Abstract
The use of a single broad-spectrum human papillomavirus (HPV) DNA-based PCR test may fail to detect lower concentrations of HPV DNA due to competition between different genotypes in mixed infections. To improve HPV detection by PCR, broad-spectrum and type-specific (TS) PCRs were combined, with a focus on HPV-16 and HPV-18. Cervical and cervicovaginal cell samples were obtained from 1,113 healthy women (age range, 15 to 25 years) participating in an HPV-16/HPV-18 candidate vaccine efficacy trial. These samples were tested by a broad-spectrum SPF10 PCR-DNA enzyme immunoassay, followed by a primer SPF10-based line probe assay (SPF10 LiPA), and HPV-16- and HPV-18-TS PCRs. The results for the majority of the HPV-16/18 SPF10 LiPA-positive samples were confirmed by TS-PCR (kappa values, 0.775 for HPV-16 and 0.785 for HPV-18). However, TS PCR revealed additional positive samples among those that contained other HPV genotypes due to competition. Conversely, SPF10 LiPA identified HPV-16 or -18 in samples that remained negative by TS PCR as a result of sampling variation. Analysis of follow-up samples from more than 1,000 women confirmed that the combination of SPF10-LiPA with additional HPV-16- and HPV-18-TS PCR diminishes the rate of false-negative diagnosis. The combination of broad-spectrum and TS PCRs resulted in a novel testing algorithm. This combination of assays is more accurate than either method alone, and the novel algorithm offers a highly accurate and effective method for the analysis of HPV infections.
Human papillomavirus (HPV) is a DNA virus that infects cutaneous and mucosal epithelia and induces epithelial proliferation. More than 40 HPV genotypes have been detected in the anogenital region; and the clinically most important types are the oncogenic (high-risk) HPV genotypes (e.g., HPV-16 and HPV-18), which are involved in the development of high-grade cervical intraepithelial neoplasias and cervical cancer (5, 23, 37). HPV DNA has been detected in 99.7% of cervical cancer tissues (35); and persistent infection with an oncogenic HPV type, particularly HPV-16 or -18, is recognized as the necessary cause of cervical cancer (34). It is estimated that cervical cancer contributes to approximately 290,000 deaths and 490,000 new cases per year worldwide (24). Vaccination against the most common oncogenic HPV genotypes, HPV-16 and HPV-18, could prevent persistent infections with those genotypes and ultimately could also prevent the development of up to 70% of cases of cervical cancer worldwide (21, 33).
Diagnosis of HPV infections is based on the detection of HPV genomic DNA in cervical cell samples or cervical biopsy specimens by molecular methods, such as liquid hybridization (e.g., the Hybrid Capture assay [Digene Corporation]) (3, 7) or PCR (8, 31). Liquid hybridization detects HPV DNA by direct probe hybridization and can distinguish between groups of high-risk and low-risk HPV genotypes, but it does not permit the identification of individual genotypes (25). PCR methods amplify parts of the HPV DNA genome, resulting in high degrees of analytical sensitivity and specificity.
The high degree of genetic heterogeneity among the different HPV genotypes complicates effective diagnosis (4). PCR can use type-specific (TS) primers for the detection of individual HPV genotypes, but this requires multiple reactions (2, 34). Also, type-specific PCRs may be hampered by the existence of uncharacterized viral variant sequences that would not efficiently match the PCR primers. Alternatively, broad-spectrum PCR primers that permit the simultaneous amplification of a range of HPV genotypes in a single PCR test can be used (10, 12, 17). Broad-spectrum PCR is based on the use of primers that target relatively well conserved genomic sequences and that match multiple HPV genotypes. Basically, three different approaches are possible (31). The first approach is to select a single forward primer and a single reverse PCR primer which perfectly match only one or a few HPV genotypes. To compensate for the mismatches with other HPV genotypes, the PCR is performed at a low annealing temperature, which permits the cross-reactivity of the primers with imperfect target HPV sequences. The GP5+/GP6+ PCR system is an example of this approach (18). Second, broad-spectrum PCR primer sets may comprise degenerate primers to compensate for the intertypic sequence variation at the priming sites. My11/My09 is an example of a degenerate PCR primer set (17). In fact, this primer set comprises an undefined mixture of many different oligonucleotides. The third option for a broad-spectrum PCR primer set is the use of a cocktail that comprises a number of distinct forward and reverse primers. This primer set does not contain degenerate primers but may contain inosine, a nucleotide analogue that matches multiple nucleotides in the opposite strand. The use of such a defined mixture of nondegenerate primers has the advantage that the cocktail of oligonucleotide primers can be synthesized with a high degree of reproducibility, and PCR can be performed at optimal annealing temperatures, increasing the sensitivity, specificity, and reproducibility. Examples of such primer sets are the PGMY primers (6, 13) and the SPF10 primers (19, 20).
Since broad-spectrum PCR primers do not have the same sensitivity and specificity for each genotype, the amplification efficiency may differ among individual genotypes. More importantly, broad-spectrum PCR is affected by competition between the different HPV genotypes present in the same sample. Due to this competition effect at the primer target level, broad-spectrum PCR underestimates the prevalence of multiple genotypes, especially minority species, which are present at low relative concentrations.
HPV vaccines are being developed to prevent HPV-16 and -18 infections that lead to cervical cancer (15, 21). Clinical trials are under way to assess the efficacy and safety of HPV vaccines, where virological end points such as incident and persistent HPV infection are important surrogates. It is essential to have established efficient methods for HPV detection and genotyping methods that yield high analytical sensitivities. This aim is clearly different from that from the use of HPV diagnosis in a clinical setting, where the sensitivity should be clinically relevant (29).
