Abstract
Bovine coronavirus (BCoV) is an etiological agent associated with winter dysentery (WD), prevalent in adult cattle during the winter. Although we previously detected, isolated, and characterized BCoV strains from adult cattle with WD (WD-BCoV strains) during the winter in South Korea, the precise epidemiology, as well as the causative agent of diarrhea in adult cattle in the warmer seasons, has not been examined. We examined 184 diarrheic fecal specimens collected from 75 herds of adult cattle from seven provinces during the spring (warm), autumn (warm), and summer (hot) seasons. Bovine coronavirus-positive reactions were detected for 107 (58.2%) diarrheic fecal samples (in 47/75 herds). Of these 107 positive samples, 90 fecal samples from 33 herds tested positive for BCoV alone and 17 fecal samples from 14 herds also tested positive for other pathogens. Biological comparisons between the 9 BCoV strains isolated in this study and the 10 previously isolated WD-BCoV strains revealed that there was no receptor-destroying enzyme (RDE) activity against mouse erythrocytes in the 9 BCoV strains but the 10 WD-BCoV strains had high RDE activity. Phylogenetic analysis of the spike (S) and hemagglutinin/esterase (HE) proteins revealed that all the Korean BCoVs clustered together regardless of season and were distinct from the other known BCoVs, suggesting a distinct evolutionary pathway for the Korean BCoVs. These and previous results revealed a high prevalence and widespread geographical distribution of BCoV, suggesting that this virus is endemic in adult cattle with diarrhea in all seasons in South Korea.
Coronaviruses (CoVs) are a genus in the family Coronaviridae. These viruses are large, enveloped, positive-strand RNA viruses that have become increasingly important causes of human and animal diseases (21). Coronaviruses are divided into three genetically distinct groups (21, 27, 42). Bovine coronavirus (BCoV) belongs to group 2 and contains five major structural proteins: the nucleocapsid (N), transmembrane (M), spike (S), small membrane (E), and hemagglutinin/esterase (HE) (20). Both the S and HE glycoproteins hemagglutinate red blood cells by binding to N-acetyl-9-O-acetylneuraminic acid as a receptor determinant (34, 45) and produce virus-neutralizing antibodies (10, 44). In addition, they are involved in determining the virus tissue and host tropism (38). The HE protein also acts as a receptor-destroying enzyme (RDE), serving as an esterase to reverse hemagglutination (HA) (26, 45).
BCoV causes severe diarrhea in newborn calves (CD), winter dysentery (WD) in adult cattle, and respiratory tract infections in calves and feedlot cattle (7, 8, 22, 29, 39). WD is characterized by a sudden onset of semiliquid, often bloody diarrhea, which rapidly affects many adult cattle and can cause a marked reduction in milk production in dairy herds (32). Winter dysentery has been reported in most cattle-producing countries, including those in Europe, North America, and East Asia (30, 31). Winter dysentery has a peak incidence in winter, presumably because BCoV is moderately sensitive to heat (31). In South Korea, WD-BCoV appears to cause nationwide diarrhea outbreaks in the winter (19). In this study, we identified WD-like diarrhea in adult dairy and beef herds in the warmer seasons in South Korea, resulting in enormous economic losses from a marked reduction in milk production by dairy herds. However, the causative agents and the precise epidemiology of this type of diarrhea have not been determined. In this study, we examined the incidence of BCoV excretion in adult cattle with diarrhea from herds in South Korea by using enzyme-linked immunosorbent assays (ELISA), reverse transcription-PCR (RT-PCR), and nested PCR, and we compared this with the excretion of other enteric pathogens, including bovine rotavirus groups A, B, and C (BRV A to BRV C), bovine torovirus (BToV), bovine enteric Nebraska-like calicivirus (BEC-NBV), bovine norovirus (BNoV), bovine viral diarrhea virus (BVDV), Salmonella spp., Clostridium spp., Campylobacter spp., Mycobacterium paratuberculosis, Coccidium spp., and Cryptosporidium spp. In addition, the previously isolated WD-BCoVs and other known BCoVs were compared biologically and genetically to the BCoV isolates recovered during the warmer seasons in this study.
MATERIALS AND METHODS
Specimens.
