Abstract
A single-round PCR assay was developed for detection and differential diagnosis of the three Entamoeba species found in humans, Entamoeba moshkovskii, Entamoeba histolytica, and Entamoeba dispar, that are morphologically identical as both cysts and trophozoites. A conserved forward primer was derived from the middle of the small-subunit rRNA gene, and reverse primers were designed from signature sequences specific to each of these three Entamoeba species. PCR generates a 166-bp product with E. histolytica DNA, a 752-bp product with E. dispar DNA, and a 580-bp product with E. moshkovskii DNA. Thirty clinical specimens were examined, and the species present were successfully detected and differentiated using this assay. It was possible to detect as little as 10 pg of E. moshkovskii and E. histolytica DNA, while for E. dispar the sensitivity was about 20 pg of DNA. Testing with DNA from different pathogens, including bacteria and other protozoa, confirmed the high specificity of the assay. We propose the use of this PCR assay as an accurate, rapid, and effective diagnostic method for the detection and discrimination of these three morphologically indistinguishable Entamoeba species in both routine diagnosis of amoebiasis and epidemiological surveys.
Infections with Entamoeba spp. can result in either a harmless colonization of the intestine or an invasion of the colon wall and damage of other host tissues such as the liver, lung, and brain (amoebiasis). In most cases, a clinical diagnosis of amoebiasis can be confirmed and usually depends on the visualization of parasites by light microscopy of a wet smear or stained specimens. This procedure is inexpensive and simple, but it has several limitations, such as being incapable of distinguishing between the cysts and trophozoites of the disease-causing species Entamoeba histolytica, the nonpathogenic species Entamoeba dispar, and the amphizoic amoeba Entamoeba moshkovskii, which occasionally infects humans. Multiple samples often have to be requested and examined, and the presence of cysts of different species of Entamoeba, Iodamoeba, or Endolimax can make the diagnosis even more difficult (4, 5). Furthermore, with the reports of sporadic cases of human infection with E. moshkovskii (3, 6) and the recent finding of a high prevalence and association of E. moshkovskii with E. histolytica and E. dispar in young children in Bangladesh (2), the differentiation of the three species in clinical samples by other means becomes of great importance both for diagnosis and for epidemiological studies.
Although several PCR methods have been applied to the detection of E. histolytica and E. dispar in stool samples (1, 11, 12, 13), there is so far only one report of PCR being used for the identification of E. moshkovskii in stool samples (2). PCR coupled with additional methods, such as PCR with restriction fragment length polymorphism analysis or riboprinting (3) and PCR with reverse line blot hybridization (14), has also been used to differentiate E. moshkovskii from other species. The sensitivity of these methods for detecting E. moshkovskii has been shown to be higher than that of microscopy. However, these methods are still relatively time-consuming and require extra and complex procedures, such as nested PCR, restriction endonuclease assays, or hybridization, to achieve their higher sensitivity (2, 3, 14).
Antibody-based methods have also been developed for the differentiation of E. histolytica and E. dispar in stool samples (7) but so far not for E. moshkovskii. Detection of antibodies to amoebae in patient sera has been reported to indicate infection by E. histolytica (9, 11). However, with serological testing it may be difficult to distinguish past from present infections in individuals who emigrate from or currently reside in areas of endemicity (8, 10).
There is a need for simpler and better tools suitable for identification of these amoebae in clinical specimens, not only for diagnostic purposes and patient care management, where E. moshkovskii/E. dispar-infected patients could be treated unnecessarily with antiamoebic chemotherapy, but also for a better understanding of the epidemiology of these parasites in the human population.
In this study, the development of a single-round PCR assay for detection and differential diagnosis of the three morphologically identical Entamoeba species found in humans, E. histolytica, E. dispar and E. moshkovskii, was performed.
MATERIALS AND METHODS
DNA samples.
DNAs isolated from axenically grown E. histolytica HM-1:IMSS, E. dispar SAW 760, and E. moshkovskii Laredo were used as positive controls in this study. All of these control DNAs were a gift from Graham Clark (London School of Hygiene and Tropical Medicine).
Clinical samples.