A broad-spectrum SPF10 PCR system amplifies a fragment of only 65 bp from the L1 region (19, 20). The SPF10 system comprises a highly sensitive broad-spectrum PCR, followed by the general detection of amplified HPV DNA to determine the presence of the virus. SPF10 amplimers from HPV-positive samples can then be genotyped by using reverse hybridization on a line probe assay (LiPA). When this SFP10-based system (referred to here as SPF10 LiPA) is used, multiple HPV genotypes have frequently been observed in cervical cell samples and biopsy specimens, and infections with multiple HPV genotypes appear to be common (19, 22). Therefore, it is expected that the competition effect will play a significant role.
The aim of the study described here was to design and develop a testing algorithm that combines the advantages of the broad-spectrum PCR with the specificity of the type-specific PCR for HPV-16 and -18 and optimize the overall efficacy for the detection of these two genotypes, minimizing competition effects. This novel testing algorithm was evaluated by testing cervical cell samples from women participating in a clinical trial of a candidate bivalent prophylactic HPV-16 and HPV-18 L1-based vaccine (15).
MATERIALS AND METHODS
Competition testing.
Competition between different genotypes in a single clinical sample was experimentally tested. Plasmids containing the complete genomic DNA of HPV genotype 11, 16, 18, or 44 were used as the amplification targets in various experiments. These plasmids were kindly provided by E.-M. de Villiers, Heidelberg, Germany (genotypes 11, 16, and 18), and A. Lorincz, Silver Spring, MD (genotype 44). Competition between HPV-16 and -44 and also between HPV-11 and HPV-18 was tested because the primer target sites of the HPV genotypes in these pairs are highly similar and the highest level of competition can be expected. Serial (10-fold) plasmid dilutions were prepared in 10 mM Tris-HCl (pH 8.0) containing 0.1 μg/μl poly(A) as a carrier. Different amounts of plasmids containing different HPV genotype genomes were mixed and tested in relative amounts ranging from 1:1 to 106:1. Each mixture was subjected to amplification by type-specific PCR for the minority species, as well as broad-spectrum SPF10 PCR followed by DNA enzyme immunoassay (DEIA) and LiPA.
DNA isolation and quality control.
Total DNA was isolated from 200 μl of a PreservCyt (Cytyc Corporation, Boxborough, MA) suspension containing the cervical cells with a MagNA Pure LC instrument (Roche Diagnostics, Almere, The Netherlands) and a total DNA isolation kit (Roche Diagnostics). DNA was eluted in 100 μl of water, and 10 μl was used for each PCR.
To determine the presence of PCR inhibitors, part of the human β-globin gene was amplified by PCR with a specific primer set, as described previously (27). In addition, each run contained positive and negative controls to monitor the DNA isolation, PCR, HPV detection, and genotyping procedures.
PCR testing.
The SPF10 PCR primer set was used to amplify a broad spectrum of HPV genotypes, as described earlier (19, 20). Briefly, this primer set amplifies a small fragment of 65 bp from the L1 region of HPV. Reverse primers contain a biotin label at the 5′ end, which enables capture of the reverse strand onto streptavidin-coated microtiter plates. The captured amplimers are denatured by alkaline treatment, and the captured strand is detected by a defined cocktail of digoxigenin-labeled probes that detect a broad spectrum of HPV genotypes. This method is designated the HPV DEIA, which provides an optical density value. If the SPF10 PCR-DEIA yielded a borderline value (75 to 100% of the cutoff value), the SFP10 PCR was repeated and the amplimer was retested by DEIA.
The same SPF10 amplimers can be used to identify the HPV genotype by reverse hybridization by an LiPA containing probes for 25 different HPV genotypes (HPV genotypes 6, 11, 16, 18, 31, 33, 34, 35, 39, 40, 42, 43, 44, 45, 51, 52, 53, 54, 56, 58, 59, 66, 68/73, 70, and 74; SPF10 HPV LiPA version 1; Labo Bio-medical Products, Rijswijk, The Netherlands). In cases in which the DEIA yielded a positive result but the HPV genotype was not identified by LiPA, the SPF10 amplimer was analyzed by direct sequence analysis with the BigDye Terminator cycle sequencing kit (Applied Biosystems, Nieuwerkerk a/d Ijssel, The Netherlands). The sequences were used as a query for screening of the sequences in the GenBank database (www.ncbi.nlm.nih.gov) with BLAST software (1). HPV genotypes were assigned when a complete match between the 22-bp interprimer region and an HPV sequence in GenBank was found.
TS PCR primer sets were used to selectively amplify HPV-16 (TS16) and HPV-18 (TS18). The primers were based on those described by Baay et al. (2) and generate amplimers of 92 and 126 bp for HPV-16 and HPV-18, respectively. Amplimers from the TS PCRs were detected by DEIA, similar to the method for β-globin and SPF10 amplimer detection.
Clinical materials.
A total of 1,113 young women (age range, 15 to 25 years) were enrolled to participate in a phase II study in Brazil, the United States, and Canada (15). The aim of the study was to investigate the safety and efficacy of a bivalent candidate vaccine against HPV-16 and HPV-18.
Health care providers obtained cervical specimens with a cervical brush and spatula (washed in PreservCyt [Cytyc Corporation]) for cytology and HPV DNA testing at screening and month 0 and subsequently every 6 months. At months 0 and 6 and subsequently every 3 months (up to 27 months), women self-obtained cervicovaginal samples with two sequential swabs (placed in PreservCyt) for HPV DNA testing (15).
The two sampling methodologies yielded a total number of 15,546 cervical cell samples. Two aliquots of cervical cell samples (1 ml each) were prepared from the total volume (20 ml) of the PreservCyt suspension containing the cervical cells. In addition, 650 negative controls (PreservCyt medium only) were prepared during the PCR aliquoting procedure.
