Abstract
The dimorphic fungus, Penicillium marneffei, is an emerging opportunistic pathogen endemic in Southeast Asia, especially for those with impaired cellular immunity such as human immunodeficiency virus-infected persons. A discriminatory and reproducible method based on the analysis of nucleotide sequences would facilitate epidemiologic investigations of this fungus. Twenty-four clinical or environmental isolates of P. marneffei obtained from China, Thailand, and Vietnam were analyzed by nucleotide sequence analysis. A total of 3,803 bp, consisting of eight nuclear gene fragments (transcription factor [AbaA], catalase [CpeA]], homodomain transcription factor [StlA], isocitrate lyase [Icl1], polyaromatic amino acid biosynthesis [PAA], NADH-dependent glutamate synthase [NGS], lovastatin nonaketide synthase [LNS], a cell wall mannoprotein [MP1], and a gene fragment of the cytochrome oxidase subunit 1 gene [COX1] of the P. marneffei mitochondrial genome) were amplified by PCR and then sequenced. No polymorphic sites within the Cox1 gene fragment were observed. Likewise, no nucleotide sequence polymorphisms were observed for three gene fragments: StlA, AbaA, and NGS. Seven single-nucleotide polymorphisms were observed for three gene fragments, Icl1, CpeA, and PAA, providing only a low degree of discriminatory power (D = 0.747). In contrast, the gene fragment for an antigenic cell wall glycoprotein, MP1, a useful immunologic marker for infection, was observed to be highly polymorphic with 12 different MP1 types (D = 0.887). Single-nucleotide polymorphisms were observed at 21 different locations in the MP1 gene fragment. Indels of 3, 21, 24, and 42 bp were observed and were in frame for protein translation. The relatively high degree of MP1 polymorphisms suggests the sequence is rapidly evolving in order to evade host immune responses. After all polymorphic gene sequences were combined, a high degree of genetic variation was observed (D = 0.949) for a total of 16 different haploid sequence types with 11 genotypes represented by single isolates. Phylogenetic analysis detected clusters composed of isolates obtained only from China or Thailand, as well as clusters with a combination of isolates from these two countries, indicating some mixing or common descent. Identical sequences were observed for isolates passed in vitro for 8 weeks, suggesting good reproducibility. The low degree of nucleotide diversity in housekeeping and regulatory genes suggests the recent emergence and spread as a species or an evolutionary bottleneck. In summary, multilocus sequence typing demonstrated a high degree of discriminatory power and reproducibility and may provide a robust and reliable adjunct method for genotyping isolates of P. marneffei and facilitating interlaboratory comparisons.
Penicilliosis of humans is a rare disease with one exception: infections caused by the opportunistic fungal pathogen Penicillium marneffei. In contrast to the worldwide and ubiquitous distribution of other species of Penicillium, P. marneffei is endemic to Southeast Asia and is responsible for causing serious disseminated opportunistic infections in persons with defects in their cell-mediated immunity such as cancer, Hodgkin's disease, and especially in human immunodeficiency virus (HIV)-infected persons (10, 25, 43, 54). Although less frequent, infections in immunocompetent persons have also been reported (12, 27). Infection with P. marneffei has emerged in the past decade as an important public health problem in Southeast Asia, where the infection often serves as an indicator disease for HIV (43, 45, 52). P. marneffei is a facultative intracellular pathogen. The primary route of infection is believed to be through inhalation of conidia into the lungs, where it can then disseminate via a hematogenous route to other body locations, especially the liver (10, 54). Clinical diagnosis is difficult since the clinical presentation of the disease is nonspecific and may include rapid weight loss, fever, and skin lesions following dissemination (10, 54). The histopathology of infected tissues reveals oval yeast-like pleomorphic cells 4 to 8 μm in diameter reproducing by fission. Infections are usually fatal if untreated and have been treated using amphotericin B and itraconazole (26, 45).
Phylogenetic analysis of mitochondrial and nuclear ribosomal gene sequences of P. marneffei detected a close relationship to Penicillium subgenus Biverticillium and Talaromyces species (31, 32). However, the fungus is still believed to be asexual since its teleomorph presently has not been identified. P. marneffei is dimorphic, that is, capable of growing in two different morphological states: either as a septated, filamentous fungus or as yeast-phase cells. Exposure of mycelial cells grown in vitro to environmental stimuli such as a temperature increase to 37°C triggers a reversible morphological transition to yeast phase cells that reproduce by schizogamy (1).
Despite being the third most common opportunistic infection in HIV-infected persons in Thailand (44) and a 10% infection rate of HIV-infected persons in Hong Kong (55), the ecology and natural reservoir of this dimorphic pathogen remain uncertain. P. marneffei has been isolated from the internal organs and soil surrounding the burrows of bamboo rats, but case-control studies detected no significant epidemiologic link between P. marneffei infections and exposure to bamboo rats (9). At present, it is not known whether bamboo rats are a major reservoir for human infections or are only part of life cycle of the fungus or from another source in the environment (21). Isolates obtained from humans and bamboo rats were observed to share a common genotype; however, the route of infection and transmission to humans has not been determined, although aerosolization of conidia has been proposed as the most likely mechanism (21, 54). Recently, typing using polymorphic microsatellite markers proposed geographic differences in population and localized dispersion of strains, but no clear epidemiologic link was observed between environmental and clinical isolates (17, 30).
Genotyping of environmental and clinical isolates may provide important answers to questions regarding routes and source of transmission, persistence of strains, relapse, and the presence of pathogenic strains. Multilocus sequence typing (MLST) was first developed to study the relatedness of bacterial isolates (33) and was later applied to investigations of pathogenic fungi (4, 11, 13, 19, 47). Both multilocus enzyme analysis and MLST analysis rely on surveying polymorphisms in housekeeping loci. MLST, however, has the advantage of the ability to also detect neutral mutations, thereby providing for a more robust method for detecting genetic variation between isolates than can be provided by multilocus enzyme analysis. MLST has been shown to be able to determine genetic relatedness between isolates and has the ability to share and standardize results compiled between laboratories that may result in the opportunity for worldwide typing databases such as proposed for Candida albicans (5).
Despite its emerging public health importance, at present, few complete P. marneffei gene sequences have been determined, nor has the genetic relatedness between isolates been examined. The goals of the present study were to determine the usefulness of a typing scheme based on nucleotide sequence variation in the P. marneffei genome and to examine the performance of the method. Differences in MLST genotypes for clinical isolates obtained from China and Thailand were investigated.
MATERIALS AND METHODS
Isolates and purification of DNA.
