Abstract
We describe the validation of an enzyme-linked immunosorbent assay (ELISA) and confirmatory immunoblotting assays based on a recombinant p30 protein (p30r) produced in insect larvae using a baculovirus vector. Such validation included the following: (i) the scaling up and standardization of p30r production and the associated immunoassays, (ii) a broad immunological analysis using a large number of samples (a total of 672) from Spain and different African locations, and (iii) the detection of the ASF virus (ASFV)-antibody responses at different times after experimental infection. Yields of p30r reached up to 15% of the total protein recovered from the infected larvae at 3 days postinfection. Serological analysis of samples collected in Spain revealed that the p30r-based ELISA presented similar sensitivity to and higher specificity than the conventional Office International des Epizooties-approved ASFV ELISA. Moreover, the p30r ELISA was more sensitive than the conventional ELISA test in detecting ASFV-specific antibodies in experimentally infected animals at early times postinfection. Both the recombinant and conventional ELISAs presented variable rates of sensitivity and specificity with African samples, apparently related to their geographical origin. Comparative analyses performed on the sequences, predicted structures, and antigenicities of p30 proteins from different Spanish and African isolates suggested that variability among isolates might correlate with changes in antigenicity, thus affecting detection by the p30r ELISA. Our estimations indicate that more than 40,000 ELISA determinations and 2,000 confirmatory immunoblotting tests can be performed with the p30r protein obtained from a single infected larva, making this a feasible and inexpensive strategy for production of serological tests with application in developing countries.
African swine fever (ASF) was initially described in Kenya at the beginning of the past century (15), and still today it remains endemic in many sub-Saharan African countries, where it represents a major threat for swine industry and commerce. Its etiological agent, the African swine fever virus (ASFV), is a large enveloped virus composed of an icosahedrical virion holding a double-stranded DNA genome of 170 to 190 kbp which was recently classified as the sole member of the Asfarviridae family (4). With ASF currently included in list A of infectious diseases of the Office International des Epizooties (OIE) (16), maintenance and transmission of the ASFV in nature imply the cycling of virus between soft ticks (Ornithodoros moubata) and wild pig populations (Phacochoerus aethiopicus and Potamochoerus porcus) (20, 23). In domestic pigs, ASFV also may be transmitted by direct contact with infected material or animals, and disease may occur in several variants, ranging from highly virulent lethal forms to chronic and subclinical infections.
At present there are no efficient vaccines against ASF, and eradication in countries where it is endemic is based only on competent diagnosis programs and sacrifice of the infected animals. However, ASF diagnosis is difficult due to the variation in pathogenesis between different isolates and the similarities of clinical symptoms between ASF and other hemorrhagic diseases (16). Recently developed real-time PCR tests proved to be useful for identifying ASFV in infected animals (24); however, detection of ASFV-specific antibodies remains a major tool for diagnosis and control of the disease.
Current OIE-approved assays for ASFV-specific antibody determination consist in a first screening of sera by enzyme-linked immunosorbent assay (ELISA) and a Western blot assay to confirm positive results. However, viral antigens included in the OIE-approved tests derive from live virus and involve manipulation of the infectious agent and the requirement of level 3 biosafety facilities for production and handling of the pathogen (16, 18, 19). Alternatively, diagnostic tests based on recombinant viral proteins may prevent the use of potentially dangerous live ASFV and allow their production in standard laboratories (6, 17).
In previous reports we described the production of the ASFV p30 protein as a recombinant antigen expressed in Sf9 insect cells and Trichoplusia ni larvae by using a baculovirus expression system, as well as its preliminary characterization for ASFV diagnosis (2, 17). Here we present a detailed study on the optimal conditions for scaling up production of the recombinant p30 antigen in T. ni larvae and its validation as a diagnosis reagent for ASFV, using a large collection of serum samples from susceptible animals collected from different locations in Spain and Africa, provided by the Centro de Investigación en Sanidad Animal (CISA, Spain), a reference laboratory of ASFV for Europe. Our results showed that the recombinant p30 antigen produced in T. ni larva extracts improved the results obtained by the current OIE-approved diagnosis tests, allowing both unambiguous interpretation of the results and simple manufacture of the kits. Assays using samples obtained in Africa produced variable results, which seemed to correlate with the geographical distribution of the ASFV isolates. Possible relation between the reduced accuracy of the assays and antigenic variation among different African ASFV isolates affecting the p30 protein is discussed.
MATERIALS AND METHODS
Recombinant baculovirus.
A recombinant baculovirus expressing the ASF virus protein p30 from the E75 strain was produced using the Bac-To-Bac system (Invitrogen). The recombinant p30 protein (p30r) coding sequence was amplified by PCR, including the BamHI and HindIII restriction sites at the 5′ and 3′ ends of the gene, respectively, cloned into the pFastBac 1 vector under control of the polyhedrin promoter, and the recombinant baculovirus (p30BAC) was produced by following the manufacturer's instructions. Additionally, a pFastBac 1 vector without an insert was used to produce the control baculovirus (wtBAC), following the same protocol. Resulting baculoviruses were amplified in Sf21 cells to reach a concentration of 108 PFU/ml, keeping a working stock at 4°C and storing aliquots at −80°C.