A total of 184 fecal specimens from 69 dairy (Holstein) and 6 Korean native beef (Hanwoo) herds of adult cattle with diarrhea were collected from seven provinces in South Korea from 2002 to 2003 during the spring (41 samples/15 herds), summer (43 samples/23 herds), and autumn (100 samples/37 herds). The ages of the cattle tested from all provinces ranged from 1 to 8 years. Upon arrival of the fecal samples, they were examined for common bacterial enteric pathogens, including Salmonella spp., Clostridium spp., and Campylobacter spp., using specific agar media, and the suspect colonies were identified based on biochemical tests. Mycobacterium paratuberculosis was detected by PCR with the specific primers P90 (5′-GAAGGGTGTTCGGGGCCGTC-3′) and P91 (5′-GAGGTCGATCGCCCACGTGAC-3′) (25). Testing for parasite eggs (Coccidium spp. and Cryptosporidium spp.) was done using standard flotation techniques. For virologic assays, fecal suspensions of each sample were prepared by diluting the feces 1:10 in 0.01 M phosphate-buffered saline (PBS), pH 7.2. The suspensions were vortexed for 30 s and then centrifuged (1,200 × g for 20 min), and the supernatants, along with the remaining bulk samples, were collected and stored at −80°C for further testing.
Detection of BCoV antigen by ELISA.
An indirect antigen capture ELISA employing monoclonal antibodies (MAbs) to BCoV as the capture antibodies was used to detect BCoV in the fecal suspensions, as previously described (6, 37). Briefly, 96-well microtiter plates were coated with a mixture of the three MAbs developed against BCoV structural proteins (HE, N, and S proteins) of the CD DB2 strain of BCoV (antibody-positive coating) or with BCoV antibody-negative mouse ascitic fluids (antibody-negative coating). After overnight incubation of wells at 4°C, 5% (wt/vol) nonfat dry milk in a solution of PBS (pH 7.4) and 0.05% Tween 20 was applied as a blocking reagent for 1 h at 20 to 22°C. Fecal suspensions (1:25 dilution) were added to duplicate wells coated with the BCoV capture MAbs or BCoV antibody-negative ascites. Fecal samples from calves positive and negative for BCoV and unknown test samples were added to duplicate wells that had antibody-positive or antibody-negative coatings and were incubated for 1 h at 20°C. After plates were washed with a solution of PBS (pH 7.4) and 0.05% Tween 20, optimally diluted guinea pig anti-BCoV hyperimmune serum was added to each well. Plates were incubated for 1 h at 25°C, and an indicator antibody consisting of optimally diluted sheep anti-guinea pig immunoglobulin G conjugated to horseradish peroxidase was added to each well. The chromogen substrate was 2,2′-azino-di-3-ethylbenzthiazoline sulfonic acid with a final concentration of 0.03% hydrogen peroxide. Plates were read at 414 nm using an ELISA reader, and the absorbances were saved as an ASCII file. A spreadsheet program was used to calculate the ELISA values for the samples by subtracting the average absorbance of the paired wells with BCoV antibody-negative coatings from the average absorbance of the paired wells with BCoV antibody-positive coatings. Samples with absorbances of 0.1 or greater were considered positive for the BCoV antigen.
RNA extraction.
RNA was extracted from 200-μl starting volumes of the centrifuged 10% fecal suspensions by using the Trizol-LS (Gibco-BRL Life Technologies, Grand Island, NY) procedure. The recovered total RNA was suspended in 50 μl of RNase-free water and stored at −80°C.
RT-PCR and nested PCR.
RT-PCR assays with different primer sets (Table 1) for the detection of BCoV, BRV A to BRV C, BEC-NBV, BNoV, BToV, and BVDV were performed using a standard one-step RT-PCR, as described by Cho et al. (6). Briefly, 5 μl of RNA was added to a tube containing 45 μl of the RT-PCR mixture, comprising 5 μl of 10× PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2, 0.01% gelatin), 5 μl of MgCl2 (25 mM), 1 μl of 10 mM deoxynucleoside triphosphates, 1 μl of the upstream primer (50 pmol), 1 μl of the downstream primer (50 pmol), 0.5 μl of avian myeloblastosis virus reverse transcriptase (5.0 U; Promega Corp., Madison, Wis.), 0.5 μl of RNasin RNase inhibitor (10 U; Promega Corp.), 0.5 μl of Taq polymerase (2.5 U; Promega Corp.), and 30.5 μl of water.
TABLE 1.
