ nontranslated region, the two envelope glycoprotein-coding regions (E1 and E2), and five nonstructural-protein-coding regions (NS2, NS3, NS4, NS5A, and NS5B) (J. Xiang, F. LaBrecque, W. N. Schmidt, D. Klinzman, D. Brashear, D. R. LaBrecque, M. J. Perino-Phillips, and J. T. Stapleton, presented at the Tenth Triennial International Symposium on Viral Hepatitis and Liver Disease, 9 to 14 April 2000, Atlanta, GA). Using these primers to detect GBV-C viremia in patients with hepatitis C virus and GBV-C coinfection, we found that RT-PCR using primers representing the E2 protein-coding region and the 5′ nontranslated region of the genome provided equal sensitivity, although the E2 primers provided more consistent results. Consequently, we and others used primers amplifying a portion of the E2 protein-coding region in several epidemiological studies of GBV-C and HIV coinfection (11, 23, 32, 33, 38).
In a recent study, different estimates of GBV-C prevalences were found when sera were tested by RT-PCR methods employing E2 and 5′-NTR primers (I. E. Souza, W. Zhang, R. S. Diaz, K. Chaloner, D. Klinzman, and J. T. Stapleton, presented at the Infectious Disease Society of America annual meeting, Boston, MA, 28 to 30 September 2004), suggesting that the specific primers selected to amplify GBV-C RNA might provide discordant results that could influence epidemiological studies. Consistent with this finding, three studies involving more than 20 laboratories evaluating the reproducibility of GBV-C RNA testing found surprisingly heterogeneous results, even among laboratories using a commercial assay (5, 12, 13). Because different testing methods for GBV-C viremia may influence both the estimated prevalence and the estimates of associations between GBV-C and HIV disease markers, we studied three populations of HIV-infected individuals with several primer sets in order to determine the optimal approach for detecting GBV-C viremia.
(This work was presented in part at the International AIDS Society meeting, 24 to 27 July 2005, in Rio de Janeiro, Brazil.)
MATERIALS AND METHODS
Patients.
Three cohorts of HIV-infected subjects (939 subjects) were studied to determine the sensitivities and specificities of test methods. Antiretroviral-naive HIV-positive Brazilian patients (n = 175) who participated in a previously described, randomized, prospective antiretroviral clinical trial were retrospectively studied (1, 25). The study participants were randomized to receive ART between 1994 and 1996 with a single drug (indinavir; group 1), two drugs (zidovudine and lamivudine; group 2), or three drugs (indinavir, zidovudine, and lamivudine; group 3). Demographic and clinical information and laboratory data (including CD4 cell counts and plasma HIV RNA levels) were obtained as previously described. The GBV-C RNA statuses of these patients were initially determined using one set of primers (E2 region) in a study designed to investigate the effect of GBV-C viremia on the response to antiretroviral therapy (25). Another population of HIV-infected individuals (n = 223) receiving care at the University of Iowa HIV clinic who had sufficient samples stored for research purposes were also studied. Patient characteristics have been previously described (11, 36). A third cohort of HIV-infected women participating in the Women's Interagency HIV Study (WIHS) were also studied (1,082 samples from 541 HIV-positive women) using a limited set of primers based on a strategy derived from the results of more extensive testing of the Brazilian and Iowa cohorts. Informed consent was obtained from all participants, and the study was approved by the University of Iowa Institutional Review Board.
GBV-C viremia and E2 antibody detection.
Plasma or serum samples were prepared as previously described and were stored at −70°C prior to this study (25, 33). All samples were identified by a unique code, and laboratory personnel were blinded to clinical data. To detect GBV-C viremia, RNA was extracted from plasma using a previously described guanidinium-isothiocyanate extraction method (the Brazilian cohort) (35) or a QIAamp Viral RNA Mini kit (the Iowa and WIHS cohorts) according to the manufacturer's instructions. RNA representing 25 μl of plasma was used in nested RT-PCRs, and the RT-PCR amplification conditions and product identification were as previously described (35). Five different sets of conserved oligonucleotide primers were designed based on alignment of published GBV-C sequences (Table 1). Negative and positive controls were included with each set of samples undergoing PCR testing.
TABLE 1.
Oligonucleotide primer sequences utilized to detect GBV-C RNA
| Region | RT-PCR product size (bp)a | GBV-C primer sequence
|
|
|---|---|---|---|
| Outer | Inner | ||
| 5′ NTR | 204 | + AAGCCCCAGAAACCGACGCC | CGGCCAAAAGGTGGTGGATG |
| − TGAAGGGCGACGTGGACCGT | GTAACGGGCTCGGTTTAACG | ||
| E2b | 745 | + GDC GYG AYT CGA ARA TMG AYG | GATATCGAARATMGAYGTGTGGAG |
| − AAGATCAACGGGACCAGCCGTGCCTCA | TTAGGTACCGCCTCAGCCAGCTTCAT | ||
| E2newb | 340 | + TGTGGGGTTCCGTDTCTTGGTT | TGGNTCWGCCAGCTGYACCATAGC |
| − RAACGTHCCRCTVGGAGGCT | DTCYCGGATCTTGGTCATGG | ||
| NS3 | 310 | + GGTRWCCCTTGATCCCACCAT | TCGGCWGAAYTGTCGATGCA |
| − CACATBGTCCGCTGAAC | ACGCCGCGHACYTTTGCCCA | ||
| NS5A | 200 | + ATGGTYTAYGGYCCTGGVCAAA | CTGGVCAAAGYGTYACCATT |
| − TACTGCARTCYTCCATGATGACAT | TTCAAGAATCCTCGCAGCATTCT | ||
| 5′ NTRc | + 5′ TACCGGTGTGAATAAGGGCC | CTCGTCGTTAAACCGAGCCCGTCAd | |
| − CGTCGTTTGCCCAGGTG | |||
Numbering is based on the sequence of the infectious GBV-C clone (GenBank accession number AF121950).
E2 primers contain restriction endonuclease sites designed for previous studies.
Primers used for real-time PCR experiments.
Probe.