Insect growth conditions and inoculation.
Trichoplusia ni (cabbage looper) larvae were reared under level 2 biosafety conditions as previously described (2). Briefly, eggs were placed into specially designed larva developmental cages containing an artificial insect diet (13, 14) and were kept in growth chambers at 22 ± 1°C under controlled humidity (50%) and light period (8 h/day) conditions.
For all experiments, fourth-instar larvae were sedated by incubation on ice for 15 min and then injected with baculovirus preparations near the proleg (forward along the body cavity). Larvae were inoculated with p30BAC or wtBAC baculoviruses to produce the p30r-containing or control antigen, respectively. Inoculated larvae were kept in growth chambers at 28°C for 48 to 96 h and then harvested and frozen immediately at −20°C until processed.
Preparation of insect protein extracts.
Frozen insect material was homogenized using an extraction buffer containing phosphate-buffered saline (PBS), pH 7.2, 0.01% Triton X-100, 1% sodium dodecyl sulfate [SDS], 2.5 mM dithiothreitol, 10 mM β-mercaptoethanol, and a protease inhibitor cocktail (Complete, Roche, Germany). The resulting suspension was then centrifuged at 10,000 × g for 10 min, and the pellet was discarded. The supernatant was filtered through Whatman papers and centrifuged once again as described above. The final preparation was divided in 1-ml aliquots, lyophilized overnight in a Telstar Cryodos freeze-dryer (Telstar, Spain), and stored at 4°C until used.
Analysis and quantification of recombinant protein production.
Crude protein larva extracts (20 μg per lane) were resolved in duplicate 12% SDS-polyacrylamide gels for Coomassie blue staining and transferred onto nitrocellulose membranes (Bio-Rad). For Western blot analysis, membranes were blocked overnight with PBS-Tween 20 (PBST) containing 4% skim milk and subsequently incubated at room temperature (RT) with a pool of polyclonal sera from pigs experimentally inoculated with ASFV and anti-pig immunoglobulin G-specific alkaline-phosphatase-labeled conjugate as a secondary antibody (Sigma). Protein bands were then detected by addition of a Nitro Blue Tetrazolium chloride-5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt substrate (Sigma). For quantification, samples were loaded in Pro260 chips (Bio-Rad) and analyzed by capillary electrophoresis using the Experion system (Bio-Rad), following the manufacturer's instructions.
Serum samples.
Pig sera used in this study comprised a collection of 672 samples obtained from Spain and different locations in Africa. Of those, 468 samples were collected from enzootic areas during the eradication programs performed in Spain until 1994 and were characterized as seropositive or seronegative based on the OIE-approved protocols during the official eradication campaigns, and 9 samples were from animals experimentally inoculated at CISA. The rest of the samples were tested during epidemiological and surveillance studies performed by CISA staff in Nigeria (n = 14, all from domestic pigs) and Uganda (n = 181, 26 from warthogs and 155 from domestic pigs).
p30r-based ELISA.
Freeze-dried samples from larvae inoculated with either the recombinant p30BAC [Ag(+)] or wtBAC baculoviruses [Ag(−)] were reconstituted in distilled H2O, and total solubilized protein was measured by the Bradford assay using a commercial kit (Bio-Rad). For both antigens, the concentration was adjusted to 6 mg of total solubilized protein/ml, and ELISA microplates (Polysorp, Nunc, Denmark) were coated with 100 μl of a 1:3,000 dilution in 50 mM carbonate/bicarbonate buffer, pH 9.6, and incubated overnight at 4°C. The next day, plates were washed with PBST three times and used immediately or stored at −20°C until use. The following incubations were performed for 1 h at 37°C under constant agitation. Plates were sequentially incubated with blocking buffer (PBST-2% bovine serum albumin, 50 μl/well) and pig serum samples diluted 1:80 in blocking buffer in duplicate wells. Each serum sample was tested against both Ag(+) and Ag(−). Plates were washed four times with PBST, and 50 μl/well of protein A-horseradish peroxidase conjugate (Sigma, Missouri), diluted 1:2,000, was finally added. For the substrate reaction, plates were washed four times, and 50 μl/well of substrate solution consisting of 1 mM 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid), 35 mM citric acid, 67 mM Na2HPO4, pH = 5.0, and 0.015% H2O2 was added to the plates. A peroxidase reaction was allowed to develop for 15 min at room temperature, and reactions were stopped and read at 405 nm in an ELISA microplate reader (Multiskan EX; Thermo Electron Corp.). Serum titers were expressed as the ratio between the mean optical density (OD) obtained for each sample in duplicate assays against Ag(+) and Ag(−) [OD for Ag(+)/OD for Ag(−)].
p30r-based immunoblotting assay.