The twenty-four isolates of P. marneffei and their sources used in this investigation are listed in Table 1. Nine isolates (B-6318 to B-6326) were provided by the West China Hospital of Sichuan University, Chengdu, China. Eight isolates, B-6466, B-6463, and the isolates labeled IFM were provided by Kazuko Nishimura, Research Center for Pathogenic Fungi and Microbial Toxicoses, Chiba University, Chiba, Japan. Isolates labeled NCPF were obtained from the National Collection of Pathogenic Fungi maintained at the PHLS Mycology Reference Laboratory, Bristol, United Kingdom. All other isolates were obtained from the culture collection maintained by the Mycotic Diseases Branch at the Centers for Disease Control and Prevention and were previously described (30). Identification to the species level was based on three criteria: the ability of the cultures to convert from yeast to mold, the production of a red pigment, and the morphological identification of colonies and conidiophores (10, 56). Yeast-phase cultures were maintained on Bacto brain heart infusion agar (Becton Dickinson, Sparks, Md.), and mold cultures were maintained on Sabouraud dextrose agar. Genomic DNA was purified from mycelial phase cells as described by Lasker and Ran (30). DNA purified from isolates subcultured in vitro for 7 to 8 weeks was described previously (30).
TABLE 1.
P. marneffei isolates, sources, and MLST genotypes
| Isolate | Source | Geographic location | Genotype
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AbaA | StlA | CPE1 | ICL1 | LNS | PAA | NGS | MPa | HSTb | |||
| ATCC 64101 | Human | China | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ATCC 64102 | Rat | China | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 |
| NCPF 4090 | Human | Hong Kong | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 3 |
| NCPF 4158 | Human | Thailand | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 4 | 4 |
| B-6318 | Human | China | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 5 | 5 |
| B-6319 | Human | China | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 6 |
| B-6320 | Human | China | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | 6 |
| B-6321 | Human | China | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 7 | 7 |
| B-6322 | Human | China | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | 8 |
| B-6323 | Human | China | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 8 | 8 |
| B-6324 | Human | China | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | 9 |
| B-6325 | Human | China | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 9 | 10 |
| B-6326 | Human | China | 1 | 1 | 1 | 1 | 1 | 4 | 1 | 9 | 11 |
| ATCC 200050 | Human | Thailand | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 10 | 12 |
| ATCC 200051 | Human | Thailand | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 10 | 13 |
| ATCC 18224 | Rat | Vietnam | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 11 | 14 |
| B-6466 | Human | Thailand | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 7 | 7 |
| B-6463 | Human | Thailand | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | 13 |
| IFM 47279 | Human | Thailand | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | 13 |
| IFM 47285 | Human | Thailand | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 10 | 15 |
| IFM 47286 | Human | Thailand | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | 13 |
| IFM 47287 | Human | Thailand | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 12 | 16 |
| IFM 47288 | Human | Thailand | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 10 | 13 |
| IFM 47289 | Human | Thailand | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 10 | 15 |
MP1 sequence genotype.
HST assignments based on analysis for the five polymorphic loci (CPE1, ICL1, LNS, PAA, and MP1).
PCR amplification and sequencing.
Oligonucleotide primer sets used for the amplification of nine gene fragments by PCR are shown in Table 2. Target loci were selected based on the availability of internal gene fragments with different cellular functions and their deposition in the GenBank database. The nucleotide sequence for the cytochrome oxidase subunit 1 (Cox1) of the mitochondrial genome was obtained from Woo et al. (58). DNA sequences for the regulatory gene transcription factor homologues, AbaA, and the homodomain transcription factor, StlA, were obtained from Borneman et al. (3) and Borneman et al. (2), respectively. DNA sequences for five housekeeping genes—catalase (CpeA [39]), isocitrate lyase (Icl1), polyaromatic amino acid biosynthesis (PAA), NADH-dependent glutamate synthase (NGS), and lovastatin nonaketide synthase (LNS)—were obtained from Yuen et al. (58). The cell wall antigen MP1 gene sequence was described by Cao et al. (8).
TABLE 2.
Gene fragments and oligonucleotide primers for amplification and sequencing
| Locus | Gene product | GenBank accession no. | Fragment size (bp) | Primer typec | Primer sequence (5′ to 3′) |
|---|---|---|---|---|---|
| Cox1 | Cytochrome oxidase | AY347307 | 419 | F | ACATTTTCCACAAGGAAAAGGTGGA |
| R | AACGAAAAACTCCAGAAGTACCTGA | ||||
| AbaA | Transcription factor | AF272838 | 411 | F | TTGGTCGGATGCATTGGA |
| R | TAATCGATGGAGGTTCGGTA | ||||
| CPE1 | Catalase | AF537129 | 312 | F | TCTCAACAGCTGGCCCGACA |
| R | AAGGTGTGACCGCCGGCAA | ||||
| NGS | NADH-dependent glutamate synthase | AL684347 | 425 | F | TGAGAGAGTAACTGCCTCATCAGGA |
| R | GAGGACCGGCAACTTCCCTGA | ||||
| ICL1 | Isocitrate lyase | AF373018 | 435 | F | TACCGACCTCCTTGCCATTGCA |
| R | TTGACGGCGCATTGCGTACCA | ||||
| LNS | Lovastatin nonaketide synthase | AL684236 | 454 | F | CTCCGGGATCAACCAATGTACAGA |
| R | ATTCGGGATCAACCAATGTACAGA | ||||
| PAA | Polyaromatic amino acid biosynthesis | AL684053 | 438 | F | CGTGAAGGTGGCTTGCGCAA |
| R | GTCCTTGGGAACGCCAGGAA | ||||
| StlA | Transcription factor | AF284062 | 459 | F | CAAAAGTCACATTCCTGCCCTA |
| R | GACTCTTCTTCGAGCGAACCAA | ||||
| MP1 | Cell wall antigen | AF009957 | 450a | F | CAAGCCCTCCAGAAAGGTATCCA |
| R | CTTTGTGGAGACCAATTCGCTGA | ||||
| 303b | F | GTTGGTGGGGGTTATGGTCCTCA | |||
| R | GAGAAGCATCACCGCCCACGTA |
Primer types F and R correspond to positions 527 to 549 and positions 954 and 976, respectively, of MP1 gene sequence for P. marneffei isolate PM4 (8).
Primer types F and R correspond to positions 386 to 408 and positions 698 and 710, respectively, of MP1 gene sequence for P. marneffei isolate PM4 (8).
F, forward; R, reverse.