Furthermore, we assessed the impact of using the testing algorithm by analyzing the combined data for the series of follow-up samples at various time points from each individual woman. The aim was to determine whether the earliest positive sample in a follow-up series was detected only by the SPF10 LiPA or only by the TS PCR, or by both methods, at the same time point.
RESULTS
Competition between genotypes.
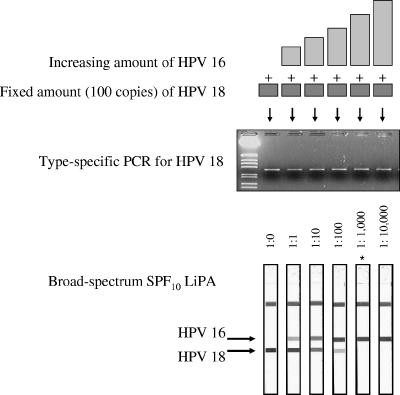
The effect of competition between different genotypes in a single clinical sample was experimentally tested with plasmid mixtures containing relative molar amounts of two HPV genotypes ranging from 1:1 to 106:1. The results are shown in Fig. 1 and Table 1. The type-specific PCR for HPV-18 was positive for all samples, irrespective of the HPV-16 plasmid concentration. The SPF10 LiPA showed both HPV-16 and HPV-18 in those samples, which contained up to a 100-fold molar excess of the HPV-16 plasmid. However, when the concentration of HPV-16 became more than 1,000-fold higher than the HPV-18 concentration, only HPV-16 was identified by the LiPA and a signal for HPV-18 was no longer visible. When a fixed amount of HPV-16 was mixed with increasing amounts of HPV-18, similar results were obtained, with a detection limit of 1:1,000. Mixtures of HPV-18 and HPV-11 and mixtures of HPV-16 and HPV-44 were also tested (Table 1), revealing the same results. Broad-spectrum PCR may underestimate the presence of minority species if the relative concentration of that minority species becomes too low. TS PCR also detects low concentrations of a minority species in a mixture of genotypes.
FIG. 1.
Competition between HPV genotypes. The top part is a graphical representation of the mixtures of the HPV-16 and HPV-18 plasmids prepared in this experiment. The middle part shows the agarose gel electrophoresis results of the type-specific HPV-18 PCR. The bottom part shows the results of SPF10 LiPA. The positions of the probes for types 16 and 18 are indicated (note that only the top part of the LiPA strips which contains the relevant probes is shown). The strip marked with as asterisk (1:1,000) showed a very weak reactivity with the HPV-18 probe line, which is not visible on the scans.
TABLE 1.
Detection of HPV genotypes in mixtures containing different ratios of input DNA from two HPV genotypes
| HPV type in sample | Genotype ratio (no. of genome copies/PCR mixture) | PCR-DEIA result
|
HPV type(s) by SPF10 LiPA | ||
|---|---|---|---|---|---|
| SPF10 | TS16 | TS18 | |||
| 11 | 100 | + | NDa | - | 11 |
| 11 and 18 | 100:100 | + | ND | + | 11 + 18 |
| 11 and 18 | 1,000:100 | + | ND | + | 11 + 18 |
| 11 and 18 | 10,000:100 | + | ND | + | 11 + 18 |
| 11 and 18 | 100,000:100 | + | ND | + | 11 |
| 11 and 18 | 1,000,000:100 | + | ND | + | 11 |
| 11 and 18 | 10,000,000:100 | + | ND | + | 11 |
| 44 | 100 | + | - | ND | 44 |
| 44 and 16 | 100:100 | + | + | ND | 16 + 44 |
| 44 and 16 | 1,000:100 | + | + | ND | 16 + 44 |
| 44 and 16 | 10,000:100 | + | + | ND | 44 |
| 44 and 16 | 100,000:100 | + | + | ND | 44 |
| 44 and 16 | 1,000,000:100 | + | + | ND | 44 |
| 44 and 16 | 10,000,000:100 | + | + | ND | 44 |
| 16 | 100 | + | + | - | 16 |
| 18 and 16 | 100:100 | + | + | + | 16 + 18 |
| 18 and 16 | 1,000:100 | + | + | + | 16 + 18 |
| 18 and 16 | 10,000:100 | + | + | + | 16 + 18 |
| 18 and 16 | 100,000:100 | + | + | + | 18 |
| 18 and 16 | 1,000,000:100 | + | + | + | 18 |
| 18 and 16 | 10,000,000:100 | + | + | + | 18 |
| 18 | 100 | + | - | + | 18 |
| 16 and 18 | 100:100 | + | + | + | 16 + 18 |
| 16 and 18 | 1,000:100 | + | + | + | 16 + 18 |
| 16 and 18 | 10,000:100 | + | + | + | 16 + 18 |
| 16 and 18 | 100,000:100 | + | + | + | 16 + 18 |
| 16 and 18 | 1,000,000:100 | + | + | + | 16 |
| 16 and 18 | 10,000,000:100 | + | + | + | 16 |
ND, not determined.
Clinical samples evaluated by HPV testing algorithm.
Since competition between different genotypes in a single sample clearly plays an important role, a group of 15,546 cervical cell samples from 1,113 women participating in a phase II HPV-16/18 vaccine trial was evaluated for the presence of HPV-16 and HPV-18.
All samples were tested by β-globin PCR and SPF10 PCR. The β-globin PCR yielded negative results for only 12 samples (0.08%). Of the total number of 15,546 samples tested, 4,577 (29.4%) were SPF10 PCR-DEIA positive; less than 1% of the samples yielded borderline DEIA results.