Total soluble proteins from Ag(+) crude larva extracts (100 μg/gel) were resolved in preparative 12% SDS-polyacrylamide gels and transferred onto nitrocellulose membranes as described above. The portion of the membranes containing proteins with molecular masses between 80 kDa and 20 kDa was excised and divided into 4.0-mm-wide strips. For Western blot assays, strips were blocked for 1 h with PBST-4% skim milk and subsequently incubated for 1 h at RT with pig sera diluted 1:50 and a protein A-horseradish peroxidase conjugate (Sigma) at a 1:2,000 dilution in blocking buffer. Finally, assays were developed using 0.3% 4-chloronaphthol solution (Sigma) as a substrate, and the reaction was stopped with distilled water after 5 min.
Conventional ELISA and immunoblotting assays (OIE-approved analyses).
Both conventional ELISA and immunoblotting assays were performed using the whole ASFV isolate Spain 1970 (E70) produced in a monkey kidney cell line (MS) as a detector antigen (18, 19), following the protocols described in the OIE manual of diagnosis (16). For the ELISA, serum samples were considered to be positive when presenting an absorbance value of more than twice the mean absorbance value of control negative sera on that plate.
Analysis of sequences.
Complete amino acid sequences of the p30 protein of ASFV corresponding to different African and European isolates were compared with that of the Spanish E70 isolate (GenBank accession no. AF462272). DNA sequences from the CP204L gene, which codes for the p30 protein, were translated, and alignments and identity scores were obtained using the Clustal-W program (European Bioinformatics Institute [http://www.ebi.ac.uk/clustalw/]) (3, 22). Comparison included the Benin 97/1 isolate (L. K. Dixon, personal communication) and Kenya 1950, Malawi Lil-20/1 1983, Mkuzi 1979, Pretorisuskop/96/4, Tengani 62, Warmbaths, and Warthog ASFV isolates (GenBank accession no. AY261360 to AY261366, respectively). Identification of protein structural features within the p30 sequences was carried out using the DisEMBL analysis tool (11) (version 1.4, EMBL [http://dis.embl.de/]), and determination of antigenicity profiles along the sequence were performed using the algorithm described by Hopp and Woods (9) (http://bioinformatics.org/JaMBW/3/1/7/index.html).
RESULTS
Optimization of the production of recombinant p30 in T. ni larvae.
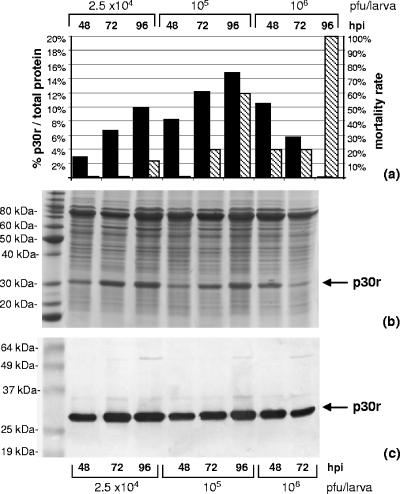
A set of experiments was carried out to establish optimal conditions for production of recombinant p30 in T. ni larvae. Fourth-instar larvae were inoculated with three different doses of the p30BAC baculovirus, and larvae were harvested and processed at different times postinoculation. Total protein extracts were obtained from infected larvae and analyzed by different electrophoretic methods. As shown in Fig. 1, the recombinant ASFV p30 protein was produced and accumulated in the inoculated larvae in a dose- and time-dependent manner. p30r was clearly identified as a single band with an electrophoretic mobility around 30 kDa in Coomassie blue-stained gels (Fig. 1b), and it was also detected in Western-blot assays using a pool of ASFV-specific sera as detectors (Fig. 1c), with a signal intensity resembling that of p30r in stained gels. A more-detailed analysis using capillary electrophoresis determined that p30r was produced as a single species with a mobility of 29.05 kDa and reaching levels up to 14.9% of total protein extracted. According to the accumulation levels of p30r and the mortality rates observed for each treatment, a dose of 105 PFU/larva and an infection period of 72 h were selected for the production of the recombinant antigen (Fig. 1a). Under these conditions, the p30r protein reached 12.2% of the total protein extracted, and 80% of the inoculated larvae were recovered for processing.
FIG. 1.
Production of the recombinant p30 produced in T. ni larvae. Fourth-instar larvae were inoculated with different doses of recombinant p30BAC, and infections were allowed to progress for 48, 72, or 96 h postinfection. Total protein extracts from each group were analyzed by (a) capillary electrophoresis: gray bars represent the percentage of the p30r protein produced out of the total extracted protein, and black bars indicate the mortality rate associated with each group; (b) Coomassie brilliant blue staining of SDS-page gels; and (c) Western blotting using a pool of swine sera from ASFV-seropositive animals as a probe. Arrows indicate the position of the recombinant p30.
Reactivity of pig sera to larva extracts.