A total of 30 clinical samples identified as E. histolytica positive by microscopic examination, including 27 fecal specimens and three liver abscess aspirates, were collected individually from 30 patients who sought medical attention for a variety of reasons at the Phramongkutklao and Ramathibodi hospitals in Bangkok, Thailand. All specimens were first examined by microscopy in the corresponding laboratories and subsequently used to evaluate the PCR method developed in this study. DNA extraction was performed directly on stool samples by using a QIAamp stool DNA extraction kit (QIAGEN, Hilden, Germany). The extracted DNA was then stored at −20°C until further use.
All DNA samples were also screened and tested for Entamoeba by using a genus-specific PCR assay (15) and tested for E. histolytica and E. dispar by using a previously described PCR assay (12).
Design of primers.
The primers were designed based on the reported E. histolytica, E. dispar, and E. moshkovskii small-subunit rRNA gene sequences (GenBank accession no. X64142, Z49256, and AF149906, respectively). The sequences were aligned using Multialin (http://prodes.toulouse.inra.fr/multalin/multalin.html). The forward primer sequence (EntaF) was derived from the central region of the small-subunit rRNA gene that was conserved in all three Entamoeba species, whereas the reverse primers EhR, EdR, and EmR, specific for E. histolytica, E. dispar, and E. moshkovskii, respectively, were designed from signature sequences on the respective small-subunit rRNA sequences that are specific to each of the three parasites. The primer sequences used were as follows: for EntaF, 5′-ATG CAC GAG AGC GAA AGC AT-3′; for EhR, 5′-GAT CTA GAA ACA ATG CTT CTC T-3′; for EdR, 5′-CAC CAC TTA CTA TCC CTA CC-3′; and for EmR, 5′-TGA CCG GAG CCA GAG ACA T-3′.
All primer sequences were compared to sequences in GenBank. This showed that the forward primer (EntaF) sequence is found only in Entamoeba and that the three reverse primer (EhR, EdR, and EmR) sequences are species specific. They are therefore suitable for species differentiation. The forward primer in combination with the appropriate reverse primer generates a 166-bp PCR product with E. histolytica DNA, a 752-bp PCR product with E. dispar DNA, and a 580-bp product with E. moshkovskii DNA.
PCR amplification for differential diagnosis.
The PCR amplification reaction was performed in a final volume of 50 μl in 0.1-ml PCR tubes by use of a Px2 thermal cycler (ThermoHybaid, United Kingdom). Reaction conditions were optimized to combine the forward primer (EntaF) with each of the three reverse primers (EhR, EdR, and EmR) in a single reaction mixture and under the same conditions.
The reaction mixture contained 200 μM of each deoxynucleoside triphosphate, 0.1 μM of each forward and reverse primer, 6 mM MgCl2, 0.5 U of Taq polymerase, 1× Taq buffer, and 10 μl of extracted DNA samples. Amplification of each species-specific DNA fragment started with an initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 7 min. Amplified products were visualized with ethidium bromide staining after electrophoresis on 1.5% agarose gels.
Determination of the sensitivity and specificity of PCR primers.
To determine the sensitivity of the assay, 10 concentrations of DNA from each Entamoeba species were prepared by twofold serial dilution from 5 ng to 0.6 pg of DNA. The sensitivity test was performed using the same protocols described above.
A mixture of DNAs from E. histolytica, E. dispar, and E. moshkovskii was tested for any cross-reaction or cross-amplification between primers designed for the three Entamoeba species. The assay was also tested for specificity against a panel of genomic DNAs from different bacterial pathogens and other protozoa. Group I contained eight different genomic DNAs obtained from organisms in culture, including human cell lines and axenic cultures of a variety of pathogens known to caused intestinal infections in humans: Escherichia coli, Salmonella spp., Shigella spp., Vibrio cholerae, Blastocystis hominis, Giardia lamblia, and Cryptosporidium spp. DNA extracted from a parasite-free fecal sample was used as a negative control. Group II consisted of DNA extracted from eight fecal samples, each containing one of the following parasites: Entamoeba coli, Endolimax nana, Blastocystis hominis, Giardia lamblia, or Cryptosporidium parvum.
RESULTS
Species-specific PCR products.