A subset of β-globin PCR- and SFP10 PCR-positive samples (n = 3,138) were also tested by TS PCRs for HPV-16 and HPV-18. The results for HPV-16 are shown in Table 2. Overall, the concordance between the results of SPF10 LiPA and those of the type-specific PCR was high (kappa = 0.775 [95% confidence interval = 0.740 to 0.810]) for the detection of HPV-16. However, there were also samples in which HPV-16 was detected by SPF10 LiPA alone (n = 47) or by TS PCR alone (n = 72). Of the 47 samples in which HPV-16 was detected by SPF10 LiPA alone, 19 (40%) contained multiple genotypes; and of the 72 samples in which HPV-16 was detected by TS PCR alone, 70 (97%) contained multiple genotypes. These proportions of multiple genotypes (40% versus 97%) are significantly different (P < 0.001).
TABLE 2.
Detection of HPV-16 by SPF10 LiPA and type-specific PCR
| TS PCR result for HPV-16 | No. of samples with the following LiPA result for HPV-16:
|
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 232 | 72 | 306 |
| Negative | 47 | 2,787 | 2,837 |
| Total | 279 | 2,859 | 3,138 |
The combined results for HPV-18 are shown in Table 3. There was a good agreement between the rate of detection by SPF10 LiPA and that by the type-specific PCR (kappa = 0.785 [95% confidence interval = 0.749 to 0.819]). However, there were also samples in which HPV-18 was detected only by SPF10 LiPA (n = 38) or only by TS PCR (n = 28). These cases were further analyzed. Of the 38 samples positive for HPV-18 by SPF10 LiPA but negative by TS PCR, 12 (32%) samples contained multiple genotypes. In contrast, of the 28 samples negative for HPV-18 by SPF10 LiPA but positive by TS PCR, 27 (96%) samples contained multiple genotypes. These proportions of multiple genotypes (32% versus 96%) are significantly different (P < 0.001).
TABLE 3.
Detection of HPV-18 by SPF10 LiPA and type-specific PCR
| TS PCR result for HPV-18 | No. of samples with the following LiPA result for HPV-18:
|
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 108 | 28 | 136 |
| Negative | 38 | 2,964 | 3,007 |
| Total | 146 | 2,992 | 3,138 |
Of the 4,577 SPF10 PCR-positive samples, 1,003 (21.9%) samples remained LiPA negative, and 857 of these were tested by TS16 and TS18. For TS16, eight samples were positive, and of these, five samples showed other HPV genotypes by sequencing and no probes were present on the LiPA for these genotypes. The remaining three samples could not be sequenced due to the low yield of the PCR product.
Similarly, four samples were positive by TS18, and of these, three samples revealed other HPV (non-LiPA) genotypes and one sample could not be sequenced.
Of the SFP10 PCR-DEIA-positive but LiPA-negative samples, the sequence of the SPF10 amplimer was determined in 477 cases. The sequences were compared with those in the GenBank database and revealed complete sequence matches with the following HPV genotypes: HPV type 3, 7, 14, 20, 26, 30, 32, 55, 61, 62, 67, 69, 72, 75, 76, 82, 83, 84, 86, 87, 89, 90, and 91. No probes for any of these genotypes are present in the current version of the LiPA. Genotypes 61, 67, 84, and 89 were the most common types in this group. Some SPF10 PCR-positive but LiPA-negative samples could not be analyzed by sequence analysis, due to the very low SPF10 amplimer yield. In several cases, no complete match (one to four mismatches) between the 22-bp interprimer SFP10 amplimer sequence and any HPV sequence in the GenBank database was found and no HPV genotype could be assigned. These sequences likely represent undefined variants of HPV genotypes, but further analysis of other parts of the viral genome would be required to confirm this hypothesis.
Finally, over 1,000 SPF10 DEIA-negative samples were also tested by the TS16 and the TS18 PCRs and did not yield any additional HPV-positive samples.
Development of HPV testing algorithm.
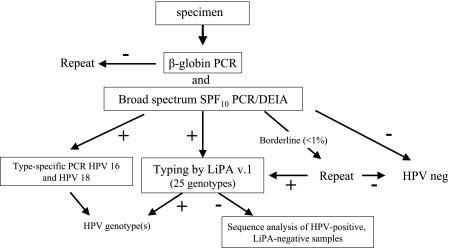
On the basis of these results, an HPV testing algorithm is proposed, as shown in Fig. 2, in which broad-spectrum PCR was combined with TS PCR for HPV-16 and HPV-18.
FIG. 2.
Novel HPV testing algorithm. DNA was isolated from cervical scrapes by use of the MagNA Pure LC instrument and subjected to both β-globin PCR and SPF10 PCR. The PCR products are analyzed by a DNA enzyme immunoassay with specific probes. If the β-globin PCR-DEIA yielded a negative result, the PCR was repeated. If the SPF10 PCR-DEIA yielded a negative result, the sample was considered HPV negative. Samples with borderline DEIA results were retested. Amplimers from SPF10 PCR-DEIA-positive samples were tested by LiPA (version 1.0, which contains probes for 25 genotypes) to determine the HPV genotype. If the LiPA yielded a negative result, SPF10 amplimers were sequenced. All samples HPV positive by SPF10 PCR-DEIA were also tested by TS16 and TS18 PCRs.
DNA from each sample is analyzed by β-globin PCR and SPF10 PCR. When the β-globin PCR is negative by DEIA, the PCR is repeated (except for the 650 processing control samples). When the SPF10 PCR is negative by DEIA, the sample is considered HPV negative (provided that the β-globin PCR of the sample yields a positive result). If the SPF10 PCR yields a DEIA borderline result (optical density value 75 to 100% of the cutoff value), the SFP10 PCR is repeated. If a sample repeatedly yields a borderline result, it is considered HPV negative, but this was an extremely rare event. If a samples remains β-globin PCR negative, it is considered either to not contain any human cells or to be unsuitable for PCR due to inhibition.