PCR mixtures consisted of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, each primer at 0.2 μM, a 0.2 mM concentration of each deoxynucleotide triphosphate, 30 ηg of genomic DNA, and 2.5 U of Taq DNA polymerase in a final volume of 30 μl. Using a GeneAmp PCR System 9700 thermal cycler, PCR amplification was performed starting with an initial denaturation step for 3 min at 95°C, followed by 30 cycles each of 30 s at 95°C, 30 s at 58°C, and 60 s at 72°C. PCR amplification products were purified by using Qiaquick PCR purification columns (QIAGEN, Chatsworth, Calif.). PCR amplification was verified by electrophoresis of samples through 1.4% (wt/vol) agarose gels. Purified DNA fragments were sequenced in both forward and reverse directions using an ABI Prism BigDye terminator cycle sequencing ready reaction kit version 1.1 (Perkin-Elmer/Applied Biosystems) by using the manufacturer's reagents and recommendations for cycle sequencing. Centricep columns (Princeton Separations, Aldelphia, N.J.) were used to remove unlabeled nucleotides as well as unincorporated dye-labeled nucleotides. Samples were dried in a Speed Vac (Savant Instruments, Holbrook, N.Y.) and then suspended overnight in 20 μl of formamide (Applied Biosystems, Inc., Foster City, Calif.). Samples were denatured at 95°C for 6 to 8 min, followed by chilling rapidly in an ice bath. Denatured samples were automatically loaded and electrophoresed through a 47-cm long, 0.75-μm-inside-diameter capillary filled with POP-6 polymer (Applied Biosystems) using an ABI Prism 310 genetic analyzer (Applied Biosystems). Parameters for capillary electrophoresis includes an injection time of 3 to 8 s, an injection voltage of 15 kV, an electrophoretic voltage of 15 kV, a temperature block set at 60°C, and a collection time of 45 min.
Sequence data analysis.
Raw sequence data were collected and then edited by using Sequencher version 4.5 software (Gene Codes Corp., Ann Arbor, MI). Multiple sequence alignments for each locus were obtained by using CLUSTAL X version 1.83 software for windows (available from ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX) as described by Thompson et al. (50). Obvious misalignments were corrected manually, and insertions and/or deletions (indels) in the MP1 gene fragments were identified. Gap information in the MP1 gene fragments was coded and added to the data matrix by the method of Simmons and Ochoterena (40) by using the SeqState program and IndelCoder software (34, 35). Phylogenetic analysis was performed by using PAUP* version 4.0 beta10 software (46). A neighbor-joining tree was generated by analysis of DNA sequences concatenated from five gene fragments (CpeA, NGS, ICL1, LNS, PAA, and MP1) for 24 isolates. Parsimony analysis was based on heuristic searches with all nucleotides equally weighted. Support and robustness of the resulting phylograms were based on bootstrap analysis (14) implemented by using PAUP* software. Mp1 types are defined as the unique nucleotide combinations obtained by comparison of MP1 gene sequences. Haploid sequence types (HST) are defined as for the unique nucleotide sequences obtained for the five polymorphic loci (CpeA, ICL1, LNS, PAA, and MP1). Discriminatory power (D), the probability that two randomly selected isolates are different, was calculated by the method of Hunter (24).
Nucleotide sequence accession numbers.
GenBank accession numbers of MP1 gene fragments are DQ088822 to DQ088845, respectively.
RESULTS
Nucleotide variability.
The nucleotide sequences for the Cox1 gene fragments of the mitochondrial genome and for three of eight nuclear gene fragments, AbaA, StlA, and NGS, showed no nucleotide sequence diversity for all of the isolates for a total of 1,714 bp (Tables 1 and 3). However, a low degree of nucleotide sequence diversity was observed for the CpeA, Icl1, LNS, and PAA gene fragments. Among the 1,639 bp of nucleotide sequence information for these four loci, only seven nucleotide substitutions were observed for a mutation frequency of 1/336 bp. For instance, one nucleotide substitution was detected at position 211 of the 312-bp CpeA gene fragment, predicting a nucleotide substitution, A→G, resulting in a nonsynonymous amino acid change (D→N). This single nucleotide substitution was observed in isolate ATCC 18224, obtained from a bamboo rat from Vietnam, and isolate NCPF 4158, a clinical isolate obtained from Thailand. Analysis of the PAA gene fragment detected three different nucleotide substitutions in three clinical isolates obtained from Thailand. Isolates B-6325 and B-6326 had a synonymous A→C nucleotide substitution at position 229 of the 418-bp fragment, whereas isolate B-6326 exhibited a synonymous T→A transversion at position 419. A third synonymous nucleotide substitution (A→G) was observed at position 263 for isolate IFM 47287. Two different nonsynonymous mutations were observed in the ICl1 gene fragment. Of 24 isolates (Table 1), 11 exhibited an A→G substitution at position 282, resulting in the predicted amino acid substitution D→N. Two isolates obtained from Thailand, IFM 47279 and IFM 47289, both exhibited an A→G nucleotide substitution at position 358, and this predicts the amino acid substitution G→E. Of the 24 isolates examined, only 1, ATCC 200050 obtained from Thailand, had a synonymous A→G nucleotide substitution at position 242 in the 454-bp LNS gene fragment. Together, only a low degree of discriminatory power (D = 0.747) and nine different genotypes were obtained when the nucleotide sequence diversity findings for the seven nuclear loci were combined.
TABLE 3.
Characteristics of eight P. marneffei sequenced gene fragments for 24 isolates
| Locus | No. of polymorphic nucleotide sites | % Polymorphic nucleotide sites | No. of genotypes | No. of nonsynonymous or synonymous changes |
|---|---|---|---|---|
| AbaA | 0 | 0 | 1 | 0 |
| CPE1 | 1 | 0.23 | 2 | 1 |
| ICL1 | 3 | 0.69 | 4 | 3 |
| LNS | 1 | 0.22 | 2 | 0 |
| PAA | 3 | 0.72 | 4 | 0 |
| StlA | 0 | 0 | 1 | 0 |
| NGS | 0 | 0 | 1 | 0 |
| MP1a | 20 | 4.68 | 16 | 2.5 |
Compared to P. marneffei PM4 and excludes indel sequences.
MP1 polymorphisms.