Because it is not possible for RT-PCR testing with a single primer set to be completely accurate, we developed stringent criteria to provide a working gold-standard (GS) definition for GBV-C viremia. Because of limitations in sample volume, four primer sets were used for the Brazilian cohort, whereas all five primer sets were used for the University of Iowa cohort. Samples had to meet one of the following criteria to meet the gold standard for GBV-C viremia. First, samples that tested positive or negative with all primer sets were considered GS positive or GS negative. For discordant samples to be considered positive for GBV-C viremia, two samples from the same individual obtained on different dates (within a 3-month window) had to reproducibly test positive by RT-PCR using at least two different primer sets. While this is not an infallible classification, it is a stringent definition which should be highly sensitive and specific.
All samples with GBV-C RNA detected using the 5′-NTR primers were quantified by one-step real-time PCR. RNA from 20 μl of plasma was amplified using 5′-NTR primers and a 6-carboxyfluorescein/6-carboxytetramethylrhodamine-labeled probe (Table 1) using the SuperScript II Platinum One-step Quantitative RT-PCR System (Invitrogen) as recommended by the manufacturer. Specifically, samples were amplified in an ABI Prism 7700 sequence detector, and the cycle conditions were 50°C for 15 min and 95°C for 2 min, followed by 40 cycles of 58°C for 15 s and 72°C for 1 min. A standard curve was generated using a positive control sample containing 5 × 106 GBV-C genome equivalents (GE) per ml as determined by repetitive terminal dilution analysis in nested RT-PCRs. GBV-C antibodies against the envelope glycoprotein E2 were detected in serum or plasma using an enzyme-linked immunosorbent assay method (μPlate anti-HGenv test; Roche Diagnostics GmbH, Penzberg, Germany) as recommended by the manufacturer (28).
Data analysis.
The prevalence of GBV-C viremia and the sensitivity, specificity, and negative and positive predicted values (NPV and PPV) were estimated for each RT-PCR test method (each primer set) based on results obtained on the initial GBV-C RNA testing compared to the final assignment of GBV-C RNA positive or negative based on the GS criteria described above.
For Brazilian subjects, the baseline HIV viral load (VL) (the RNA concentration in genome equivalents per ml plasma) and CD4+ T-cell count and the average changes in the HIV VL and in the square root of the CD4+ T-cell count at weeks 24, 32, and 48 combined were analyzed according to the GBV-C viremia status based on testing with each primer set. The baseline GBV-C VL was measured in those who tested positive using the 5′-NTR primer set. Multiple-regression analysis, including the treatment effect, GBV-C viremia (yes/no), GBV-C VL (for those positive by 5′ NTR), baseline CD4+ T-cell count, baseline HIV VL, and other covariates (age, gender, transmission mode, CCR2, and CCR5) (24), was used to examine the effects of these covariates on the change in the HIV VL and CD4 T-cell counts in multivariate analyses as previously described (25) (the baseline minus the average of weeks 24, 32, and 48 combined). Because the lower limit of HIV RNA detection for the Brazilian cohort was 500 copies per ml, a value of 0.5 × log10 (500) copies/ml was imputed for undetectable measurements of the HIV VL in the primary analysis (25) and 0.5 × log10 (1,000) for an undetectable GBV-C VL. The GBV-C VL was also categorized into high (>106 copies/ml), low (≤106 copies/ml), and negative (negative by GS). For the Iowa cohort, the HIV VL and CD4+ T-cell count were analyzed based on baseline data (upon entry into the clinic) and data from the same day as the GBV-C testing, and a viral-load set point was determined (using the median VL for each patient obtained when the patient was not receiving antiretroviral medication).
Validation.
Based on the results from the methods and analyses described above, an algorithm with high sensitivity and specificity but simultaneously keeping the number of RT-PCR tests small was developed and recommended for clinical use. The algorithm was implemented on the 1,082 samples from the WIHS, and the number of RT-PCRs required for implementation was recorded.
RESULTS
GBV-C viremia in the HIV-infected patient cohorts.
Among the Brazilian subjects studied, 37 (21.1%) had concordant positive results and 108 (61.71%) had concordant negative results in RT-PCRs using all four sets of primers. Forty-nine subjects (28.0%) had discrepant results when tested with different primer sets. Sixteen of these subjects tested negative when E2 primers were used, yet GBV-C RNA was amplified by all three of the other primer sets. In 5 of these 16 subjects, repeat testing using the E2 primer set was positive. These subjects were by definition GS GBV-C RNA positive. The overall estimated prevalence of GBV-C viremia in this population was 30.3% (53/175) based on the GS definition (see Materials and Methods). The rate of GBV-C viremia detection when samples were tested with each of the four individual primer sets ranged from 21.1% to 38.3%. The PPV obtained using the E2 primer set was 100% in this cohort, although the sensitivity was only 69.8%. The 5′ NTR, NS5A, and NS3 primer pairs individually or together detected all of the Brazilian subjects who were classified as GBV-C RNA positive (GS). However, the specificities of these three primer sets were poor, only 89.3%, 90.9%, and 88.5%, respectively (Table 2).
TABLE 2.
GBV-C viremia prevalence and test characteristics for combined Brazilian and U.S. cohorts (n = 398)
| Primera | Prevalence | Sensitivity (%) | Specificity (%) | PPV | NPV | κb |
|---|---|---|---|---|---|---|
| 5′ NTR | 34.4 | 100 | 90.9 | 81.0 | 100 | 0.85 |
| E2 | 21.4 | 76.6 | 100 | 100 | 91.7 | 0.83 |
| E2newc | 22.4 | 79.4 | 97.6 | 92.0 | 93.1 | 0.80 |
| NS3 | 34.7 | 88.3 | 86.1 | 71.0 | 95.0 | 0.69 |
| NS5A | 34.0 | 95.5 | 89.9 | 78.5 | 98.1 | 0.80 |
| GS | 27.9 |
Primer set used to amplify GBV-C RNA.
κ, kappa coefficient.
Data only from U.S. (Iowa) cohort.