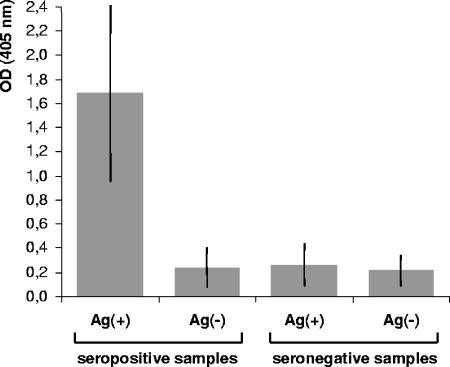
The specific and background reactivity of pig sera against crude protein extracts from T. ni larvae was tested by ELISA. A large collection of ASFV-seropositive and -seronegative samples, previously characterized during the Spanish ASFV eradication program, was tested in plates coated with Ag(+) or Ag(−). The optimum amount of antigen bound to the plates for detection of ASFV-specific antibodies was experimentally determined using serial dilutions of the crude protein extracts in carbonate-bicarbonate buffer (data not shown). The final protein concentration was adjusted to 200 ng per well, bearing ∼25 ng of p30r in Ag(+) preparations. As shown in Fig. 2, ASFV-positive samples (n = 165) produced OD readings significantly higher than those obtained with the control antigen. In the same way, ASFV-seronegative samples (n = 303) presented significantly lower OD readings against both Ag(+) or Ag(−) than those of the ASFV-positive sera with the positive antigen. These results indicated that positive signals in ELISA were specific to the p30r-containing extracts and that pig sera presented no significant background reactivity against larva protein extracts.
FIG. 2.
Reactivity of porcine sera in ELISA assays based on insect-derived antigens. Samples from ASFV-seropositive (n = 165) or seronegative (n = 303) animals, previously characterized by the OIE-approved tests, were tested by ELISA using crude extracts from larvae infected with p30BAC [Ag (+)] or wtBAC [Ag(−)] as detector antigens. Bars represent the mean OD obtained for each group, and standard deviations are indicated in each case by transversal lines.
Comparative study between conventional and p30r-based ELISAs using Spanish field serum samples.
Results obtained using this pool of Spanish ASF field serum samples were used to establish the cutoff values for the p30r-based ELISA. Based on the previous results showing the absence of significant background of porcine sera against control larvae extracts [Ag(−)], we decided to express results for the p30r ELISA as the ratio between the absorbancies obtained for each sample against the Ag(+) and Ag(−) preparations. All samples were also analyzed by the confirmatory Western-blot assay based on the larva-produced p30r.
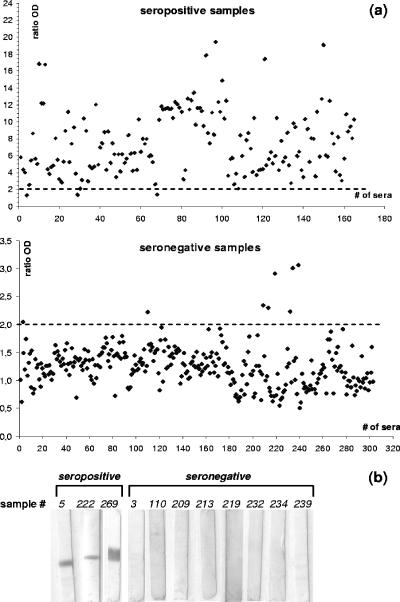
Setting an OD ratio of 2.0 as the cutoff level for the p30r ELISA, 162 out of 165 seropositive samples were positive for the assay (Fig. 3a). All three samples with OD ratios below 2 were confirmed as positive by the p30r-based immunoblotting (Fig. 3b). A very similar proportion was detected with the OIE-approved ELISA (163 out 165), resulting in a sensitivity of 98.2% for p30r-based assay and 98.8% for the conventional ELISA. However, analysis of seronegative samples clearly favored the recombinant ELISA. While only 266 out of 303 were determined as seronegative by the ASFV-based assay, 295 samples from the total of 303 were classified as seronegative using the recombinant ELISA. The resulting specificities were 87.8% for the OIE-approved ELISA and 97.4% for the p30r ELISA. Also, in this case, those samples above the cutoff value were confirmed as seronegative using the p30r-based Western blot (Fig. 3b).
FIG. 3.
Detection of ASFV-seropositive and -seronegative samples using the p30r-based ELISA and Western blotting. (a) Ratio ODs obtained by ELISA from seropositive and seronegative samples. Values refer to the media of duplicate analyses. Cutoff values are shown as dotted lines on each panel, and assays were carried out as described in Methods. (b) Western blots performed with all the seropositive and seronegative samples that failed to be detected by p30r ELISA.
Detection of ASFV-specific antibodies in infected animals.
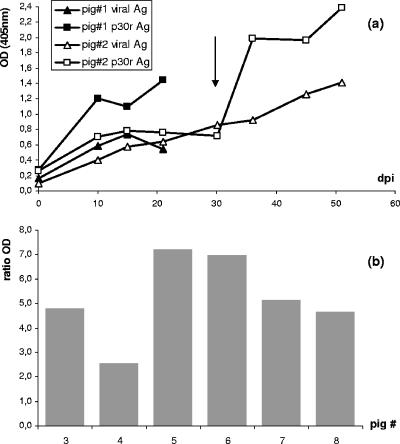
The capacity of the p30r-based ELISA for early detection of ASFV antibodies was assessed in experimentally infected animals. Two animals (pigs 1 and 2) were inoculated with the attenuated ASFV strain E75 CV1-4, and serum samples were taken at 10, 14, and 21 days postinfection (dpi). A comparative study showed that the ELISA based on the p30r was able to detect ASFV antibodies with more sensitivity than the conventional assay at early times postinfection (Fig. 4a). Pig 2 was reinoculated with the homologous virulent virus E75L8, and interestingly these differences in sensitivity increased after inoculation (Fig. 4a). The p30r-based assay was also used to detect ASFV-specific humoral responses in sera from six other pigs (pigs 3 to 8) 30 days after being infected with the E75 CV1-4 attenuated virus. OD ratios resulted in positive results in all cases, reaching values from 2.6 to 7.2 (Fig. 4b).