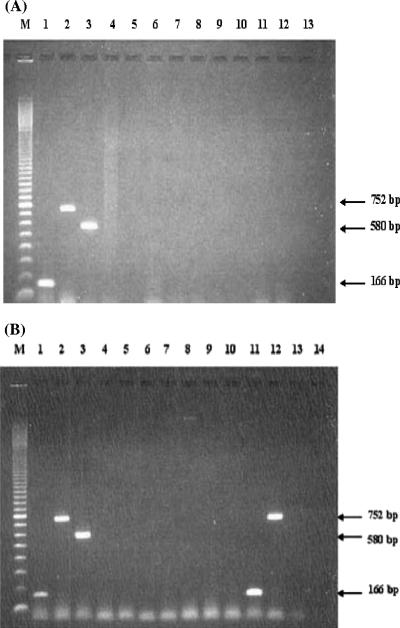
By using the designed primers, E. histolytica primers (EntaF/EhR) amplified DNA from the HM-1:IMSS strain but not from E. dispar SAW 760 or E. moshkovskii Laredo; the E. dispar primers (EntaF/EdR) and the E. moshkovskii primers (EntaF/EmR) also showed the expected specificities. Amplifications produced fragments of 166, 580, and 752 bp corresponding to the expected products from E. histolytica, E. moshkovskii, and E. dispar, respectively. Similar results were also observed when the primers for E. histolytica, E. dispar, and E. moshkovskii were mixed together and amplified in a single reaction. Amplification of the expected products for E. histolytica, E. dispar, and E. moshkovskii was again observed (Fig. 1A, lanes 1 to 3), and the intensity of each fragment was found to be similar to that obtained with the separate reactions. No interference or cross-amplification was observed when other organisms were tested with these three Entamoeba-specific primers (Fig. 1A and B).
FIG. 1.
(A) Specificity of PCR with primers EntaF and combined primers EhR, EdR, and EmR in a single reaction mixture by using DNAs of various organisms extracted from pure or axenic cultures. Lane M, molecular weight marker (100-bp ladder); lane 1, E. histolytica DNA; lane 2, E. dispar DNA; lane 3, E. moshkovskii DNA; lane 4, human DNA; lane 5, Escherichia coli DNA; lane 6, Salmonella sp. DNA; lane 7, Shigella sp. DNA; lane 8, Vibrio cholerae DNA; lane 9, Blastocystis hominis DNA; lane 10, Giardia lamblia DNA; lane 11, Cryptosporidium sp. DNA; lane 12, DNA extracted from healthy human feces; lane 13, negative control. (B) Specificity of the designed primers by using DNA extracted from fecal samples containing various organisms. Lane M, molecular weight marker (100-bp ladder); lane 1, E. histolytica DNA; lane 2, E. dispar DNA; lane 3, E. moshkovskii DNA; lanes 4 and 5, Entamoeba coli DNA; lanes 6 and 7, Endolimax nana DNA; lanes 8 and 9, Giardia lamblia DNA; lanes 10 and 13, Cryptosporidium parvum; lane 11, E. histolytica; lane 12, E. dispar; lane 14, negative control.
Similar results were also observed when DNAs of E. histolytica, E. dispar, and E. moshkovskii were mixed together in a single reaction and amplified in the PCR assay. Amplification of each corresponding fragment for E. histolytica, E. dispar, and E. moshkovskii was successful (Fig. 1B). The intensities of all of the corresponding fragments were found to be similar to the amplification of an individual DNA, as seen in Fig. 1A. No interference or cross-amplification was observed to occur between each of the paired forward and reverse primers towards the DNA from the other species of Entamoeba.
Sensitivity of the PCR assay.
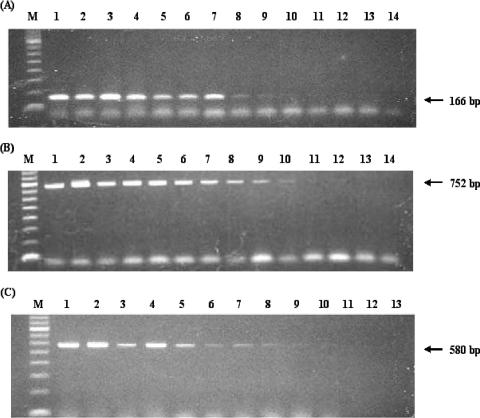
The sensitivity of this assay was evaluated using series of twofold serially diluted DNA samples containing known concentrations of E. histolytica, E. dispar, and E. moshkovskii DNA. The result showed that the assay is able to detect as little as 19 pg for E. histolytica or E. moshkovskii DNA (Fig. 2A and C, lanes 9) and 9.5 pg of E. dispar DNA (Fig. 2B, lane 10). The minimum detection level of the assay suggests that it is sensitive enough for accurate identification of the three species in clinical specimens.