To determine if the reason for the lower rate of detection with the E2 primer set was related to a decrease in the sensitivity of the assay, the end point dilution RNA detection was determined for each primer set using a set of control samples. The amount of RNA required for amplification by the E2 primer set was 16-fold greater than that required for the NS3 and NS5A primer sets and 8-fold higher than that required for the 5′-NTR primers (Fig. 1). Thus, the reduced rate of detection of GBV-C viremia by the E2 region primers appears to be related to its reduced sensitivity, at least in part. An alternative hypothesis is that the GBV-C E2 sequence differs significantly between viruses infecting Brazilian subjects and subjects from the United States, as the primers were previously tested in U.S. populations. Consequently, we analyzed 223 subjects followed in the University of Iowa HIV clinic. In addition to the four sets of primers used to study the Brazilian cohort, we designed and validated an additional set of primers from the E2 region (E2new) (Table 1). The primer sequences were chosen based on alignment of the E2 regions from 40 GBV-C isolates representing all five GBV-C genotypes. In addition, the length of the GBV-C genome that was amplified was shorter to improve sensitivity. Using control samples, the sensitivities of these primers were comparable to those of the 5′-NTR primers. Isolated discordant samples were retested under conditions without reverse transcriptase in the reaction mixtures. These samples were negative, suggesting that false-positive results may not be related to carryover contamination of PCR products (data not shown).
FIG. 1.
Sensitivities of GBV-C RNA detection using different primer sets. RNA was extracted from GBV-C RNA positive control serum. The control RNA template was diluted 1:800 (initial dilution [1]) and in serial twofold dilutions as indicated. RT-PCR was performed on each dilution using the four designated primer sets. The NS3 and NS5A primers required 8- and 16-fold less RNA than did the 5′-NTR and E2 primers (respectively) to amplify GBV-C RNA.
Using these five primer sets to amplify GBV-C RNA from samples from the subjects in the Iowa cohort, the estimated prevalences ranged from 21.5% to 33.2% with individual primers, and using our GS definition, 26% were ultimately classified as GBV-C viremic. Similar to the Brazilian cohort, the 5′-NTR primers were the most sensitive in amplifying GBV-C RNA; however, the specificity was 92.1%, and the NS3 and NS5A primers were less specific (82.4% and 90.9%, respectively). The E2 primers remained 100% specific but did not detect 17.3% of the GBV-C RNA-positive individuals defined by the GS criteria. Although the new primer set amplifying sequences from the GBV-C E2 region (E2new) had an analytical sensitivity comparable to that of the 5′-NTR primer set using the control RNA preparation, these primers did not perform as well as the original primers (sensitivity, 77.6%; specificity, 97.6%), suggesting that the reduced sensitivity with control RNA preparations observed with the E2 primers does not totally explain the poor sensitivity observed in the two cohorts (Table 2). However, the fact that the new E2 primers did not perform better than the original E2 primers, despite improved sensitivity with control RNA, suggests that the reduced analytical performance of the E2 primers may be related to nucleotide sequence variability in this region. Studies are under way to address this hypothesis.
Combining the results from the Brazilian and Iowa cohorts, GBV-C estimated prevalences ranged from 21.4% to 34.4% when individual primers were used. The most specific primers were those from the E2 region (100%), although the sensitivity was only 76.6%. The most sensitive primers were those amplifying the 5′-NTR region (100%), although their specificity was only 90.9%. The prevalence, sensitivity, specificity, and PPV and NPV summary data for the combined Brazilian and U.S. cohorts are shown in Table 2. Storage conditions may reduce the quality of RNA in clinical samples with a resultant decrease in RT-PCR sensitivity. Thus, we compared GBV-C prevalence results and VL values among the Iowa cohort samples stored for more than 5 years with those stored for less than 3 years and did not find any differences, suggesting that specimen storage differences between samples did not significantly influence our results (data not shown).
GBV-C viremia, clinical outcomes, and HIV surrogate markers.
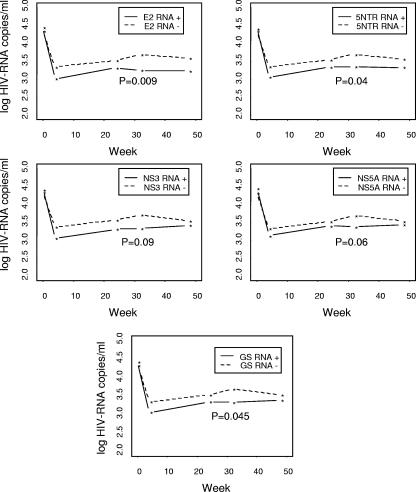
Because different primer sets yielded false-positive and false-negative results, it is possible that misclassification of GBV-C viremia based on the testing methods used might influence studies investigating potential associations between GBV-C viremia and HIV clinical findings. To illustrate this concern, we examined the effects of testing GBV-C RNA using different primer sets on markers of HIV replication (HIV VL) or pathogenesis (CD4 depletion) in the Brazilian and U.S. HIV-infected populations. In contrast to some reports (14, 30), no association was identified between the GBV-C VL and the baseline HIV VL or baseline CD4 T-cell counts for the Brazilian or Iowa populations, regardless of which primer set was used (data not shown). However, HIV VL reduction following initiation of their first ART regimen was greater in GBV-C RNA-positive subjects than in GBV-C-negative Brazilians (Fig. 2). The statistical significances of the results differed depending upon which primer set was used. Specifically, when GBV-C coinfection was determined by the E2 primers, 5′-NTR primers, or the gold-standard definition (PCR positive with two or more primer sets), the reduction in HIV RNA was statistically significant at P values of <0.05. However, using the primer sets with low specificity (NS3 and NS5A) to determine GBV-C viremia, the change in the HIV VL did not reach statistical significance at the 0.05 level. Thus, not only did the estimated rates of GBV-C viremia detection vary when different primers were used, but the interpretation of the clinical-outcome measurements also depended on which primer set was used to detect GBV-C RNA.
FIG. 2.
Effect of primer selection on HIV RNA reduction following ART among 175 ART treatment-naïve Brazilians. HIV RNA reductions 24, 36, and 48 weeks after initiation of ART were significantly (P < 0.05) greater among GBV-C RNA-positive subjects than among GBV-C RNA-negative subjects when tested with either the E2 or 5′-NTR primer set and with the GS definition (see Materials and Methods). When GBV-C RNA was detected by the NS3 and NS5A primers (which yielded higher rates of false-positive results), a greater reduction in HIV RNA was also demonstrated; however, the results were no longer statistically significant at a P level of <0.05.