FIG. 4.
Detection of ASFV-specific antibodies from experimentally infected pigs using the p30r-based ELISA. (a) Pigs 1 and 2 were inoculated with the attenuated strain E75 CV1-4, and serum samples were taken at 10, 14, and 21 dpi and analyzed by the OIE-approved and p30r-based ELISAs. Pig 1 was sacrificed at 21 dpi, and pig 2 was reinoculated (arrow) with the E75L8 virulent strain; (b) ratio ODs obtained using the p30r-based ELISA at 30 dpi for six additional animals (no. 3 to no. 8) infected once with the attenuated E75 CV1-4 strain.
Performance of the p30r ELISA using serum samples from Africa.
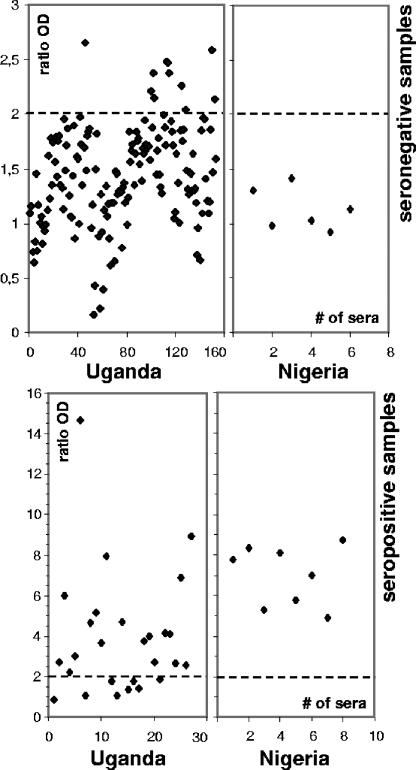
A smaller number of African serum samples, which were previously characterized using the OIE-approved immunoblotting assay, were analyzed using conventional and p30r-based ELISAs. Samples (n = 195) were obtained from domestic pigs in Nigeria (n = 14) and Uganda (n = 181), except for 26 seropositive samples taken from warthogs in Uganda. While samples from Nigeria were characterized with 100% of specificity (n = 6) and sensitivity (n = 8) using the p30-based ELISA, samples from Uganda showed lower levels of specificity and sensitivity (93% [143 out of 154] and 70% [19 out 27], respectively), similar to those of the conventional ELISA (99% [152 out of 154] and 67% [18 out of 27], respectively). Recombinant and conventional ELISA produced coincident results in 92% of seronegative samples (142 out of 154) and in only 44% of the seropositive samples (12 out of 27).
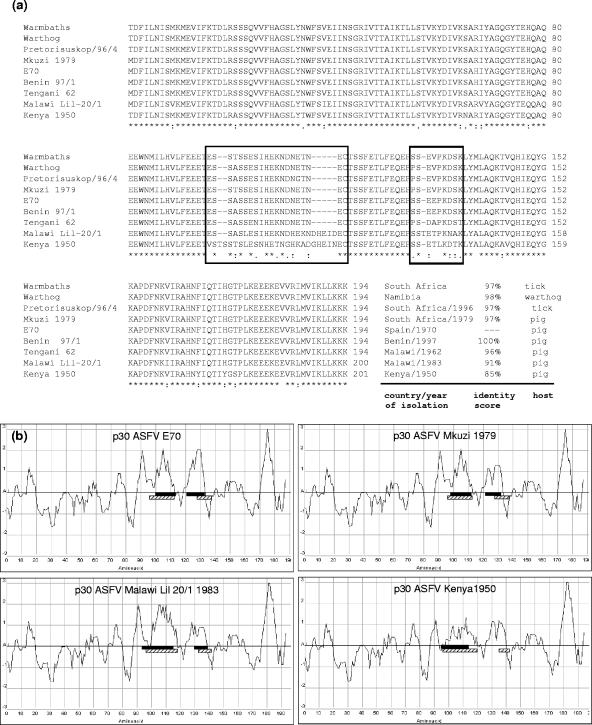
In order to study whether these findings may be related to antigenic differences existing between African and European ASFV isolates, we decided to compare sequences of a Spanish isolate (E70) to others from Europe and Africa, an analysis never performed for p30. While all Spanish isolates analyzed (E75, 608, and Ba71) showed 100% identity (data not shown), results were variable for African isolates depending on geographical origin. As shown in Fig. 5a, the total length for the p30 protein was 194 residues in all the isolates, except for the Malawi Lil 20/1 1983 and Kenya 1950 sequences, with 200 and 201 amino acids, respectively. Consistent with this, identity scores in relation to the E70 strain decreased from 100% for a Western isolate (Benin 97/1) to 98% and 96% for Southern isolates (Mkuzi 1979, Warmbaths, Pretorisuskop/96/4, Warthog, and Tengani 62), finally dropping to 91% and 85% for Eastern isolates (Malawi Lil 20/1 1983 and Kenya 1950, respectively) (Fig. 6a), in a pattern very similar to that described for the ASFV p72 structural protein (12). Additionally, p30 sequences were analyzed for identification of the following: (i) antigenic fragments within the primary structure and (ii) regions with intrinsic structural disorder, as established by the Hopp and Woods algorithm (9) and the DisEMBL analysis tool (11), respectively. Interestingly, we observed that those regions with the highest sequence variability also presented high antigenic indexes and theoretical low structural order. Figure 6b shows these analyses applied to the E70 isolate and to three African isolates with different degrees of identity with the Spanish strain, although the same results were observed for all the isolates mentioned above.