FIG. 2.
Sensitivity of the PCR assay with twofold serial dilutions of E. histolytica DNA (A), E. dispar DNA (B), and E. moshkovskii DNA (C). Lane M, 100-bp ladder DNA marker; lane 1, 5 ng of DNA; lane 2, 2.5 ng of DNA; lane 3, 1.25 ng of DNA; lane 4, 0.625 ng of DNA; lane 5, 0.3125 ng of DNA; lane 6, 0.156 ng of DNA; lane 7, 0.078 ng of DNA; lane 8, 0.039 ng of DNA; lane 9, 0.019 ng of DNA; lane 10, 0.0095 ng of DNA; lane 11, 0.00475 ng of DNA; lane 12, 0.002375 ng of DNA; lane 13, 0.001188 ng of DNA; lane 14, 0.000594 ng of DNA.
Specificity of the PCR assay.
No cross-amplification was observed when the mixed primers were tested against human DNA (Fig. 1, lane 4) or against any genomic or infected stool DNA containing different bacterial or protozoal pathogens (Fig. 1A, lanes 5 to 11, and B, lanes 4 to 10 and 13). In contrast, two fecal samples from Thai patients infected with either E. histolytica or E. dispar, as confirmed by both microscopic examination and another PCR assay (12, 15), were successfully amplified. No cross-amplification or additional bands were observed (Fig. 1B, lanes 11 and 12). Moreover, parasite-free fecal samples found to be negative for Entamoeba cysts and trophozoites by microscopic examination and confirmed negative for all Entamoeba species (15) showed no PCR product under the same conditions (Fig. 1A, lane 12).
Evaluation of the PCR assay in fecal samples from Thai patients.
Thirty clinical samples collected from Thai patients were used to evaluate the PCR assay, including both fecal specimens and liver abscess aspirates. All specimens were reported as positive for Entamoeba cysts or trophozoites by microscopic examination. After being tested with a genus-specific PCR assay (15), 25 samples were positive for Entamoeba spp. whereas 5 samples were negative (Table 1) . Since the primers used in the genus-specific PCR assay for Entamoeba were designed from conserved regions, DNA of all Entamoeba species should be amplified. Therefore, the five samples which were negative by this assay but reported as positive for Entamoeba cysts by microscopy are likely the result of misdiagnosis because of other cysts being mistakenly identified and reported as being from Entamoeba species.
TABLE 1.
Summary of PCR results for E. histolytica, E. dispar, and E. moshkovskiia
| Primers | Specificity | Product size (bp) | Detection rateb (%) |
|---|---|---|---|
| Entam 1 and Entam 2c | All Entamoeba species | 550 | 25/30 (83.3) |
| P11 and P12d | E. histolytica | 101 | 4/30 (13.3) |
| P13 and P13d | E. dispar | 101 | 6/30 (20) |
| EntaF and EhRe | E. histolytica | 166 | 4/30 (13.3) |
| EntaF and EdRe | E. dispar | 752 | 6/30 (20) |
| EntaF and EmRe | E. moshkovskii | 580 | 0/30 (0) |
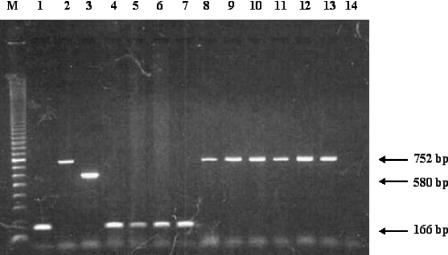
By using the PCR assay developed in this study, we successfully identified the species present in a total of 10 of the 30 clinical samples tested: 4 were positive for E. histolytica and 6 for E. dispar (Fig. 3, lanes 4 to 13). The same results were obtained when previously described E. histolytica-specific and E. dispar-specific primers were used (12). No amplification of E. moshkovskii was observed with any specimens.