GBV-C quantification.
Quantification of GBV-C RNA in the Brazilian cohort was performed using real-time RT-PCR, and the average reduction in the HIV VL was studied in relation to the GBV-C VL. Figure 3 demonstrates a relationship between GBV-C VL and HIV VL reduction following therapy. The effect of the GBV-C VL was well fitted by a linear trend, with the log10 HIV VL reduction increasing by approximately 0.24 copy/ml for a one-category increase (P = 0.03). The fit was not significantly improved by fitting a separate effect for each category (P = 0.74), and there was no significant treatment by GBV-C category interaction (P = 0.90).
FIG. 3.
Relationship between GBV-C RNA concentration and response to ART. The average reduction in HIV-RNA from baseline in the Brazilian subjects (n = 166) is shown when stratified by GBV-C RNA concentration. Subjects classified as GBV-C RNA negative using the criteria defining the gold standard (neg) were compared with those with “low” GBV-C RNA concentrations (≤106 copies/ml GBV-C RNA) or “high” GBV-C RNA concentrations (>106 copies/ml). The estimated increased reduction in log HIV RNA for one category increase in GBV-C RNA (neg = 1, low = 2, and high = 3) was 0.236 copies/ml (P = 0.03). Each box is between the upper and lower quartiles, with the bar denoting the median. Hatched lines represent 2 deviations from the mean. The whiskers represent the largest and smallest datum points unless there are outliers, which are shown as dots. Trt grp, treatment group.
Overall, GBV-C RNA levels ranged from 2.3 × 103 to 6.5 × 108 GE per ml (Brazilian) and 6.5 × 108 to 5.8 × 108 GE/ml (Iowa). The sensitivity of the real-time method was lower than that of the nested-RT-PCR methods, and 6.5% of the GBV-C RNA-positive subjects did not have detectable GBV-C RNA using the real-time PCR method. A previously published German study suggested a weak inverse correlation between the GBV-C RNA concentration and the HIV RNA concentration (R = −0.33) (30), and another longitudinal study of HIV-infected individuals found that the rate of the increase in HIV VL was significantly lower over 5 to 6 years of follow-up in GBV-C RNA-positive subjects than in GBV-C RNA-negative subjects (33). However, a relationship between GBV-C and HIV RNA levels was not observed at the pre-ART visit (Brazilian) or at the viral set point (Iowa) (Fig. 4): the estimated correlation coefficient was −0.01 (P = 0.92), and a moderate-to-large correlation was ruled out by the 95% confidence interval (−0.22 to 0.20). Similarly, in the Iowa cohort, the HIV VL did not correlate with GBV-C RNA levels at baseline or on the date of GBV-C RNA testing (data not shown).
FIG. 4.
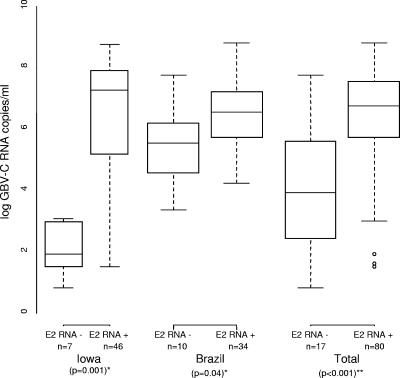
Relationship between baseline log10 GBV-C RNA copies/ml and detection by RT-PCR using the E2 primer set. The subjects were those who were GBV-C RNA positive by the GS definition and for whom log10 GBV-C RNA was available (n = 97). A value of 0.5 log10 (1,000) was imputed for 1 Brazil and 18 Iowa subjects whose GBV-C RNAs were below the lower limit of detection (*, P values from two-sample t test, for each study; **, P value adjusted for study). Each box is between the upper and lower quartile, with the bar denoting the median. The whiskers represent the largest and smallest datum points.
Because the E2 primer set required a higher GBV-C titer in order to amplify GBV-C RNA (Fig. 1), we compared the GBV-C VLs among Brazilian subjects whose GBV-Cs were classified based on the GS (E2 true-positive results) but who were negative when tested with the E2 primer set (E2 false-negative results). The Brazilians who tested negative with the E2 primers had significantly lower GBV-C VLs (5.60 log10) than those who tested positive with the E2 primers (6.47 log10; P = 0.039). This difference was more significant when the Iowa cohort was analyzed, as the mean GBV-C VL among those samples that were negative by the E2 primers was 1.68 log10 compared to 6.16 log10 among those positive by the E2 primers (P < 0.01).
GBV-C E2 antibody: prevalence and relationship to GBV-C viremia.
Each sample from the Brazilian and Iowa study subjects was tested for antibody to the GBV-C envelope glycoprotein (E2). Consistent with previous studies, the prevalence of detection of antibody to the GBV-C E2 protein was high, with E2 antibodies detected in 35% (136/391) of the entire study population tested (25% of the Brazilians and 43% of the U.S. cohort). Among those without GBV-C viremia, the overall rate of E2 antibody was 42% (Table 3). Evidence for exposure to GBV-C (RNA or antibody) was less common in Brazilians (54%) than in the U.S. cohort (63%). Concurrent GBV-C viremia and E2 antibody detection was found in 12% of the subjects. Among subjects with GBV-C infection as defined by the GS criteria, the frequency of E2 antibody was greater among those who tested negative for GBV-C RNA using the E2 primer set (data not shown). The GBV-C VL was lower among viremic individuals with E2 antibody (mean, log10 2.80 GE/ml versus log10 5.89 GE/ml; P = 0.0001), suggesting that E2 antibody indicates that subjects are in the process of clearing GBV-C viremia or, perhaps, that these antibodies select for less fit neutralization escape variants. Further studies to address this question are under way.
TABLE 3.
GB virus C E2 antibody prevalence in the Brazilian and Iowa HIV-infected cohorts
| GBV-C RNA status | GBV-C E2 antibody results
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Brazil
|
Iowa
|
Combined
|
||||||
| Negative | Positive | Negative | Positive | QNSa | Negative | Positive | QNS | |
| Positive | 51 | 2 | 43 | 11 | 4 | 94 | 13 | 4 |
| Negative | 81 | 41 | 80 | 82 | 3 | 161 | 123 | 3 |
| Total | 132 | 43 | 123 | 93 | 7 | 255 | 136 | 7 |
QNS, quantity not sufficient for testing.