FIG. 5.
Analysis of ASFV African serum samples using the p30r-based ELISA. Samples from Eastern (Uganda) and Western (Nigeria) African countries, previously characterized by the OIE-approved immunoblotting, were analyzed by the p30r-based ELISA. Charts show ratio ODs obtained by p30r ELISA from seropositive and seronegative samples from these two countries. Values refer to the media of duplicate analyses. Cutoff values are shown as dotted lines on each panel, and assays were carried out as described in Materials and Methods.
FIG. 6.
Sequence analysis of p30 proteins from different ASFV isolates. (a) Alignment of p30 sequences obtained from different African locations, including the Spanish E70 isolate. The lower row represents the degree of identity between the Spanish and the different African isolates, and boxed fragments indicate the most variable positions within these sequences. Additional information, including the identity score regarding the E70 virus, country, and year of isolation and hosts from which they were obtained, is included at the end of each of the sequences. *, identical residues in all sequences; :, highly conserved column; ., weakly conserved column. (b) Correlation between antigenicity, structure, and variability in the p30 protein from different isolates. Charts display the variation of the antigenic index as a function of amino acid position for the Spanish E70 and African isolates: Malawi Lil 20/1 1983, Mkuzi 1979, and Kenya 1950. Horizontal black bars indicate regions of predicted intrinsic disorder, and hatched bars mark those fragments presenting greater variability among the isolates as determined in the sequence alignments.
DISCUSSION
Still today no effective treatment or vaccine has been developed for ASF, and disease can be controlled and eradicated only through enhancing regional and farm-level biosecurity measures. These are basically built on consistent knowledge of the epidemiological patterns and the management of the risk of introduction and spread of the disease. In this context, development of efficient and low-priced diagnostic tests are essential as molecular tools for epidemiological studies. Following this idea, assays based on recombinant proteins produced on inexpensive platforms, such as baculovirus and insect larvae, are ideal candidates.
The conditions applied to the production of the p30r protein in T. ni larvae allowed yields between 0.9 to 1.25 mg of p30r per larvae to be obtained in 72 h with ∼20% mortality among the inoculated insects. Recombinant ELISA and immunoblotting assays performed using protein extracts without further purification presented very a low level of unspecific reactivity against both ASFV-negative sera and control larvae antigen. The amount of p30r utilized in the assay was ∼25 ng/well for the ELISA and ∼500 ng per immunoblotting strip, which would yield approximately 40,000 ELISA and 2,000 Western-blot determinations from a single larva.
ELISA tests were set up using extracts from larvae infected with wtBAC baculovirus as negative controls [Ag(−)], and ELISA results were expressed as the ratio between ODs obtained against both antigens [OD for Ag(+)/OD for Ag(−)]. This contributed to the consistency of the assay, since larva extracts represent an abundant source of reagent, which may be better monitored for uniformity and stability than controls based on OD values from ASFV-negative sera. Furthermore, lyophilized crude larva extracts can be stored either at 4°C or RT for more than 2 years without a significant loss of activity (D. M. Pérez-Filgueira, unpublished data).
The cutoff level for the recombinant ELISA (OD ratio = 2.0) was established using a pool of 468 samples comprising ASFV-seronegative and -seropositive pig sera. With these conditions, the sensitivity of the p30r-based assay was very similar to that of the conventional ELISA (98.2% versus 98.8%); however, specificity was greatly improved with recombinant antigen coated on the plates (97.4% versus 87.8%). This enhanced specificity of the p30r ELISA is particularly important for eradication programs in developing countries, where false-positive results are frequent due to the deficient conditions of sera transport and storage, one of the most common reasons for unspecific results in ELISA (5). Additionally, accurate detection of seronegative animals is critical for reduction of the slaughtering in the susceptible population, especially in economically depressed areas, where individual animals have high relative values. It is important to observe that analysis of the whole pool of sera by recombinant immunoblotting was 100% coincident with OIE-recommended diagnostic methodologies, a result that indicates the high level of confidence of the whole test.