FIG. 3.
PCR amplification for detection and differentiation of E. histolytica, E. dispar, and E. moshkovskii by using primers EntaF, EhR, EdR, and EmR. Lane M, 100-bp ladder DNA marker; lane 1, E. histolytica DNA; lane 2, E. dispar DNA; lane 3, E. moshkovskii DNA; lanes 4 to 7, amplified products (166 bp) which indicate the E. histolytica-positive specimens; lanes 8 to 13, amplified products (752 bp) which indicate the E. dispar-positive specimens; lane 14, negative control.
DISCUSSION
In this report, we describe the development of a single-round PCR-based approach for differential diagnosis of three species of Entamoeba, E. moshkovskii, E. histolytica, and E. dispar, which share identical morphology as both cysts and trophozoites. This simple diagnostic PCR technique does not require extra steps, as is the case with nested PCR (2), PCR-restricted fragment length polymorphism (13), and PCR with reverse line blot hybridization (14).
The assay successfully amplified the positive controls and samples from E. histolytica- and E. dispar-infected individuals that had been confirmed by other PCR methods (12, 15), as shown in Table 1. We also showed that the assay is sensitive, detecting as little as 91 pg of DNA for E. histolytica and E. moshkovskii and 9.5 pg of DNA for E. dispar. Even though the positive bands of E. histolytica and E. moshkovskii at 19 pg DNA seen in Fig. 2 (Fig. 2A and C, lanes 9) are rather faint, they were clearly observed from the real gels and they occurred consistently when several repeats of the experiment were performed. As no similar assay that differentiates all three species in a single-round PCR has been developed, we are unable to fully compare the relative sensitivity of our PCR assay. Nevertheless, when our assay is compared with the other PCR assays used (2, 12, 15) the results suggest that its sensitivity and reliability are sufficient for accurate identification of all three species in clinical specimens. The high specificity of our assay is clearly shown since no amplified PCR products were detected when DNAs from other intestinal protozoa, bacteria, or humans were tested.
By direct comparison with existing PCR assays (12), we showed that our assay was able to detect and differentiate all 10 E. histolytica and E. dispar infections in the 30 microscopy-positive clinical samples examined. Our assay showed that only one-third (10/30) of the suspected Entamoeba cases were actually positive for either E. histolytica or E. dispar (Table 1). This result clearly indicates the difficulty faced by technicians in morphologically differentiating the cysts of Entamoeba and other species by using microscopy for routine diagnosis. This could significantly affect estimates of the true prevalence of Entamoeba infections in the Thai population.
Based on our results, the prevalence ratio of E. histolytica to E. dispar in Thailand is about 2:3. However, only one out of the four E. histolytica-positive samples was from feces, the other three samples being from liver abscess. Therefore, the ratio of infections is more likely to be 1:6 rather than 2:3.
The 15 samples negative for both E. histolytica and E. dispar by our PCR assay and also by the other PCR assay (12) may belong to other Entamoeba species, as shown by a positive Entamoeba genus-specific PCR result (15). However, this speculation should be proven by the further development of molecular diagnosis for other nonpathogenic Entamoeba species commonly found in humans, such as E. coli and E. hartmanni.
There have been no E. moshkovskii-positive cases found in Thailand so far. The negative result may be from the small sample size used in this study. Since this is the first investigation of E. moshkovskii in Thailand, it is not easy to estimate the sample size without its prevalence. Therefore, if more suspected samples are available and investigated we may be able to show the presence of E. moshkovskii in the Thai population. Certainly, it will be useful to include the E. moshkovskii set of primers in any epidemiological studies in this area.
In conclusion, we propose the application of this new PCR assay as an alternative tool in routine diagnosis and in epidemiological studies of amoebiasis. It is hoped that this will provide better epidemiological data and a greater understanding of infections with these three amoebae in humans.
Acknowledgments
This work was supported by Mahidol University, Bangkok, Thailand.