SPF10 PCR-positive samples were tested by LiPA to determine the HPV genotype. If the LiPA yields a negative result, the SPF10 amplimers are sequenced. All samples HPV positive by SPF10 PCR-DEIA are also tested by the TS16 PCR and the TS18 PCR.
Retrospective analyses of clinical samples at various collection time points.
The results for samples from 1,040 women for all study visits, comprising a total of 11,323 samples (average, 10.9 samples per woman), were analyzed. Overall, among these 1,040 women, 637 (61.3%) were found to be HPV positive by SPF10 PCR-DEIA at one or more time points, and 145 (22.7%) of these 637 women were positive for HPV-16 and/or HPV-18 at one or more time points.
HPV-16 was found in 90 women, and the rates of detection by SPF10 LiPA and TS PCR were further analyzed, as shown in Table 4. During follow-up, the first HPV-16-positive sample was detected by SPF10 LiPA in 18 (20%) of the women and by TS PCR in 26 (29%) of the women, whereas in 46 (51%) of the women detection was simultaneous by both methods.
TABLE 4.
Detection of HPV-16 and HPV-18 during follow-up analysis by SPF10 LiPA and TS PCR
| HPV type and characteristic | No. (%) of samples
|
||
|---|---|---|---|
| Total | Positive at a single time point | Positive at multiple time points | |
| HPV-16 (n = 90) | |||
| Earliest detection by SPF10 LiPA | 18 (20) | 13 | 5 |
| Earliest detection by TS PCR | 26 (29) | 11 | 15 |
| Earliest detection simultaneously by SPF10 LiPA and TS PCR | 46 (51) | 13 | 33 |
| HPV-18 (n = 61) | |||
| Earliest detection by SFP10 LiPA | 23 (38) | 15 | 8 |
| Earliest detection by TS PCR | 11 (18) | 6 | 5 |
| Earliest detection simultaneously by SPF10 LiPA and TS PCR | 27 (44) | 6 | 21 |
Similarly, HPV-18 was found in 61 women (Table 4). The earliest HPV-18-positive sample was detected by SPF10 LiPA in 23 (38%) women and by TS PCR in 11 (18%) women, whereas in 27 (44%) of the women detection was simultaneous.
Of the women for whom the earliest detection of HPV-16 or HPV-18 was simultaneous by SPF10 LiPA and TS PCR, the majority showed HPV-16 or HPV-18 positivity at multiple time points during follow-up. In contrast, of the women for whom the earliest detection of HPV-16 or HPV-18 was by either SPF10 PCR or TS PCR, the majority were positive for HPV-16 or HPV-18 at only a single time point (Table 4).
DISCUSSION
Detection and identification of HPV genotypes strongly depend on the accuracy and the precision of the methods used. As vaccine trials include surrogate virological end points (incident and persistent infections) to assess the efficacies of candidate prophylactic HPV vaccines, it is crucial that highly reliable and robust testing methods be used (15).
The present study investigated a novel testing algorithm that comprised a combination of broad-spectrum and type-specific PCRs. The results clearly showed that broad-spectrum PCR with a mixture of defined primers followed by reverse hybridization is a very useful tool for the identification of HPV genotypes in clinical samples but may underestimate the prevalence of multiple genotypes compared to the prevalence obtained by type-specific PCR. If two HPV genotypes are present but one genotype is present in great molar excess over the other, it is likely that the minor genotype will be not be detected during broad-spectrum PCR and will remain unidentified. This is clearly illustrated by the results obtained with the plasmid mixtures. When the concentration of the HPV-16 plasmid exceeded a low concentration (100 copies) of the HPV-18 plasmid more than 100-fold, HPV-18 was no longer detected by broad-spectrum PCR, whereas it was still detected by the type-specific PCR. These experiments were not repeated in sufficient replicates to calculate an accurate detection limit for each type in the mixture.
The analytical sensitivity of a broad-spectrum PCR for the detection of multiple genotypes will be influenced by the genotypes present. Since different HPV genotypes contain different nucleotide sequences at the primer target regions, each genotype is preferentially amplified by a subset of PCR primers from the available broad-spectrum primer pool. Thus, the amplification efficacy of every broad-spectrum PCR will be type dependent to some degree (6, 32). It is noteworthy that this competition phenomenon is observed only in samples that contain multiple HPV genotypes by the LiPA detection system. In samples containing only a single HPV genotype, it is adequately detected by the general PCR primer as well as the type-specific primer sets (32). More than 1,000 SPF10 PCR-DEIA-negative samples were also tested by TS PCR for HPV-16 and HPV-18 and did not yield any additional positive samples (data not shown). This indicates that the SFP10 PCR has a very high sensitivity for the detection of HPV DNA in general and can be effectively used to screen for HPV positivity. These data again confirm that the SPF10 PCR-DEIA system has a very high level of specificity, and false-positive signals are not observed when only human DNA is used as the target (19, 20).
As described here for the SPF10 primer set, the efficacy of any general HPV primer set (such as My09/My11, PGMY, and GP5+/GP6+) will depend on the intrinsic degree of type specificity of the primer set and the competition between multiple genotypes in a complex mixture in the same sample (13, 26). It has been reported earlier that there are significant differences between the My09/My11 (degenerate primers) and the PGMY (a mixture of defined primers) PCR primer sets, although they target exactly the same region of the HPV genome (6). Similarly, a comparison between SPF10 LiPA and PGMY PCR-line blot assay showed a very high degree of concordance, but for some genotypes, the two systems showed differences (32). Therefore, if one is interested only in the detection of a particular subset of HPV genotypes, type-specific PCRs may be more suitable than a broad-spectrum general PCR. However, this requires individual or multiplex testing for each HPV genotype, which is not suitable for routine diagnostic applications and which limits the information obtained for each sample.