DISCUSSION
As shown in previous studies, the estimated prevalence of GBV-C viremia is high among HIV-positive individuals (30% in the Brazilian study and 26% in the Iowa study). An additional 34% of the Brazilians and 51% of the Iowa cohort without viremia had evidence of prior GBV-C infection (E2 antibody). Published GBV-C prevalences have relied upon RT-PCR testing systems that amplify either the 5′ NTR or the E2, NS3, or NS5A region of the genome, and seldom have samples been tested by more than one set of primers. In addition, the use of nested RT-PCR for amplification of clinical samples is known to have problems with PCR carryover, leading to false-positive results. Thus, it is not too surprising that previous quality assurance studies (5, 12, 13) found poor test performance and significant variation between laboratories. Although prior studies identified problems with the reproducibility of GBV-C RNA detection, these studies did not systematically determine the validity of their test results by using multiple primer sets to amplify GBV-C RNA.
The E2 primer set resulted in the most specific detection of GBV-C RNA (Table 2). However, this primer set proved to be significantly less sensitive than the other four primer sets and frequently did not detect GBV-C RNA in subjects with VLs of <20,000 GE/ml. Nevertheless, these primers have been used to detect GBV-C in studies of HIV-infected people (4, 25, 38), and thus, some GBV-C-infected individuals were likely misclassified as GBV-C RNA negative. Testing positive with the E2 primer set is a marker of high GBV-C VL, and if higher GBV-C RNA concentrations are related to improved outcomes in HIV-positive individuals, use of the E2 primer set would increase the likelihood that a difference between GBV-C viremic and nonviremic individuals would be detected. This may explain the greater reduction in HIV VL among Brazilian subjects positive for GBV-C by the E2 system than by the other primer sets (Fig. 2), which is further supported by the relationship between the GBV-C VL and treatment response. Because the beneficial effect of zidovudine is transient due to development of drug resistance and poor tolerability, achieving a similar reduction in the HIV VL via infection with a nonpathogenic virus that is constantly in the bloodstream, frequently at high levels, strongly supports a clinically relevant beneficial effect related to our observation.
Conversely, the 5′ NTR, NS3, and NS5A primers appeared to be overly sensitive (Table 2). Our data do not exclude the possibility that the NS3 and NS5A primers may be accurate, detecting very low levels of GBV-C RNA. To maximize analytical accuracy, samples testing only positive with one of these primers were considered negative (by the gold-standard definition), and using these primers in clinical studies would dilute any potential effect of GBV-C viremia on HIV disease markers (Fig. 2). Using the E2, 5′-NTR, and gold-standard criteria, the reduction in HIV RNA following ART initiation was significantly greater in those with GBV-C viremia (P = 0.009, 0.04, and 0.045, respectively), by approximately 0.5 log10, than in those without GBV-C viremia, and there was a correlation between the reduction in HIV VL following initiation of ART and the GBV-C RNA concentration (Fig. 4). While the level of HIV VL reduction seems modest and on an individual level would not be significant, previous studies conclusively demonstrated that for each 0.5 log10 copies/ml reduction in HIV VL achieved with zidovudine monotherapy, the relative risk for progression to AIDS was reduced by approximately 33% (P < 0.001) (18). Because the beneficial effect of zidovudine is transient due to the rapid development of drug resistance, achieving a similar reduction in the HIV VL via infection with a nonpathogenic virus that is constantly in the bloodstream strongly supports a clinically relevant beneficial effect related to our observation. While it is true that current combination antiretroviral therapy is many times more potent, the additive effect of a 0.5 log10 reduction is not trivial.
Unlike some studies (14, 30), we did not demonstrate a higher baseline CD4 count or lower HIV RNA in those with GBV-C viremia than in those without it in either the Brazilian or the Iowa cohort. These data are limited by the fact that the duration of HIV infection is not known for subjects who participated in either of these studies. Furthermore, we also did not detect an inverse relationship between the GBV-C VL and the HIV VL (Fig. 4). Taken together, these data indicate that GBV-C RNA quantification is not a critical measurement in epidemiological studies of GBV-C and HIV disease progression but suggest that more accurate detection of viremia may be necessary for optimal investigation of potential interactions between these two virus infections.
These data suggest that neither RT-PCR detection of GBV-C using any of the four primer sets individually nor detection using the real-time RT-PCR method we used is sufficient to accurately classify GBV-C viremia status in clinical samples. Based on the sensitivity of the 5′-NTR primer set, we propose screening samples with these primers and testing all positive samples again using the E2 primer set. Samples testing positive with both sets of primers would be classified as true positives, and discordant samples would be tested further. Ideally, a new aliquot of the sample would be used to prepare a new RNA, or if this was not available, the remaining RNA from the first extraction would be used, and this RNA would be tested with all four primer sets (5′ NTR, E2, NS3, and NS5A). Those with repeat positives of 5′ NTR and positive by at least one of the other two primer sets would be classified as positive. Using this approach, we screened 1,082 samples from 541 women participating in the WIHS study, 21% of which (n = 231) tested positive by the 5′-NTR primers. Among the 231 5′-NTR-positive samples, 78% (n = 180) tested positive with the E2 primer set. Twenty-seven of the 51 discordant samples tested negative when the NS3 and NS5A primer sets were used. Thus, 204 of the 1,082 samples (19%) were classified as positive using the gold-standard criteria, and a total of 1,517 RT-PCRs were required to test 1,082 samples. Unfortunately, the sensitivity of the real-time RT-PCR method was poor, and more than 10% of the samples that were positive by qualitative RT-PCR were negative by real-time testing. Although the sensitivity was poor, the specificity was excellent, with >98% of positives confirmed by the nested-RT-PCR methods (data not shown). The development of a more sensitive real-time GBV-C RNA detection system is needed, and this may require the use of multiplex real-time primers. Further, development of an optimized real-time method would help decrease false-positive results and would facilitate comparisons of data among different laboratories.