We thank Graham Clark (London School of Hygiene and Tropical Medicine) for the control DNA samples, valuable suggestions, and comments and Somsri Kajorndechakiate and Niramol Thima, Department of Protozoology, Faculty of Tropical Medicine, Mahidol University, for their technical assistance.
REFERENCES
- 1.Acuna-Soto, R., J. Samuelson, P. De-Girolami, et al. 1994. Application of the polymerase chain reaction to the epidemiology of pathogenic and nonpathogenic Entamoeba histolytica. Am. J. Trop. Med. Hyg. 48:58-70. [DOI] [PubMed] [Google Scholar]
- 2.Ali, I. K. M., M. B. Hossain, S. Roy, P. F. Ayeh-Kumi, W. A. Petri, Jr., R. Haque, and C. G. Clark. 2003. Entamoeba moshkovskii infections in children, Bangladesh. Emerg. Infect. Dis. 9:580-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark, C. G., and L. S. Diamond. 1991. The Laredo strain and other Entamoeba histolytica-like amoebae are Entamoeba moshkovskii. Mol. Biochem. Parasitol. 46:11-18. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez-Ruiz, A., R. Haque, A. Aguirre, et al. 1994. Value of microscopy in the diagnosis of dysentery associated with invasive Entamoeba histolytica. J. Clin. Pathol. 47:236-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haque, R., I. K. M. Ali, S. Akther, and W. A. Petri, Jr. 1998. Comparison of PCR, isoenzyme analysis, and antigen detection for diagnosis of Entamoeba histolytica infection. J. Clin. Microbiol. 36:449-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haque, R., I. K. M. Ali, C. G. Clark, and W. A. Petri, Jr. 1998. A case report of Entamoeba moshkovskii infection in a Bangladeshi child. Parasitol. Int. 47:201-202. [Google Scholar]
- 7.Haque, R., K. Kress, S. Wood, et al. 1993. Diagnosis of pathogenic Entamoeba histolytica infection using a stool ELISA based on monoclonal antibodies to the galactose-specific adhesin. J. Infect. Dis. 167:247-249. [DOI] [PubMed] [Google Scholar]
- 8.Haque, R., L. M. Neville, P. Hahn, and W. A. Petri, Jr. 1995. Rapid diagnosis of Entamoeba infection by using Entamoeba and Entamoeba histolytica stool antigen detection kits. J. Clin. Microbiol. 33:2558-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haque, R., I. K. M. Ali, and W. A. Petri, Jr. 1999. Prevalence and immune response to Entamoeba histolytica infection on preschool children in Bangladesh. Am. J. Trop. Med. Hyg. 60:1031-1034. [DOI] [PubMed] [Google Scholar]
- 10.Mirelman, D., Y. Nuchamowitz, and T. Stolarsky. 1997. Comparison of enzyme-linked immunosorbent assay-based kits and PCR amplification of rRNA genes for simultaneous detection of Entamoeba histolytica and Entamoeba dispar. J. Clin. Microbiol. 35:2405-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pillai, D. R., J. S. Keystone, D. C. Shepard, et al. 1999. Entamoeba histolytica and Entamoeba dispar: epidemiology and comparison of diagnostic methods in a setting of nonendemicity. Clin. Infect. Dis. 29:1315-1318. [DOI] [PubMed] [Google Scholar]
- 12.Tachibana, H., S. Kobayashi, M. Takekoshi, and S. Ihara. 1991. Distinguishing pathogenic isolates of Entamoeba histolytica by polymerase chain reaction. J. Infect. Dis. 164:825-826. [DOI] [PubMed] [Google Scholar]
- 13.Tannich, E., and G. D. Burchard. 1991. Differentiation of pathogenic from nonpathogenic Entamoeba histolytica by restriction fragment analysis of a single gene amplified in vitro. J. Clin. Microbiol. 29:250-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verweij, J. J., D. Laeijendecker, E. A. T. Brienen, et al. 2003. Detection and identification of Entamoeba species in stool samples by a reverse line hybridization assay. J. Clin. Microbiol. 41:5041-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verweij, J. J., A. M. Polderman, and C. G. Clark. 2001. Genetic variation among human isolates of uninucleated cyst-producing Entamoeba species. J. Clin. Microbiol. 39:1644-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]