In contrast to the paucity in the nucleotide sequence variation that was observed for seven genomic loci, the MP1 gene fragment was observed to be robustly polymorphic. One source of nucleotide polymorphisms was single-nucleotide substitutions compared to the MP1 sequence obtained from isolate MP4 from a patient in Hong Kong (accession number AF009957). Nucleotide substitutions were observed at 21 different positions, ranging from gene fragment positions 22 to 435 (Table 4). Single-nucleotide substitutions in the MP1 gene fragment consisted of 6 synonymous and 15 nonsynonymous changes (ratio = 2.5). A second source of nucleotide diversity detected in the MP1 gene fragment was by sequence length variation due to the presence of indels. By using the MP1 nucleotide sequence of isolate PM4 (8) as the reference sequence for comparisons, three isolates—B-6321, B-6324, and B-6466—were observed to harbor a 21-bp insertion starting at nucleotide position 67 of the MP1 gene fragment (Table 5, MP1 type 7). This 21-bp indel predicts a tandem duplication of the KREATKV amino acid motif that is in frame for protein translation. In contrast to the MP1 sequence for Mp1 type 7 isolates, seven isolates belonging to Mp1 types 1, 3, 9, 11, and 12, as well as isolate PM4, were observed to harbor MP1 gene sequences with only a single copy of the KREATKV motif (Table 5). When compared to the MP1 nucleotide sequence for isolate PM4, 11 clinical isolates belonging to Mp1 types 5, 8, and 10 were observed to harbor a 42-bp deletion starting at position 67. This indel predicts the loss of a 14-amino-acid motif consisting of EATKVKREATKVQ but remaining in the proper reading frame for translation. A similar indel was observed for an isolate obtained from a bamboo rat in China, ATCC 64102, but with an indel in its MP1 sequence at a different 5′ start that predicts the deletion of the 14 amino acids: KREATKVKREATKV (Table 5, Mp1 type 2). Interestingly, for three isolates, B-6319 and B-6320 obtained from China and isolate NCPF 4158 obtained from Thailand, the MP1 gene fragment could not be amplified under a variety of experimental conditions using the PCR primer set used previously to amplify a 450-bp fragment of the MP1 gene (Table 2). Therefore, a second PCR primer set was designed to amplify a 303-bp fragment of the MP1 gene (Table 2). The new MP1 gene primer set was designed to amplify approximately 100-bp upstream of the binding site of the forward primer for the 450-bp fragment and to overlap by 203 bp with the amplified MP1 gene fragment. Nucleotide sequences of the overlapping contigs were combined and used to reconstruct the MP1 gene sequence for these three isolates. The lack of PCR amplification for these three isolates by the primer set for the 450-bp fragment could be explained by detecting significant nucleotide substitutions and indels within the complementary binding site of the first primer set. For example, the first eight amino acids of the MP1 gene sequence for isolates B-6319 and B-6320 were different from the first eight amino acids observed for the other 22 isolates of P. marneffei. In these two isolates the first eight amino acids QALQKGIQ observed in 22 other isolates are replaced by an insertion of the amino acid motif KAIQDGIN (Table 5, Mp1 type 6). In addition, the MP1 nucleotide sequences of isolates B-6319, B-6320, and NCPF 4158 predict a second indel of 18 bp. This 18-bp indel predicts an amino acid motif of HTGSRR that replaces a five-amino-acid motif of ISARQ conserved in 21 of 24 isolates (Table 5). The 42-bp deletion of the KREATKV amino acid motif is also observed in these three isolates. A third indel was identified and consists of a 3-bp insertion of CAC codon at nucleotide 34 and predicts the insertion of a histidine amino acid residue. All MP1 sequences are predicted to be translated in the proper reading frame. A conserved amino acid motif consisting of four amino acids, ATKV, was detected at least once in all 24 isolates. Two copies (Mp1 types 1, 3, 9, 11, and 12) or three copies (Mp1 type 7) of the ATKV motif were observed (Table 5). In summary, a total of 12 different Mp1 types were observed for a relatively high degree of discriminatory power (D = 0.877).
TABLE 4.
Distribution of polymorphic nucleotide sites for the MP1 locus in comparison to P. marneffei isolate PM4a
| MP1 type (no. of isolates)c | Nucleotide at positionb:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | 60 | 64 | 65 | 103 | 105 | 116 | 118 | 170 | 202 | 206 | 227 | 230 | 231 | 248 | 258 | 412 | 414 | 426 | 435 | |
| 1 (1) | - | - | - | - | - | - | - | - | - | G | C | - | - | - | T | - | - | - | T | - |
| 2 (1) | G | - | - | - | C | - | - | G | - | - | - | - | - | - | - | - | - | - | - | T |
| 3 (1) | - | - | C | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 4 (1) | - | - | - | - | - | C | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 5 (1) | - | - | - | - | - | - | - | - | - | - | G | - | - | C | - | T | - | - | - | T |
| 6 (1) | - | - | - | - | - | - | - | - | - | - | - | G | A | C | - | - | - | - | - | - |
| 7 (1) | - | - | - | - | C | - | G | G | - | - | - | - | - | - | A | - | - | - | - | - |
| 8 (1) | G | T | - | - | C | - | - | G | - | - | - | - | - | - | - | - | - | - | - | - |
| 9 (1) | - | - | C | G | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| 10 (1) | G | T | - | - | C | - | G | G | - | - | - | - | - | T | - | - | - | - | - | - |
| 11 (1) | - | - | C | G | C | - | G | G | A | - | - | - | - | - | - | - | - | - | - | - |
| 12 (1) | - | - | - | - | C | - | G | G | - | - | - | - | - | - | - | - | A | - | - | - |
Only variable nucleotide sites that differ from the isolate PM4 sequence are shown. A dash denotes a nucleotide identical to that observed for isolate PM4.
Numbers along the top corresponds to nucleotide positions for the MP1 gene fragment for isolate PM4.
The numbers within parentheses are the numbers of isolates with an identical MP1 sequence.
TABLE 5.
Comparison of predicted amino acid sequences for the MP1 gene fragment and maximized alignment of matched residues
| MP1 type | No. of isolates | Predicted amino acid sequencea |
|---|---|---|
| MP7 | 3 | QALQKGIQAFSISARQ--ATKVKREATKVKREATKVQRDISAFQKVIRDISLAVN |
| MP9 | 2 | QALQKGIQAFSISARQ--ATKVKREATKV-------QRDISAFKKVIRDISLAVN |
| MP11, MP12 | 2 | QALQKGIQAFSISARQ--ATKVKREATKV-------QRDISAFQKVIRDISLAVN |
| PM4, MP1, MP3 | 3 | QALQKGIQAFSISARQ--ATKVKREATKV-------QRDISAFKKVIRNISLAVN |
| MP5 | 1 | QALQKGIQAFSISARQ--ATKVK--------------RDISAFKKVIRNISLAVN |
| MP8 | 2 | QALQKGIEAFSISARQ--ATKVK--------------RDISAFQKVIRDISLAVN |
| MP10 | 8 | QALQKGIEAFSISARQ--ATKV---------------RDISAFQKVIRDISLAVN |
| MP2 | 1 | QALQKGIEAFSISARQ--ATKVK-------------QRDISAFQKVIRDISLAVN |
| MP4 | 1 | QALQKGIQAFSHTGSRRATKVK--------------QRDISAFQKVIRDISLAVN |
| MP6 | 2 | KAIQDGINAFSHTGSRRATKV---------------QRDISAFKKVIQNISLAVN |
| Consensus | QALQKGIQAFSISARQ--ATKV--------------QRDISAFQKVI-DISLAVN |
Amino acid substitutions different from the consensus are underlined. The position of the indels are presented as dashed lines or indicated by boldface with underlining.