Post-PCR detection methods also may yield different detection rates of multiple HPV genotypes (30). Analysis of broad-spectrum PCR products by direct sequencing lacks sensitivity and specificity for the analysis of complex mixtures of multiple genotypes and identifies only the most predominant types. An individual genotype should constitute at least 20% of the mixture to permit adequate identification by direct sequencing. In contrast, reverse hybridization (such as LiPA) is much more sensitive and allows the identification of minority genotypes, even if their DNA represents less than 1% of the total amount of HPV DNA, as shown by the results of the plasmid mixture experiment.
The specificity of the LiPA genotyping method was further assessed by sequencing SPF10 amplimers from samples that remained negative by the LiPA. A probe was not present in the LiPA for any of the HPV genotypes identified by sequence analysis in this group, confirming the high specificity of this reverse hybridization assay. Combining the genotyping data of the LiPA and sequencing showed a total of more than 50 HPV genotypes, illustrating the truly broad-spectrum amplification by SPF10 primers. Since multiple samples were obtained from the same women over time, the epidemiological information regarding genotype distribution based on these selected analyses is limited.
Within the SPF10 system, the same PCR amplimer is used for both detection of HPV positivity by DEIA and subsequent genotyping by LiPA. The DEIA uses a cocktail of probes that recognize more than 50 different HPV genotypes, whereas the LiPA identifies only the 25 genotypes for which probes are present on the strip. Therefore, if the DEIA were omitted, the overall sensitivity of HPV detection would decrease substantially. Also, use of a reverse hybridization strip assay as a screening method is not economically efficient. Moreover, if a different PCR were used to screen for HPV positivity, reamplification of these HPV-positive samples would be required, which could lead to discrepant results between the two PCRs. Therefore, it is important to use only a single, well-characterized PCR for the initial detection of HPV as well as for subsequent genotyping.
Another important aspect of molecular diagnosis is the effect of sampling variation due to sample heterogeneity. Sampling variation is particularly relevant when nonhomogeneous clinical materials, such as cervical cell suspensions or cervical biopsy specimens, are used (11, 14, 16). Also, sampling variation may become relevant when only a small volume of the extracted nucleic acid is included in the actual PCR vial, and consequently, the test outcome may not properly represent the true status of the sample. Especially when samples contain low viral loads, sampling variation may lead to false-negative results.
To evaluate the effect of the algorithm testing, samples from a phase II HPV-16 and -18 bivalent candidate HPV-16/HPV-18 vaccine trial were analyzed. The cervical cells had been resuspended in PreservCyt medium for transport and long-term storage. The proportion of cell suspensions that yielded negative β-globin PCR results was very low (<0.1%), indicating that inhibition is a very rare event and confirming the usefulness of the PreservCyt medium for PCR-based analyses.
The comparative results of the SPF10 and the type-specific PCRs clearly confirmed the added value of the combined testing approach. The discrepant results between the SPF10 LiPA and the type-specific PCR could be explained by a combination of the competition effect and the sample heterogeneity effect. Virtually all type-specific PCR-positive but LiPA-negative samples contained multiple genotypes, whereas the proportion of multiple-genotype infections was significantly lower among the type-specific PCR-negative but LiPA-positive samples. For the latter group, sample heterogeneity played a major role. This was further substantiated by quantitative PCR analysis of a subset of the discrepant samples (data not shown). In general, samples with discrepant results (LiPA positive and TS PCR negative or vice versa) contained lower viral loads than samples with concordant positive results (LiPA positive and TS PCR positive), which is consistent with sampling heterogeneity effects.
The benefits of the novel HPV testing algorithm was evaluated with samples from a group of women participating in a trial of a candidate HPV-16/HPV-18 vaccine and clearly revealed that the use of the algorithm during the monitoring of individual women resulted in the earlier and more accurate detection of HPV-16 and/or HPV-18 than when either SPF10 LiPA or TS PCR alone was used. The earliest detection depends on several factors, such as the viral load and the presence of other HPV genotypes.
A higher number of different HPV genotypes was detected in women for whom the TS PCR detected HPV-16 and/or HPV-18 earlier than SPF10 LiPA. This would be concordant with the hypothesis that detection by SPF10 LiPA is influenced by the competition effect. The more genotypes that are present in a woman, the higher the chance that HPV-16 and/or HPV-18 is missed by SPF10 LiPA due to a competition effect.
In the majority of women for whom detection by SPF10 LiPA and TS PCR was simultaneous, HPV-16 and/or HPV-18 was detected at multiple time points. This is compatible with the hypothesis that women with HPV-16 and/or HPV-18 positivity at multiple time points contain higher viral loads, resulting in detection by both SPF10 LiPA and TS PCR. Thus, the testing algorithm will have the biggest impact in detecting incident infections, in which HPV is only transiently detectable at lower viral loads. During persistent infections, HPV is generally present at higher viral loads and is therefore readily detectable by either SPF10 LiPA or TS PCR or by both methods (9, 28, 36).
In conclusion, the present study has provided a reliable and highly effective testing algorithm for the molecular diagnosis of HPV infections. This algorithm yields superior results compared to those obtained by the use of any single PCR test. Therefore, the use of the algorithm can substantially contribute to the accuracy of analysis in vaccination trials and epidemiological studies.
Acknowledgments
GlaxoSmithKline Biologicals, Rixensart, Belgium, funded and coordinated this clinical trial (trial 580299/001).