In summary, these studies demonstrate that the primers used to amplify GBV-C RNA in nested RT-PCRs provide high rates of discordant results, and no one primer set consistently provides accurate results. To be confident in RT-PCR detection of GBV-C RNA, amplification using at least two different primer sets should be performed. Differences in the sensitivities and specificities of testing methods can affect epidemiological studies, as shown by the difference in statistical significance of the HIV RNA reduction in the Brazilian cohort following initiation of ART (Fig. 2). Finally, although a correlation between the GBV-C VL and the HIV VL set point was not observed, the plasma concentration of GBV-C RNA correlated with the extent of reduction in HIV RNA following ART (Fig. 3), and subjects with the highest level of GBV-C RNA had the greatest reduction in HIV RNA 48 weeks after therapy was initiated.
Acknowledgments
We expressly thank those individuals who participated in the Brazilian, Iowa, and WIHS cohort studies. Without their generous donation of time and specimens, this work would not have been possible. We thank the WIHS investigators, including Kathryn Anastos, Howard Minkoff, Mary Young, Alexandra Levine, Ruth Greenblatt, and Mardge Cohen. We also thank Conceicao Aparecida Accetturi for clinical assistance with the Brazilian cohort, Jeffery Meier and Kris Davis for assistance with the University of Iowa cohort, Wei Zhang for assistance with some of the statistical analyses, and Stephanie O'Conner for secretarial assistance. Data in the article were collected by the WIHS Collaborative Study Group with centers (principal investigators) in the New York City/Bronx Consortium (Kathryn Anastos); Brooklyn, NY (Howard Minkoff); the Washington DC Metropolitan Consortium (Mary Young); the Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt); the Los Angeles County/Southern California Consortium (Alexandra Levine); the Chicago Consortium (Mardge Cohen); and the Data Coordinating Center (Stephen Gange).
This work was supported by VA Merit Review grants (J.T.S. and J.X.), NIAID grants AI587401 and AI50478, and a subcontract from the Women's Interagency Health Survey to the Iowa City VA (J.T.S.) and a grant from the National Security Agency (K.C.). The work was also supported in part by a research fund from FAPESP, Sao Paulo, Brazil (grant 1996-5709-0); Pronex, Brazilian Ministry of Science and Technology, Brazil (grant 41.96.0943.00); and Merck Research Laboratories. The WIHS is funded by the National Institute of Allergy and Infectious Diseases, with supplemental funding from the National Cancer Institute and the National Institute on Drug Abuse (UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590). Funding is also provided by the National Institute of Child Health and Human Development (grant UO1-CH-32632) and the National Center for Research Resources (grants MO1-RR-00071, MO1-RR-00079, and MO1-RR-00083). None of the authors have any conflict of interest except J. T. Stapleton and J. Xiang, who are the holders of a patent (U.S. patent 6,870,043) for an infectious GB virus C clone, and D. Zdunek and G. Hess, who work for Roche Diagnostics and who developed the E2 antibody test kit.
REFERENCES
- 1.Accetturi, C. A., R. Pardini, G. H. N. Pinto, G. Turcato, Jr., D. S. Lewi, and R. S. Diaz. 2000. Effects of CCR5 genetic polymorphism and HIV-1 subtype in antiretroviral response in Brazilian HIV-1-infected patients. J. Acquir. Immune Defic. Syndr. 24:399-400. [DOI] [PubMed] [Google Scholar]
- 2.Alter, H. J. 1997. G-pers creepers, where'd you get those papers? A reassessment of the literature on the hepatitis G virus. Transfusion 37:569-572. [DOI] [PubMed] [Google Scholar]
- 3.Antonucci, G., E. Girardi, A. Cozzi-Lepri, M. R. Capobianchi, G. Morsica, P. Pizzaferri, L. Signhinolfi, A. Chiodera, M. Solmone, E. Lalle, G. Ippolito, A. Monforte, and the HepaICoNA Study Group. 2005. Response to HAART and GB virus type C coinfection in a cohort of antiretroviral-naive HIV-infected individuals. Antivir. Ther. 10:109-117. [PubMed] [Google Scholar]
- 4.Bisson, G., B. Strom, R. Gross, D. Weissman, D. Klinzman, W. T. Hwang, J. Kostman, D. Metzger, J. T. Stapleton, and I. Frank. 2005. Effect of GB virus C viremia on HIV acquisition and HIV set-point. AIDS 19:1910-1912. [DOI] [PubMed] [Google Scholar]
- 5.Bogard, M., C. Buffet-Janvresse, J. F. Cantaloube, P. Biagini, G. Duverlie, S. Castelain, J. Izopet, M. Dubois, C. Defer, I. Lepot, J. Coste, P. Marcellin, M. Martinot-Peignoux, P. Halfon, V. Gerolami, L. Frangeul, J. M. Pawlotsky, F. Roudot-Thoraval, E. Dussaix, P. Loiseau, N. Ravera, P. Lewin, J. Lamoril, J. Lerable, and P. Lebon. 1997. GEMHEP multicenter quality control study of PCR detection of GB virus C/hepatitis G virus RNA in serum. J. Clin. Microbiol. 35:3298-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brumme, Z. L., K. J. Chan, W. W. Y. Dong, T. Mo, B. Wynhoven, R. S. Hogg, J. S. G. Montaner, M. V. O'Shaughnessy, and P. R. Harrigan. 2002. No association between GB virus-C viremia and virological or immunological failure after starting initial antiretroviral therapy. AIDS 16:1929-1933. [DOI] [PubMed] [Google Scholar]
- 7.Feucht, H.-H., B. Zollner, S. Polywka, and R. Laufs. 1996. Vertical transmission of hepatitis G. Lancet 347:615-616. [PubMed] [Google Scholar]
- 8.Fischler, B., C. Lara, M. Chen, A. Sonnerborg, A. Nemeth, and M. Sallberg. 1997. Genetic evidence for mother-to-infant transmission of hepatitis G virus. J. Infect. Dis. 176:281-285. [DOI] [PubMed] [Google Scholar]
- 9.Heringlake, S., J. Ockenga, H. L. Tillmann, C. Trautwein, D. Meissner, M. Stoll, J. Hunt, C. Jou, N. Solomon, R. E. Schmidt, and M. P. Manns. 1998. GB virus C/hepatitis G virus infection: a favorable prognostic factor in human immunodeficiency virus-infected patients? J. Infect. Dis. 177:1723-1726. [DOI] [PubMed] [Google Scholar]
- 10.Hwang, S. J., C. W. Chu, R. H. Lu, K. H. Lan, J. C. Wu, Y.-J. Wang, F. Y. Chang, and S. D. Lee. 2000. Seroprevalence of GB virus C/hepatitis G virus-RNA and anti-envelope antibody in high-risk populations in Taiwan. J. Gastroenterol. Hepatol. 15:1171-1175. [DOI] [PubMed] [Google Scholar]
- 11.Klinzman, D., L. Katz, S. L. George, Q. Chang, J. Xiang, D. Zdunek, G. Hess, W. N. Schmidt, D. R. LaBrecque, and J. T. Stapleton. 2004. GB virus C/hepatitis G virus: does clearance of viremia differ based on mode of transmission?, p. 488-490. In A. R. Jilbert, E. V. L. Grgacic, K. Vickery, C. J. Burrl, and E. E. Cossart (ed.), 11th International Symposium on Viral Hepatitis and Liver Disease. Australian Center for Hepatitis Virology, Melbourne, Australia.