Another interesting feature of the p30r ELISA was its ability for early detection of ASFV-specific antibodies after experimental infection. The recombinant assay was able to clearly identify ASFV-specific antibodies approximately 1 week earlier than the conventional assay. This same effect was observed after reinfection with a virulent virus strain, with ELISA signals rapidly increasing within a week postchallenge. Most probably, the assay takes advantage of specific biological attributes associated with the p30. On one hand, p30 as a main ASFV-induced protein which is incorporated in the virus particle carries strong immunogenic determinants and consequently represents a very efficient inducer of humoral responses (7, 8, 10). On the other hand, this protein is expressed and secreted at early times after infection (1, 21), which would explain its rapid immune recognition after the infection.
Analysis of serum samples from Africa using the p30r-based and conventional ELISAs resulted in variable percentages of sensitivity and specificity. Samples were obtained from Western (Nigeria) and Eastern (Uganda) locations of the continent and grouped according their ASFV serology. Our results show that while samples taken from Nigeria presented 100% specificity and sensitivity with both assays, results for sera from Uganda showed lower specificity (93% for p30r ELISA and 98% for the OIE-approved ELISA) and especially much lower sensitivity (70% and 67% for the p30r and conventional ELISA, respectively).
Comparison of the p30 sequences from Spanish isolates and different isolates obtained at Western, Southern, and Eastern Africa showed that identity scores decreased from West to East, with those of the Southern isolates intermediate between these two. A similar pattern was reported for the ASFV p72 structural protein (12) in a study which actually included sequences from Nigeria and Uganda: genotypes from Western countries showed results identical to those of European, South American, and Caribbean viruses (genotype I), Eastern isolates were more genotypically distant and more variable, and the situation in Southern Africa was in the middle of these two extremes. Interestingly, our analyses also show that regions predicted to contain antigenic determinants and fragments of p30 with putative intrinsic structural disorder overlapped with the two areas where variability among isolates was greater. Together these results might suggest that isolates from Eastern and Southern countries of Africa may be antigenically different regarding the p30 protein and that these changes in antigenicity are produced, at least to some extent, through variations that occur in areas of the protein with low relevance in structure or function and which are thus more permissive to sequence modifications.
Also, it is important to note that 26 out of 27 seropositive samples from Uganda were obtained from wild suids (Phacochoerus aethiopicus), a fact that may also have affected the accuracy of the ELISAs. Besides this possibility, our analyses showed that while efficiency of the OIE and p30r ELISAs was very similar, the percentage of warthog samples detected by both ELISAs was very low (12 out of 26 [46%]). Assuming that antigenic determinants recognized by antibodies in each ELISA are only partially coincident, antigenic variation affecting different ASFV proteins might lead to this lack of coincidence between the OIE-approved ELISA—based on a mixture of ASFV-derived antigens—and the p30r ELISA, based only on this protein.
In any case, whether these preliminary findings reflect, in fact, antigenic differences between isolates awaits further studies, including greater numbers of p30 sequences and serum samples from different regions of Africa, as well as samples from both domestic and sylvatic hosts.
In conclusion, our results demonstrate that production of the ASFV p30 protein in insect larvae can be reliably scaled up and that crude extracts can be directly used for both ELISA and immunoblotting without further purification. According to the procedures described in this report, a single T. ni larva could produce sufficient recombinant p30 to perform approximately 40,000 individual ELISA determinations or 2,000 confirmatory Western blot assays. The p30r-based ELISA improved the specificity of the OIE-recommended assay as well as its sensitivity to detect early humoral responses after experimental challenge. Studies using African samples showed that the ELISA based on a recombinant p30 derived from a Spanish isolate was very accurate when tested with a limited number of samples from Western Africa but performed with less effectiveness with samples from Eastern Africa. If these results are in fact related to antigenic variations among isolates, it would be necessary to produce additional versions of p30r from those ASFV serotypes more distant to genotype I and include them in the antigen preparation. In any case, this report indicates that this technology can be of great use, offering a very inexpensive means for producing ASFV detection kits using crude extracts and more importantly without the need of highly specific biological containment facilities, a key attribute regarding its possible application in resource-limited areas of the world.
Acknowledgments
The present work was supported by grants AGL2004-07857 from Ministerio de Educación y Ciencia of Spain and The Wellcome Trust (grant 075813).