Cluster analysis.
Since no nucleotide sequence variation was observed in the AbaA, Cox1, StlA and NGS loci, for all 24 P. marneffei isolates analyzed, haploid genotypes (HGT) were defined based on assignments using the nucleotide sequences obtained from five polymorphic loci: CpeA, Icl1, LNS, PAA, and MP1. Each HGT assignment was represented by a unique allelic combination. Using nucleotide sequences for these five gene fragments, a relatively high degree of discriminatory power (D = 0.949) was observed. For 24 isolates, 16 different HGT were detected, with 11 HST types represented by single isolates.
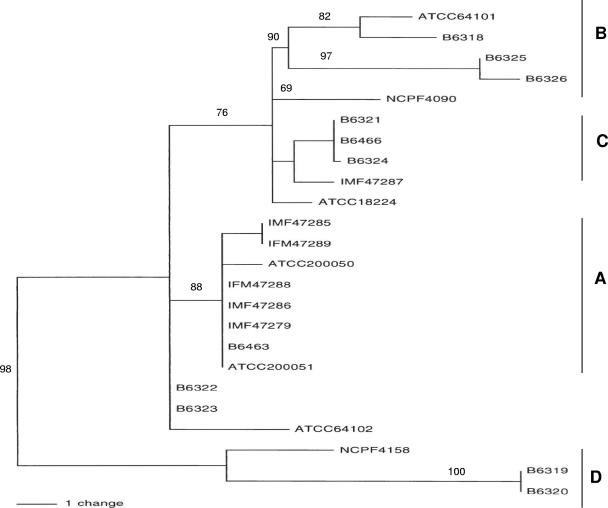
Phylograms for individual genes were not constructed or compared due to the paucity of nucleotide sequence diversity. To better analyze the genetic diversity of the isolates, a neighbor-joining tree was constructed using nucleotide sequence data obtained from the set of five polymorphic gene fragments and is shown in Fig. 1. Eight clinical isolates obtained from Thailand were clustered (cluster A) and showed a high degree of support (bootstrap values of 94), and this suggests a high degree of genetic relatedness. A subcluster designated B, composed of five isolates obtained from China, was observed. Two other subclusters, designated C and D, were observed to be composed of isolates obtained from both China and Thailand. A low degree of genetic relatedness was observed between the two isolates obtained from bamboo rats: isolate ATCC 64102 obtained from China and isolate ATCC 18824 from Vietnam.
FIG. 1.
Representation of the consensus data set by neighbor-joining analysis for 24 isolates of P. marneffei. Branch lengths are equivalent to distance. Numbers denote the bootstrap values for the appropriate node based on 1,000 replicates. Only bootstrap values of >50 are shown.
To confirm the accurate clustering by neighbor joining, isolates were also clustered by using PAUP* software to construct a tree using parsimony. The tree, shown in Fig. 2, consists of 2,116 characters resulting in a consistency index of 0.8846 and a retention index of 0.9390. Overall, isolates clustered by parsimony analysis were very similar to the clustering obtained by neighbor joining. For instance, isolates observed in clusters A, B, C, and D by both neighbor-joining and parsimony analysis contained identical isolates.
FIG. 2.
Consensus tree based on parsimony analysis for 24 isolates of P. marneffei based on five loci. The bottom scale bar represents distance. Numbers denote the bootstrap values for the appropriate node based on 100 replicates. Only bootstrap values of >50 are shown.
The in vitro reproducibility of MLST analysis was evaluated by analysis of DNA sequences obtained from DNA purified from mycelial cells grown in broth for 7 to 8 weeks by passing the cells in fresh broth every 2 to 3 days (30). In all cases no differences were detected between the nucleotide sequences obtained from the original DNA preparations from the nucleotide sequences obtained from the DNA preparations from cultures transferred in vitro.
DISCUSSION
Nucleotide sequence analysis or MLST analysis usually rely on the detection and analysis of nucleotide sequence polymorphisms found in several housekeeping genes of a given microorganism. MLST was originally successfully used to characterize isolates of bacterial pathogens, and the method was shown to have a high degree of discrimination among isolates of Neisseria meningitidis (33). Subsequently, MLST has been shown to be a useful method for the characterization of isolates for fungal pathogens such as Aspergillus flavus (19), C. albicans (4, 11), C. glabrata (13), and Coccidioides immitis (29), as well as for elucidating the phylogenetic relationship of isolates of Histoplasma capsulatum (28) and Cryptococcus neoformans (59). When MLST was applied to the analysis of P. marneffei isolates here, a low degree of nucleotide sequence diversity was observed in seven nuclear genes (Tables 1 and 3). Low sequence diversity has been observed in other microorganisms such as C. parapsilosis, C. orthopsilosis, and C. metapsilosis (18, 48) and the Mycobacterium tuberculosis complex (42). The lack of genetic diversity may be due to events such as recent speciation, selective sweeps, or population bottle necks. In comparison, genetic diversity in another fungal pathogen, C. albicans, was found to range from 1.5% for the gene for acetyl coenzyme A carboxylase to 4.0% for the gene for the vacuolar protein sorting protein (4). In C. albicans, an increase in the level of discrimination could also be achieved by scoring for nucleotide sequence heterozygosity, which may be expected in an organism with a diploid genome. For P. marneffei, no sequence heterozygosity was observed for all of the loci analyzed in this investigation, and this result is consistent for a microorganism believed to have a haploid genome (60).
Molecular epidemiologic investigations using MLST may provide several advantages over other methods for the molecular subtyping of isolates. One advantage of MLST is based on the exact identification of the potential alleles in a locus that is not possible by using a traditional method for subtyping, such as multilocus enzyme analysis (33). MLST can also detect rapid changes in the genome such as microevolution, identify geographically distinct isolates, and provide reproducible genomic characters in sequential isolates. Of importance is the ability of MLST to directly compare and compile large databases of shared DNA sequence information, thereby facilitating collaborative investigations and data sharing. Disadvantages of MLST analysis include the requirement for specialized equipment, expensive reagents, and highly trained personnel and the fact that the method may not be amenable to the analysis of all microorganisms. Low sequence diversity may preclude its usefulness to distinguish between isolates and makes it more difficult to accurately assess the genetic relatedness between isolates (48, 49).
Two loci, AbaA and StlA, analyzed in this investigation are regulatory gene sequences, whereas five loci—Cpe1, Icl1, LNS, PAA, and NGS—code for housekeeping functions. A low degree of sequence diversity was observed for both the regulatory and the housekeeping gene sequences. Although most investigations utilize sequence information obtained only from housekeeping genes, a relatively high degree of sequence variation has been reported in regulatory genes. For example, the level of sequence diversity for the regulatory sequences was shown to be comparable to the level of sequence diversity observed for other nuclear loci (37).