- 12.Kunkel, U., M. Hohne, T. Berg, U. Hopf, A. S. Kekule, G. Frosner, G. Pauli, and E. Schreier. 1998. Quality control study on the performance of GB virus C/hepatitis G virus PCR. J. Hepatol. 28:978-984. [DOI] [PubMed] [Google Scholar]
- 13.Lefrère, J. J., J. Lerable, M. Mariotti, M. Bogard, V. Thibault, L. Frangeul, P. Loiseau, F. Bouchardeau, S. Laperche, J. M. Pawlotsky, J. F. Cantaloube, P. Biagini, X. de Lamballerie, J. Izopet, C. Defer, I. Lepot, J. D. Poveda, E. Dussaix, V. Gerolami, P. Halfon, C. Buffet-Janvresse, C. Ferec, B. Mercier, P. Marcellin, M. Martinot-Peignoux, and M. Gassain. 2000. Lessons from a multicentre study of the detectability of viral genomes based on a two-round quality control of GB virus C (GBV-C)/hepatitis G virus (HGV) polymerase chain reaction assay. J. Virol. Methods 85:117-124. [DOI] [PubMed] [Google Scholar]
- 14.Lefrère, J. J., F. Roudot-Thoraval, L. Morand-Joubert, J.-C. Petit, J. Lerable, M. Thauvin, and M. Mariotti. 1999. Carriage of GB virus C/hepatitis G virus RNA is associated with a slower immunologic, virologic, and clinical progression of human immunodeficiency virus disease in coinfected persons. J. Infect. Dis. 179:783-789. [DOI] [PubMed] [Google Scholar]
- 15.Lefrère, J. J., F. Roudot-Thoraval, R. Roudot-Thoraval, P. Loiseau, J. F. Cataloube, P. Biagini, M. Mariotti, G. LeGac, and B. Mercier. 1999. GBV-C/hepatitis G virus (HGV) RNA load in immunodeficient individuals and in immunocompetent individuals. J. Med. Virol. 59:32-37. [DOI] [PubMed] [Google Scholar]
- 16.Lefrère, J.-J., F. Roudot-Thoraval, L. Morand-Joubert, Y. Brossard, F. Parnet-Mathieu, M. Mariotti, F. Agis, G. Rouet, J. Lerable, G. Lefevre, R. Girot, and P. Loiseau. 1999. Prevalence of GB virus type C/hepatitis G virus RNA and of anti-E2 in individuals at high or low risk for blood-borne or sexually transmitted viruses: evidence of sexual and parenteral transmission. Transfusion 39:83-94. [DOI] [PubMed] [Google Scholar]
- 17.Linnen, J., J. Wages, Z.-Y. Zhang-Keck, K. E. Fry, K. Z. Krawczynski, H. Alter, E. Koonin, M. Gallagher, M. Alter, S. Hadziyannis, P. Karayiannis, K. Fung, Y. Nakatsuji, J. W. K. Shih, M. Piatak, C. Hoover, J. Fernandez, S. Chen, J.-C. Zou, T. Morris, K. C. Hyams, S. Ismay, J. D. Lifson, G. Hess, S. K. H. Foung, H. Thomas, D. Bradley, H. Margolis, and J. P. Kim. 1996. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science 271:505-508. [DOI] [PubMed] [Google Scholar]
- 18.O'Brien, W. A., P. M. Hartigan, E. S. Daar, M. S. Simberkoff, J. D. Hamilton, et al. 1997. Changes in plasma HIV RNA levels and CD4+ lymphocyte counts predict both response to antiretroviral therapy and therapeutic failure. Ann. Intern. Med. 126:939-945. [DOI] [PubMed] [Google Scholar]
- 19.Polgreen, P. M., J. Xiang, Q. Chang, and J. T. Stapleton. 2003. GB virus type C/hepatitis G virus: a nonpathogenic flavivirus associated with prolonged survival in HIV-infected individuals. Microbes Infect. 5:1255-1261. [DOI] [PubMed] [Google Scholar]
- 20.Rey, D., J. Vidinic-Moularde, P. Meyer, C. Schmitt, S. Fritsch, J. M. Lang, and F. Stoll-Keller. 2000. High prevalence of GB virus C/hepatitis G virus RNA and antibodies in patients infected with human immunodeficiency virus type 1. Eur. J. Clin. Microbiol. Infect. Dis. 19:721-724. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez, B., I. Woolley, M. M. Lederman, D. Zdunek, G. Hess, and H. Valdez. 2003. Effect of GB virus coinfection on response to antiretroviral treatment in human immunodeficiency virus-infected patients. J. Infect. Dis. 187:504-507. [DOI] [PubMed] [Google Scholar]
- 22.Rubio, A., C. Rey, A. Sanchez-Zuijano, M. Leal, J. A. Pineda, E. Lissen, and G. Hess. 1997. Is hepatitis G virus transmitted sexually? JAMA 277:532-533. [DOI] [PubMed] [Google Scholar]
- 23.Schuval, S., J. C. Lindsey, J. T. Stapleton, R. B. Van Dyke, P. Palumbo, L. M. Mofenson, J. M. Oleske, J. Cervia, A. Kovacs, W. N. Dankner, E. Smith, B. Nowak, G. Ciupak, N. Webb, M. Eagle, D. Smith, R. Hennessey, M. Goodman-Kerkau, D. Klinzman, G. Hess, D. Zdunek, M. J. Levin, and the Pediatric AIDS Clinical Trials Group Protocol 1028S Team. 2005. GB virus C infection in children with perinatal human immunodeficiency virus infection. Pediatr. Infect. Dis. J. 24:417-422. [DOI] [PubMed] [Google Scholar]