REFERENCES
- 1.Afonso, C. L., C. Alcaraz, A. Brun, M. D. Sussman, D. V. Onisk, J. M. Escribano, and D. L. Rock. 1992. Characterization of p30, a highly antigenic membrane and secreted protein of African swine fever virus. Virology 189:368-373. [DOI] [PubMed] [Google Scholar]
- 2.Barderas, M. G., A. Wigdorovitz, F. Merelo, F. Beitia, C. Alonso, M. V. Borca, and J. M. Escribano. 2000. Serodiagnosis of African swine fever using the recombinant protein p30 expressed in insect larvae. J. Virol. Methods 89:129-136. [DOI] [PubMed] [Google Scholar]
- 3.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon, L. K., J. V. Costa, J. M. Escribano, D. L. Rock, E. Vinuela, and P. J. Wilkinson. 2000. Family Asfarviridae, p. 159-165. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 5.Escribano, J. M., M. J. Pastor, and J. M. Sánchez-Vizcaíno. 1989. Antibodies to bovine serum albumin in swine sera: implications for false positive reactions in the serodiagnosis of African swine fever. Am. J. Vet. Res. 50:1118-1122. [PubMed] [Google Scholar]
- 6.Gallardo, C., E. Blanco, M. J. Rodríguez, A. L. Carrascosa, and J. M. Sanchez-Vizcaino. 2006. Antigenic properties and diagnostic potential of African swine fever virus protein pp62 expressed in insect cells. J. Clin. Microbiol. 44:1489-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gómez-Puertas, P., F. Rodriguez, J. M. Oviedo, F. Ramiro-Ibañez, F. Ruiz-Gonzalvo, C. Alonso, and J. M. Escribano. 1996. Neutralizing antibodies to different proteins of African swine fever virus inhibit both virus attachment and internalization. J. Virol. 70:5689-5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gómez-Puertas, P., F. Rodríguez, J. M. Oviedo, A. Brun, C. Alonso, and J. M. Escribano. 1998. The African swine fever virus proteins p54 and p30 are involved in two distinct steps of virus attachment and both contribute to the antibody-mediated protective immune response. Virology 243:461-471. [DOI] [PubMed] [Google Scholar]
- 9.Hopp, T. P., and K. R. Woods. 1981. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA 78:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kollnberger, S. D., B. Gutierrez-Castaneda, M. Foster-Cuevas, A. Corteyn, and R. M. Parkhouse. 2002. Identification of the principal serological immunodeterminants of African swine fever virus by screening a virus cDNA library with antibody. J. Gen. Virol. 83:1331-1342. [DOI] [PubMed] [Google Scholar]
- 11.Linding, R., L. J. Jensen, F. Diella, P. Bork, T. J. Gibson, and R. B. Russell. 2003. Protein disorder prediction: implications for structural proteomics. Structure 11:1453-1459. [DOI] [PubMed] [Google Scholar]
- 12.Lubisi, B. A., A. D. S. Bastos, R. M. Dwarka1, and W. Vosloo1. 2005. Molecular epidemiology of African swine fever in East Africa. Arch. Virol. 150:2439-2452. [DOI] [PubMed] [Google Scholar]
- 13.Medin, J. A., L. Hunt, K. Gathy, R. Evans, and M. S. Coleman. 1990. Efficient, low-cost protein factories: expression of human adenosine deaminase in baculovirus-infected insect larvae. Proc. Natl. Acad. Sci. USA 87:2760-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medin, J. A., K. Gathy, and M. S. Coleman. 1995. Expression of foreign proteins in Trichoplusia ni larvae, p. 265-275. In C. D. Richardson (ed.), Methods in molecular biology, vol. 39: baculovirus expression protocols. Humana Press, Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 15.Montgomery, R. E. 1921. On a farm of swine fever occurring in British East Africa (Kenya colony). J. Comp. Pathol. Ther. 34:159-191, 243-264. [Google Scholar]
- 16.Office International des Epizooties. 2004. African swine fever, p. 1178. In Manual of diagnostic tests and vaccines for terrestrial animals, 5th ed. Office International des Epizooties, Paris, France.
- 17.Oviedo, J. M., F. Rodríguez, P. Gómez-Puertas, C. Alonso, and J. M. Escribano. 1997. High level expression of the major antigenic African swine fever virus proteins p54 and p30 in baculovirus and their potential use as diagnostic reagents. J. Virol. Methods 64:27-35. [DOI] [PubMed] [Google Scholar]
- 18.Pastor, M. J., M. D. Laviada, J. M. Sanchez-Vizcaino, and J. M. Escribano. 1989. Detection of African swine fever virus antibodies by immunoblotting assay. Can. J. Vet. Res. 53:105-107. [PMC free article] [PubMed] [Google Scholar]
- 19.Pastor, M. J., M. Arias, and J. M. Escribano. 1990. Comparison of two antigens for use in an enzyme-linked immunosorbent assay to detect African swine fever antibody. Am. J. Vet. Res. 51:1540-1543. [PubMed] [Google Scholar]
- 20.Plowright, W., J. Parker, and M. A. Pierce. 1969. The epizootiology of African swine fever in Africa. Vet. Rec. 85:668-674. [PubMed] [Google Scholar]
- 21.Prados, F. J., E. Vinuela, and A. Alcami. 1993. Sequence and characterization of the major early phosphoprotein p32 of african swine fever virus. J. Virol. 67:2475-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple alignment through sequence weighting. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomson, G. R. 1985. The epizootiology of African swine fever: the role of the free-living hosts in Africa. Onderstepoort. J. Vet. Res. 52:201-209. [PubMed] [Google Scholar]
- 24.Zsak, L., M. V. Borca, G. R. Risatti, A. Zsak, R. A. French, Z. Lu, G. F. Kutish, J. G. Neilan, J. D. Callahan, W. M. Nelson, and D. L. Rock. 2005. Preclinical diagnosis of African swine fever in contact-exposed swine by a real-time PCR assay. J. Clin. Microbiol. 43:112-119. [DOI] [PMC free article] [PubMed] [Google Scholar]