Several genotypic methods for the molecular subtyping of P. marneffei isolates have been reported. The first reported method was by restriction endonuclease analysis using HaeIII digest. Two HaeIII types for 46 isolates were reported (53). Randomly amplified polymorphic DNA and macrorestriction profiles have been utilized for typing, but these methods are difficult to reproduce or it is difficult to compare results between laboratories (23, 26, 51). Recently, microsatellite markers have been shown to provide high degree of discriminatory power and reproducibility for typing P. marneffei isolates (15, 16, 17, 21, 30). Whereas microsatellite analysis may provide more variable loci than can be obtained by MLST, the method has been shown to be susceptible to problems due to homoplasy, and monomorphic loci may confound analysis of genetic relatedness due to the hypervariability of some loci (49).
Of potential clinical relevance was the high degree of nucleotide sequence divergence observed for the MP1 locus. MP1 encodes an abundant cell wall mannoprotein originally cloned and sequenced in isolate PM4 by Cao et al. (8). Compared to the original nucleotide sequence for isolate PM4, nucleotide substitutions were detected at 21 different positions, many of which are predicted to result in amino acid substitutions (Table 4). As shown in Table 5, indels were also identified as major contributors to MP1 sequence diversity. For instance, a 21-bp indel resulted in the tandem duplication of a seven-amino-acid motif of KREATKV in three isolates obtained from China, and seven isolates contained a single copy of this motif. However, the KREATKV motif was completely deleted in the majority of the isolates, and this suggests the motif has no obligate biological or structural function. A six-amino-acid motif SISARQ, was conserved in 21 of 24 isolates but was replaced by a putative insertion of amino acids HTGSRR in three isolates(B-6319, B-6320, and NCPF 4158). The replacement of this amino acid block may be due to a more complex mechanism than by nucleotide substitution. In P. marneffei the molecular mechanisms for indel formation have not been investigated but may involve unequal crossover of repeats, movement of mobile elements, replication slippage, or recombination. Sequence diversity of MP1 gene may be driven by the accessibility of the MP1 glycoprotein to the immune system. Antigenic variation may provide this pathogen with a survival mechanism to better evade host acquired immunity during infection. Sequence changes especially by indels may be providing the organism with a mechanism for the rapid generation of new alleles in response to host defenses. Tandem repeat motifs detected in other surface glycoproteins of other microbial pathogens, such as the ALS1 adhesion glycoprotein of C. albicans (22) and malarial parasite proteins of Plasmodium species (38), may function in host evasion or binding host receptors. The MP1 protein is 462 amino acid residues and was not sequenced in entirety in this investigation and therefore offers the potential for detecting additional sequence divergence. MP1 antigenic variability may be of clinical relevance since a single purified MP1 gene product (Mp1p) and anti-Mp1p antibody have been used in an enzyme-linked immunosorbent assay for the serodiagnosis of P. marneffei infections (6, 7, 57).
Overall, a relatively high degree of discriminatory power was observed using five polymorphic loci (D = 0.949) with the bulk of nucleotide diversity due to polymorphisms detected in the MP1 gene. Although it may be possible to subtype isolates based solely on the MP1 gene fragment, a higher degree of reliability and discriminatory power may be achieved by analysis at five loci. Overall, analysis detected 16 HST for 24 isolates. Reproducibility was confirmed by the direct comparison of DNA sequences obtained from genomic DNA preparations from unpassaged cells and DNA preparations obtained from cells grown for 7 to 8 weeks (30).
Phylogenetic analysis using neighbor-joining and parsimony showed a high degree of concordance in detecting geographically enriched clusters composed of isolates obtained from either Thailand or China (Fig. 1 and 2, clusters A and B). Geographic partitioning was previously observed using polymorphic microsatellites to analyze P. marneffei genetic variation (15, 16, 30). However, dependence on absolute fragment size alone as done in these studies for assessing genealogical relationships was found to be less accurate than by using DNA sequencing (36). Geographically mixed clusters were also observed (Fig. 1 and 2, clusters C and D). For instance, isolates B-6319 and B-6320 obtained from China and NCPF 4158 obtained from Thailand, were highly related and shared a unique pattern of indels of identical length, as well as position (Table 5). Likewise, isolates B-6321 and B-6324 from China and isolate B-6466 from Thailand were highly related and harbor an identical tandem repeat (KREATKV) in the MP1 sequence. Together, these results indicate that these isolates may have descended from a common ancestor that later diverged, as would be expected for an asexual mode of reproduction. However, this analysis does not exclude the possibility of parallel insertion events from different lineages. One potential criticism of this investigation is the lack of sufficient nucleotide diversity and the reliance on a single hypervariable gene under selection by the host. However, by the combination of the nucleotide variations observed in four other loci, a relatively high degree of discriminatory power was observed. Also, indels have been shown to be reliable and important characters for constructing phylogenies since they are able to retain the historical information of their sequences in their descendants and homoplasy is often low (20, 40, 41). The nucleotide sequences for the two isolates obtained from bamboo rats, ATCC 18224 and ATCC 64101, showed extensive sequence divergence from each other but were composed of genotypes that were related to clinical isolates. This result is consistent with the investigations by Gugnani et al. (21) and Fisher et al. (16), who observed identical microsatellite genotypes obtained from human and bamboo rat isolates. At present, the importance of isolates obtained from bamboo rats or from their burrows to transmission to humans is not known (17, 21).
In this investigation DNA sequence analysis provided a robust, reproducible, and highly discriminatory method for typing isolates of P. marneffei. The expansion of a sequence database, the availability of new genetic loci for analysis, and improvements in DNA sequencing technology may provide for greater opportunities to examine the genetic diversity within this important fungal pathogen. Nucleotide sequence-based analysis may provide a useful and important method for the molecular epidemiology and surveillance of clinical and environmental isolates.