The laboratory contribution from Quest Diagnostics Inc., Teterboro, NJ, is appreciated. S. L. Wieting contributed to technical writing aspects.
HPV Vaccine Study Group received clinical study support from Anne Schuind, GSK Biologicals, King of Prussia, PA. The principal investigators and coinvestigators of the HPV Vaccine Study Group are P. Colares de Borba, P. Naud, C. M. Roteli-Martins, N. S. De Carvalho and J. C. Teixeira in Brazil (506 patients); F. Aoki B. Ramjattan, B. Romanowski, R. M. Shier, and R. Somani in Canada (45 patients); and J. Adelglass, S. Barbier, M. M. Blatter, C. Chambers, D. G. Ferris, S. A. Gall, M. Gerardi, F. Guerra, D. M. Harper, J. Hedrick, D. Henry, W. Hood, A. P. Korn, A. B. Moscicki, B. Sullivan, C. Thoming, L. B. Twiggs, S. K. Tyring, R. Watson, C. Wheeler, and M. Cabeszas-Mijuste in the United States (562 patients).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baay, M. F., W. G. V. Quint, J. Koudstaal, H. Hollema, J. M. Duk, M. P. Burger, E. Stolz, and P. Herbrink. 1996. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J. Clin. Microbiol. 34:745-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozzetti, M., B. Nonnenmacher, I. Mielzinska, L. Villa, A. Lorincz, V. Breitenbach, and J. Prolla. 2000. Comparison between Hybrid Capture II and polymerase chain reaction results among women at low risk for cervical cancer. Ann. Epidemiol. 10:466. [DOI] [PubMed] [Google Scholar]
- 4.Chan, S. Y., H. Delius, A. L. Halpern, and H. U. Bernard. 1995. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J. Virol. 69:3074-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cogliano, V., R. Baan, K. Straif, Y. Grosse, B. Secretan, and G. F. El. 2005. Carcinogenicity of human papillomaviruses. Lancet Oncol. 6:204. [DOI] [PubMed] [Google Scholar]
- 6.Coutlee, F., P. Gravitt, J. Kornegay, C. Hankins, H. Richardson, N. Lapointe, H. Voyer, and E. Franco. 2002. Use of PGMY primers in L1 consensus PCR improves detection of human papillomavirus DNA in genital samples. J. Clin. Microbiol. 40:902-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox, J. T., A. T. Lorincz, M. H. Schiffman, M. E. Sherman, A. Cullen, and R. J. Kurman. 1995. Human papillomavirus testing by hybrid capture appears to be useful in triaging women with a cytologic diagnosis of atypical squamous cells of undetermined significance. Am. J. Obstet. Gynecol. 172:946-954. [DOI] [PubMed] [Google Scholar]
- 8.Cuzick, J., P. Sasieni, P. Davies, J. Adams, C. Normand, A. Frater, M. van Ballegooijen, and E. van den Akker. 1999. A systematic review of the role of human papillomavirus testing within a cervical screening programme. Health Technol. Assess. 3:1-201. [PubMed] [Google Scholar]
- 9.Dalstein, V., D. Riethmuller, J. L. Pretet, C. K. Le Bail, J. L. Sautiere, J. P. Carbillet, B. Kantelip, J. P. Schaal, and C. Mougin. 2003. Persistence and load of high-risk HPV are predictors for development of high-grade cervical lesions: a longitudinal French cohort study. Int. J. Cancer 106:396-403. [DOI] [PubMed] [Google Scholar]
- 10.de Roda Husman, A. M., J. M. Walboomers, A. J. van den Brule, C. J. Meijer, and P. J. Snijders. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 76:1057-1062. [DOI] [PubMed] [Google Scholar]
- 11.Gravitt, P. E., J. V. Lacey, L. A. Brinton, W. A. Barnes, J. R. Kornegay, M. D. Greenberg, S. M. Greene, O. C. Hadjimichael, L. McGowan, R. Mortel, P. E. Schwartz, R. Zaino, and A. Hildesheim. 2001. Evaluation of self-collected cervicovaginal cell samples for human papillomavirus testing by polymerase chain reaction. Cancer Epidemiol. Biomarkers Prev. 10:95-100. [PubMed] [Google Scholar]
- 12.Gravitt, P. E., and M. M. Manos. 1992. Polymerase chain reaction-based methods for the detection of human papillomavirus DNA. IARC Sci. Publ. 119:121-133. [PubMed]
- 13.Gravitt, P. E., C. L. Peyton, T. Q. Alessi, C. M. Wheeler, F. Coutlée, A. Hildesheim, M. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gravitt, P. E., C. L. Peyton, R. J. Apple, and C. M. Wheeler. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 36:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper, D. M., E. L. Franco, C. Wheeler, D. G. Ferris, D. Jenkins, A. Schuind, T. Zahaf, B. Innis, P. Naud, N. S. De Carvalho, C. M. Roteli-Martins, J. Teixeira, M. M. Blatter, A. P. Korn, W. Quint, and G. Dubin. 2004. Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human papillomavirus types 16 and 18 in young women: a randomised controlled trial. Lancet 364:1757-1765. [DOI] [PubMed] [Google Scholar]
- 16.Heinmoller, E., B. Renke, K. Beyser, W. Dietmaier, C. Langner, and J. Ruschoff. 2001. Piffalls in diagnostic molecular pathology—significance of sampling error. Virchows Arch. 439:504-511. [DOI] [PubMed] [Google Scholar]