- 24.Simons, J. N., T. P. Leary, G. J. Dawson, T. J. Pilot-Matias, A. S. Muerhoff, G. G. Schlauder, S. M. Desai, and I. K. Mushahwar. 1995. Isolation of novel virus-like sequences associated with human hepatitis. Nat. Med. 1:564-569. [DOI] [PubMed] [Google Scholar]
- 25.Souza, I. E., W. Zhang, R. S. Diaz, K. Chaloner, D. Klinzman, and J. T. Stapleton. 2006. Effect of GB virus C on response to antiretroviral therapy in HIV infected Brazilians. HIV Med. 7:25-31. [DOI] [PubMed] [Google Scholar]
- 26.Stapleton, J. T. 2003. GB virus type C/hepatitis G virus. Semin. Liver Dis. 23:137-148. [DOI] [PubMed] [Google Scholar]
- 27.Stapleton, J. T., C. F. Williams, and J. Xiang. 2004. GB virus C: a beneficial infection? J. Clin. Microbiol. 42:3915-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tacke, M., S. Schmolke, V. Schlueter, S. Sauleda, J. I. Esteban, E. Tanaka, K. Kiyosawa, H. J. Alter, U. Schmitt, G. Hess, B. Ofenloch-Haehnle, and A. M. Engel. 1997. Humoral immune response to the E2 protein of hepatitis G virus is associated with long-term recovery from infection and reveals a high frequency of hepatitis G virus exposure among healthy blood donors. Hepatology 26:1626-1633. [DOI] [PubMed] [Google Scholar]
- 29.Thomas, D. L., D. Vlahov, H. J. Alter, R. Marshall, J. Astemborski, and K. E. Nelson. 1998. Association of antibody to GB virus C (hepatitis G virus) with viral clearance and protection from reinfection. J. Infect. Dis. 177:539-542. [DOI] [PubMed] [Google Scholar]
- 30.Tillmann, H. L., H. Heiken, A. Knapik-Botor, S. Heringlake, J. Ockenga, J. C. Wilber, B. Goergen, J. Detmer, M. P. Manns, M. Stoll, R. E. Schmidt, and M. P. Manns. 2001. Infection with GB virus C and reduced mortality among HIV-infected patients. N. Engl. J. Med. 345:715-724. [DOI] [PubMed] [Google Scholar]
- 31.Tillmann, H. L., S. Heringlake, C. Trauwein, D. Meissner, B. Nashan, H. J. Schlitt, J. Kratochvil, J. Hunt, X. Qiu, C. Lou, R. Pichlmayr, and M. P. Manns. 1998. Antibodies against the GB virus C envelope 2 protein before liver transplantation protect against GB virus C de novo infection. Hepatology 28:379-384. [DOI] [PubMed] [Google Scholar]
- 32.Weintrob, A. C., J. D. Hamilton, C. Hahn, D. Klinzman, G. Moyo, D. Zdunek, G. Hess, D. K. Benjamin, Jr., and J. T. Stapleton. 2004. Active or prior GB virus C infection does not protect against vertical transmission of HIV in co-infected women from Tanzania. Clin. Infect. Dis. 38:e46-e48. [DOI] [PubMed] [Google Scholar]
- 33.Williams, C. F., D. Klinzman, T. E. Yamashita, J. Xiang, P. M. Polgreen, C. Rinaldo, C. Liu, J. Phair, J. B. Margolick, D. Zdunek, G. Hess, and J. T. Stapleton. 2004. Persistent GB virus C infection and survival in HIV-infected men. N. Engl. J. Med. 350:981-990. [DOI] [PubMed] [Google Scholar]
- 34.Wu, J. C., W. Y. Sheng, Y. H. Huang, S. J. Hwang, and S. D. Lee. 1997. Prevalence and risk factor analysis of GBV-C/HGV infection in prostitutes. J. Med. Virol. 52:83-85. [PubMed] [Google Scholar]
- 35.Xiang, J., D. Klinzman, J. McLinden, W. N. Schmidt, D. R. LaBrecque, R. Gish, and J. T. Stapleton. 1998. Characterization of hepatitis G virus (GB-C Virus) particles: evidence for a nucleocapsid and expression of sequences upstream of the E1 protein. J. Virol. 72:2738-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang, J., S. Wunschmann, D. J. Diekema, D. Klinzman, K. D. Patrick, S. L. George, and J. T. Stapleton. 2001. Effect of coinfection with GB virus C (Hepatitis G virus) on survival among patients with HIV infection. N. Engl. J. Med. 345:707-714. [DOI] [PubMed] [Google Scholar]
- 37.Yeo, A. E. T., A. Matsumoto, M. Hisada, J. W. Shih, H. J. Alter, and J. J. Goedert. 2000. Effect of hepatitis G virus infection on progression of HIV infection in patients with hemophilia. Ann. Intern. Med. 132:959-963. [DOI] [PubMed] [Google Scholar]
- 38.Zander, C., J. Anderson, J. Blackard, W. Lin, D. Zdunek, G. Hess, M. Peters, M. Koziel, K. Sherman, G. K. Robbins, R. T. Chung, et al. 2003. GB virus-C is frequent in HIV/HCV coinfected patients receiving interferon and ribaviran treatment but does not adversely affect HIV-1 viremia. Hepatology 38:669A. [Google Scholar]
- 39.Zhang, W., K. Chaloner, H. L. Tillmann, C. F. Williams, and J. T. Stapleton. 2006. Effect of early and late GBV-C viremia on survival of HIV infected individuals: a meta-analysis. HIV Med. 7:173-180. [DOI] [PubMed] [Google Scholar]