REFERENCES
- 1.Andrianopoulos, A. 2002. Control of morphogenesis in the human fungal pathogen Penicillium marneffei. Int. J. Med. Microbiol. 292:331-347. [DOI] [PubMed] [Google Scholar]
- 2.Borneman, A. R., M. J. Hynes, and A. Andrianopoulos. 2000. The abaA homologue of Penicillium marneffei participates in two developmental programs: conidiation and dimorphic growth. Mol. Microbiol. 38:1034-1047. [DOI] [PubMed] [Google Scholar]
- 3.Borneman, A. R., M. J. Hynes, and A. Andrianopoulos. 2001. An STE12 homolog from the asexual, dimorphic fungus Penicillium marneffei complements the defect in sexual development of an Aspergillus nidulans steA mutant. Genetics 157:1003-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bougnoux, M.-E., S. Morand, and C. d'Enfert. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bougnoux, M.-E., D. M. Aanensen, S. Morand, M. Théraud, B. G. Spratt, and C. d'Enfert. 2004. Multilocus sequencing typing of Candida albicans: strategies, data exchange and applications. Infect. Genet. Evol. 4:243-252. [DOI] [PubMed] [Google Scholar]
- 6.Cao, L., K. M. Chan, D. Chen, N. Vanittanakom, C. Lee, C. M. Chan, T. Sirisanthana, D. N. C. Tsang, and K. Y. Yuen. 1999. Detection of cell wall mannoprotein Mp1p in culture supernatants of Penicillium marneffei and in sera of penicilliosis patients. J. Clin. Microbiol. 37:981-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, L., L. Chen, C. Lee, C. M. Chan, K. M. Chan, N. Vanittanakom, D. N. C. Tsang, and K. Y. Yuen. 1998. Detection of specific antibodies to an antigenic mannoprotein for diagnosis of Penicillium marneffei penicilliosis. J. Clin. Microbiol. 36:3028-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, L., C.-M. Chan, C. Lee, S. S.-Y. Wong, and K.-Y. Yuen. 1998. MP1 encodes an abundant and highly antigenic cell wall mannoprotein in the fungus Penicillium marneffei. Infect. Immun. 66:966-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chariyalertsak, S., T. Sirisanthana, K. Supparatpinyo, J. Praparattanapan, and K. E. Nelson. 1997. Case-control study of risk factors for Penicillium marneffei infection in human immunodeficiency virus-infected patients in Northern Thailand. Clin. Infect. Dis. 24:1080-1086. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, C. R., Jr., and M. R. McGinnis. 1997. Pathology of Penicilium marneffei. Arch. Pathol. Lab. Med. 121:798-804. [PubMed] [Google Scholar]
- 11.Cowen, L. E., C. Sirjusingh, R. C. Summerbell, S. Walmsley, S. Richarson, L. M. Kohn, and J. B. Anderson. 1999. Multilocus genotypes and DNA fingerprints do not predict variation in azole resistance among clinical isolates of Candida albicans. J. Clin. Microbiol. 43:2930-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng, Z., J. L. Ribas, D. W. Gibson, and D. H. Connor. 1988. Infections caused by Penicillium marneffei in China and Southeast Asia: review of eighteen cases and report of four more Chinese cases. Rev. Infect. Dis. 10:640-652. [DOI] [PubMed] [Google Scholar]
- 13.Dodgson, A. R., C. Pujol, D. W. Denning, D. R. Soll, and A. J. Fox. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J. Clin. Microbiol. 41:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felsenstein, J. Confidence limits on phylogenies: an approach using bootstrap. Evolution 39:783-791. [DOI] [PubMed]
- 15.Fisher, M. C., D. Aanensen, S. de Hoog, and N. Vanittanakom. 2004. Multilocus microsatellite typing system for Penicillium marneffei reveals special structured populations. J. Clin. Microbiol. 42:5065-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher, M. C., W. P. Hanage, S. de Hoog, E. Johnson, M. D. Smith, N. J. White, and N. Vanittanakom. 2005. Low effective dispersal of asexual genotypes in heterogeneous landscapes by the endemic pathogen Penicillium marneffei. PLOS Pathogens 1:159-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisher, M. C., S. D. Hoog, and N. Vanittanakom. 2004. A highly discriminatory multilocus microsatellite typing (MLMT) system for Penicillium marneffei. Mol. Ecol. Notes 4:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fundyga, R. E., R. J. Kykendall, W. Lee-Yang, and T. J. Lott. 2004. Evidence for aneuploidy and recombination in the human commensal yeast Candida parapsilosis. Infect. Genet. Evol. 4:37-43. [DOI] [PubMed] [Google Scholar]
- 19.Geiser, D. M., J. I. Pitt, and J. W. Taylor. 1998. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc. Natl. Acad. Sci. USA 95:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giribet, G., and W. C. Wheeler. 1999. On gaps. Mol. Phylogenet. Evol. 13:132-143. [DOI] [PubMed] [Google Scholar]
- 21.Gugnani, H., M. C. Fisher, A. Paliwal-Joshi, N. Vanittanakom, I. Singh, and P. S. Yadav. 2004. Role of Cannomys badius as a natural animal host of Penicillium marneffei in India. J. Clin. Microbiol. 42:5070-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoyer, L. L., S. Scherer, A. R. Shatzman, and G. P. Livi. 1995. Candida albicans ALS1: domains related to a Saccharomyces cerevisiae sexual agglutinin separated by a repeating motif. Mol. Microbiol. 15:39-54. [DOI] [PubMed] [Google Scholar]
- 23.Hsueh, P.-R., L.-J. Teng, C.-C. Hung, J.-H. Hsu, P.-C. Yang, S.-W. Ho, and K.-T. Luh. 2000. Molecular evidence for strain dissemination of Penicillium marneffei: an emerging pathogen of Taiwan. J. Infect. Dis. 181:1706-1712. [DOI] [PubMed] [Google Scholar]
- 24.Hunter, P. R. 1990. Reproducibility indices of discriminatory power of microbial typing systems. J. Clin. Microbiol. 28:1903-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imwidthaya, P., A. S. Sekhon, T. D. Mastro, A. K. Garg, and E. Ambrosie. 1997. Usefulness of a microimmunodiffusion test for the detection of Penicillium marneffei antigenemia, antibodies, and exoantigens. Mycopathologia 138:51-55. [DOI] [PubMed] [Google Scholar]
- 26.Imwidthaya, P., K. Thipsuvan, A Chaiprasert, S. Danchaivijitra, and J. Jearanaisilavong. 2001. Penicillium marneffei: types and drug susceptibility. Mycopathologia 149:109-115. [DOI] [PubMed] [Google Scholar]
- 27.Jayanetra, P., P. Nitiyanant, L. Ajello, A. A. Padye, S. Lolekha, V. Atichartakarn, and P. Vathesatogit. 1984. Penicilliosis marneffei in Thailand: report of five human cases. Am. J. Trop. Med. Hyg. 33:637-644. [DOI] [PubMed] [Google Scholar]