- 17.Hildesheim, A., M. H. Schiffman, P. E. Gravitt, A. G. Glass, C. E. Greer, T. Zhang, D. R. Scott, B. B. Rush, P. Lawler, M. E. Sherman, R. J. Kurman, and M. M. Manos. 1994. Persistence of type-specific human papillomavirus infection among cytologically normal women. J. Infect. Dis. 169:235-240. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, M. V., P. J. Snijders, A. J. van den Brule, T. J. Helmerhorst, C. J. Meijer, and J. M. Walboomers. 1997. A general primer GP5+/GP6+-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleter, B., L. J. van Doorn, L. Schrauwen, A. Molijn, S. Sastrowijoto, J. ter Schegget, J. Lindeman, B. ter Harmsel, and W. G. V. Quint. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 37:2508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleter, B., L. J. van Doorn, J. ter Schegget, L. Schrauwen, C. van Krimpen, M. P. Burger, B. ter Harmsel, and W. G. V. Quint. 1998. A novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am. J. Pathol. 153:1731-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutsky, L. A., K. A. Ault, C. M. Wheeler, D. R. Brown, E. Barr, F. B. Alvarez, L. M. Chiacchierini, and K. U. Jansen. 2002. A controlled trial of a human papillomavirus type 16 vaccine. N. Engl. J. Med. 347:1645-1651. [DOI] [PubMed] [Google Scholar]
- 22.Levi, J. E., B. Kleter, W. G. Quint, M. C. Fink, C. L. Canto, R. Matsubara, I. Linhares, A. Segurado, B. Vandenborght, J. E. Neto, and L. J. van Doorn. 2002. High prevalence of human papillomavirus infections and high frequency of multiple genotypes in HIV-infected women in Brazil. J. Clin. Microbiol. 40:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munoz, N., F. X. Bosch, S. De Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 24.Parkin, D. M., F. Bray, J. Ferlay, and P. Pisani. 2005. Global cancer statistics, 2002. CA Cancer J. Clinicians 55:74-108. [DOI] [PubMed] [Google Scholar]
- 25.Perrons, C., R. Jelley, B. Kleter, W. Quint, and N. Brink. 2005. Detection of persistent high risk human papillomavirus infections with hybrid capture II and SPF10/LiPA. J. Clin. Virol. 32:278-285. [DOI] [PubMed] [Google Scholar]
- 26.Qu, W., G. Jiang, Y. Cruz, C. J. Chang, G. Y. Ho, R. S. Klein, and R. D. Burk. 1997. PCR detection of human papillomavirus: comparison between MY09/MY11 and GP5+/GP6+ primer systems. J. Clin. Microbiol. 35:1304-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 28.Schlecht, N. F., A. Trevisan, E. Duarte-Franco, T. E. Rohan, A. Ferenczy, L. L. Villa, and E. L. Franco. 2003. Viral load as a predictor of the risk of cervical intraepithelial neoplasia. Int. J. Cancer 103:519-524. [DOI] [PubMed] [Google Scholar]
- 29.Snijders, P. J., A. J. van den Brule, and C. J. Meijer. 2003. The clinical relevance of human papillomavirus testing: relationship between analytical and clinical sensitivity. J. Pathol. 201:1-6. [DOI] [PubMed] [Google Scholar]
- 30.van den Brule, A. J., R. Pol, N. Fransen-Daalmeijer, L. M. Schouls, C. J. Meijer, and P. J. Snijders. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40:779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Doorn, L. J., B. Kleter, and W. G. Quint. 2001. Molecular detection and genotyping of human papillomavirus. Expert Rev. Mol. Diagn. 1:394-402. [DOI] [PubMed] [Google Scholar]
- 32.van Doorn, L. J., W. G. V. Quint, B. Kleter, A. C. Molijn, B. Colau, M. T. Martin, Kravang-In, N. Torrez-Martinez, C. L. Peyton, and C. M. Wheeler. 2002. Genotyping of human papillomavirus by the PGMY-line blot assay and the SPF10 line probe assay in liquid cytology cervical specimens. J. Clin. Microbiol. 40:979-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villa, L. L., R. L. Costa, C. A. Petta, R. P. Andrade, K. A. Ault, A. R. Giuliano, C. M. Wheeler, L. A. Koutsky, C. Malm, M. Lehtinen, F. E. Skjeldestad, S. E. Olsson, M. Steinwall, D. R. Brown, R. J. Kurman, B. M. Ronnett, M. H. Stoler, A. Ferenczy, D. M. Harper, G. M. Tamms, J. Yu, L. Lupinacci, R. Railkar, F. J. Taddeo, K. U. Jansen, M. T. Esser, H. L. Sings, A. J. Saah, and E. Barr. 2005. Prophylactic quadrivalent human papillomavirus (types 6, 11, 16, and 18) L1 virus-like particle vaccine in young women: a randomised double-blind placebo-controlled multicentre phase II efficacy trial. Lancet Oncol. 6:271-278. [DOI] [PubMed] [Google Scholar]
- 34.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 35.Woodman, C. B., S. Collins, H. Winter, A. Bailey, J. Ellis, P. Prior, M. Yates, T. P. Rollason, and L. S. Young. 2001. Natural history of cervical human papillomavirus infection in young women: a longitudinal cohort study. Lancet 357:1831-1836. [DOI] [PubMed] [Google Scholar]
- 36.Ylitalo, N., P. Sorensen, A. M. Josefsson, P. K. Magnusson, P. K. Andersen, J. Ponten, H. O. Adami, U. B. Gyllensten, and M. Melbye. 2000. Consistent high viral load of human papillomavirus 16 and risk of cervical carcinoma in situ: a nested case-control study. Lancet 355:2194-2198. [DOI] [PubMed] [Google Scholar]
- 37.zur Hausen, H., and O. De Barahona. 1994. Human papillomaviruses. Annu. Rev. Microbiol. 48:427-447. [DOI] [PubMed] [Google Scholar]