- 28.Kasuga, T., J. W. Taylor, and T. J. White. 1999. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J. Clin. Microbiol. 37:653-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koufopanou, V., A. Burt, and J. W. Taylor. 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 94:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lasker, B. A., and Y. Ran. 2004. Analysis of polymorphic microsatellite markers for typing Penicillium marneffei isolates. J. Clin. Microbiol. 42:1483-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LoBuglio, K. F., J. I. Pitt, and J. W. Taylor. 1993. Phylogenetic analysis of two ribosomal DNA regions indicates multiple independent losses of a sexual Talaromyces state among asexual Penicillium species in subgenus Biverticillium. Mycologia 85:592-604. [Google Scholar]
- 32.LoBuglio, K. F., and J. W. Taylor. 1995. Phylogeny and PCR identification of the human pathogenic fungus Penicillium marneffei. J. Clin. Microbiol. 33:85-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller, K. 2006. Incorporating information from length-mutational events into phylogenetic analysis. Mol. Phylogenet. Evol. 38:667-676. [DOI] [PubMed]
- 35.Müller, K. 2005. SeqState primer design and sequence statistics for phylogenetic DNA datasets. Appl. Bioinformatics 4:65-69. [DOI] [PubMed] [Google Scholar]
- 36.Ortí, G., D. E. Pearse, and J. C. Avise. 1997. Phylogenetic assessment of length variation at a microsatellite locus. Proc. Natl. Acad. Sci. USA 94:10745-10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purugganan, M. D. 2000. The molecular population genetics of regulatory genes. Mol. Ecol. 9:1451-1461. [DOI] [PubMed] [Google Scholar]
- 38.Ramasamy, R. 1991. Repeat regions of malaria parasite proteins: a review of structure and possible role in the biology of the parasite. Indian J. Malariology 28:73-81. [PubMed] [Google Scholar]
- 39.Pongpom, P., C. R. Cooper, Jr., and N. Vanittanakom. 2005. Isolation and characterization of a catalase-peroxidase gene from the pathogenic fungus Penicillium marneffei. Med. Mycol. 43:403-411. [DOI] [PubMed] [Google Scholar]
- 40.Simmons, M. P., and H. Ochoterena. 2000. Gaps as characters in sequence-based phylogenetic analysis. Syst. Biol. 49:369-381. [PubMed] [Google Scholar]
- 41.Simmons, M. P., H. Ochoterena, and T. G. Carr. 2001. Incorporation, relative homoplasy, and effect of gap characters in sequence-based phylogenetic analysis. Syst. Biol. 50:454-462. [PubMed] [Google Scholar]
- 42.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in Mycobacterium tuberculosis complex indicates evolutionary recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Supparatpinyo, K., S. Chiewchanvit, P. Hirunsri, C. Uthammachai, K. E. Nelson, and T. Sirisanthana. 1992. Penicillium marneffei infection in patients with human immunodeficiency virus. Clin. Infect. Dis. 14:871-874. [DOI] [PubMed] [Google Scholar]
- 44.Supparatpinyo, K., S. Khamwan, V. Baosoung, K. E. Nelson, and T. Sirisanthana. 1994. Disseminated Penicillium marneffei infection in Southeast Asia. Lancet 344:110-113. [DOI] [PubMed] [Google Scholar]
- 45.Supparatpinyo, K., J. Perriens, K. E. Nelson, and T. Srisanthana. 1998. A controlled trial of intraconazole to prevent relapse of Penicillium marneffei infections in patients infected with human immunodeficiency virus. N. Engl. J. Med. 339:1739-1743. [DOI] [PubMed] [Google Scholar]
- 46.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony (* and other methods). Sinauer Associates, Sunderland, Mass.
- 47.Tavanti, A., A. D. Davidson, E. M. Johnson, M. C. J. Maiden, D. J. Shaw, N. A. R. Gow, and F. C. Odds. 2005. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J. Clin. Microbiol. 43:5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tavanti, A., A. D. Davidson, N. A. R. Gow, M. C. J. Maiden, and F. C. Odds. 2005. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J. Clin. Microbiol. 43:284-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor, J. W., and M. C. Fisher. 2003. Fungal multilocus typing: it's not just for bacteria. Curr. Opin. Microbiol. 6:351-356. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trewatcharegon, S., S. Sirisinha, A. Romai, B. Eampokalap, R. Teanpaisan, and S. C. Chaiyaroj. 2001. Molecular typing of Penicillium marneffei isolates from Thailand by NotI macrorestriction and pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:4544-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsang, D. N. C., P. C. K. Li, M. S. Tsui, Y. T. Lau, K. F. Mak, and E. K. Yeoh. 1991. Penicilliosis marneffei: another pathogen to consider in patients infected with human immunodeficiency virus. Rev. Infect. Dis. 13:766-767. [DOI] [PubMed] [Google Scholar]
- 53.Vanittanakom, N., C. R. Cooper, Jr., S. Chariyalertsak, S. Youngchim, K. E. Nelson, and T. Sirisanthana. 1996. Restriction endonuclease analysis of Penicillium marneffei. J. Clin. Microbiol. 34:1834-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanittanakom, N., C. R. Cooper, Jr., M. C. Fisher, and T. Sirisanthana. 2006. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin. Microbiol. Rev. 19:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong, S. S. Y., L. Cao, and K. Y. Yuen. 1998. Management of penicilliosis marneffei. JAMA Southeast Asia 14:7-9. [Google Scholar]
- 56.Wong, S. S. Y., T. Y. C. Ho, A. H. Y. Ngan, P. C. Y. Woo, T.-L. Que, amd K.-Y. Yuen. 2001. Biotyping of Penicillium marneffei reveals concentration-dependent growth inhibition by galactose. J. Clin. Microbiol. 39:1416-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong, L. P., P. C. Woo, A. Y. Wu, and K. Y. Yuen. 2002. DNA immunization using a secreted cell wall antigen Mp1p is protective against Penicillium marneffei infection. Vaccine 20:2878-2886. [DOI] [PubMed] [Google Scholar]
- 58.Woo, P. C. Y., H. Zhen, J. J. Cai, J. Yu, S. K. P. Lau, J. Wang, J. L. L. Teng, S. S. Y. Wong, R. H. Tse, R. Chen, H. Yang, B. Liu, and K.-Y. Yuen. 2003. The mitochondrial genome of the thermal dimorphic fungus Penicillium marneffei is more closely related to those of molds than yeasts. FEBS Lett. 555:469-477. [DOI] [PubMed] [Google Scholar]
- 59.Xu, J., R. Vilgalys, and T. G. Mitchell. 2000. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 9:1471-1481. [DOI] [PubMed] [Google Scholar]
- 60.Yuen, K.-Y., G. Pascal, S. S. Y. Wong, P. Glaser, P. C. Y. Woo, F. Kunst, J. J. Cai, E. Y. L. Cheung, C. Médigue, and A. Danchin. 2003. Exploring the Penicillium marneffei genome. Arch. Microbiol. 179:339-353. [DOI] [PubMed] [Google Scholar]