RT-PCR and nested-PCR primers for detection of target viruses in fecal samples from adult cattle with diarrhea
| Target virus | Target genea | Sequence (5′→3′)b | Region | Source or reference |
|---|---|---|---|---|
| BCoV | N | F, GCAATCCAGTAGTAGAGCGT | 13-712 | Cho et al. (6) |
| R, CTTAGTGGCATCCTTGCCAA | ||||
| nF, GCCGATCAGTCCGACCAATG | 152-551 | |||
| nR, AGAATGTCAGCCGGGGTAG | ||||
| Group A rotavirus | VP7 | F, GCCTTTAAAAGCGAGAATTT | 3-1062 | Chang et al. (5) |
| R, GGTCACATCATACAAYTC TA | ||||
| nF, TTTCTAACATCAACACT | 274-930 | C. Jeong and K. O. Cho, unpublished data | ||
| nR, TTGCCACCATTTTTTCCAAT | ||||
| Group B rotavirus | VP7 | F, GGAAATAATCAGAGATG | 1-795 | Barman et al. (2) |
| R, CTACTCGTTTGGCTCCCTCC | ||||
| Group C rotavirus | VP6 | F, TCAAGAAATGGWATGCAACC | 334-918 | C. Jeong and K. O. Cho, unpublished data |
| R, CATAGCMGCTGGTCTWATCA | ||||
| BToV | M | F, TTCTTACTACACTTTTTGGA | 98-700 | C. Jeong and K. O. Cho, unpublished data |
| R, ACTCAAACTTAACACTAG AC | ||||
| nF, TATGTACTATGTTTCCAGCT | 152-560 | C. Jeong and K. O. Cho, unpublished data | ||
| nR, CCAACACAAATCCGCAACGC | ||||
| BNoV | RdRp | F, AGTTAYTTTTCCTTYTAYGGBGA | 4543-5074 | Smiley et al. (36) |
| R, AGTGTCTCTGTCAGTCATCTTCAT | ||||
| nF, GTCGACGGYCTKGTSTTCCT | 4690-5014 | C. Jeong and K. O. Cho, unpublished data | ||
| nR, CACAGCGACAAATCATGAAA | ||||
| BEC-NBV | RdRp-MCP | F, TTTCTAACYTATGGGGAYGAYG | 4518-5066 | Smiley et al. (36) |
| R, GTCACTCATGTTTCCTTCTCTAAT | ||||
| nF, CGCTCCGTGTGGGATCACGA | 4788-4981 | C. Jeong and K. O. Cho, unpublished data | ||
| nR, GCACGGGCTTCTTCTAGAGA | ||||
| BVDV | Polyprotein | F, ACAAACATGGTTGGTGCAACTGGT | 1424-2244 | Givens et al. (14) |
| R, CAGACATATTTGCCTAGGTTCCA |
RdRp, RNA-dependent RNA polymerase; MCP, major capsid protein.
F, forward primer for RT-PCR; R, reverse primer for RT-PCR; nF, forward primer for nested PCR; nR, reverse primer for nested PCR.
Nested-PCR assays with the primer pairs specific for BCoV, BRV A, BToV, BNoV, and BEC-NBV were performed to increase the sensitivity and specificity of the RT-PCR (Table 1) (6). Briefly, 5 μl of the diluted RT-PCR products (1:100) was added to a tube containing 45 μl of the PCR mixture (final dilution, 1,000 times). If the predicted band was absent by RT-PCR, 5 μl of the undiluted RT-PCR product was subjected to nested PCR. The PCR mixture consisted of 5 μl of 10× buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2, 0.01% gelatin), 5 μl of MgCl2 (25 mM), 1 μl of 10 mM deoxynucleoside triphosphates, 1 μl of the nested-PCR upstream primer (50 pmol), 1 μl of the nested-PCR downstream primer (50 pmol), 0.5 μl of Taq polymerase (Promega Corp.), and 31.5 μl water.
As a negative control, RNA was extracted from normal feces of a mock-infected, colostrum-deprived calf. The amplification products were analyzed by 1.5 or 2% agarose gel electrophoresis and visualized by UV irradiation of the ethidium bromide-stained samples.
BCoV isolation.
Monolayers of human rectal tumor (HRT-18G) cell cultures grown in 6-well plates were used to isolate the virus, as described elsewhere (3, 43). Briefly, the cells were washed with Eagle's minimal essential medium and inoculated into duplicate wells along with the selected filtered (pore size, 0.20 μm) fluids from fecal suspensions that had been found positive for BCoV by RT-PCR, nested PCR, or ELISA. The fluids from the fecal suspensions were adsorbed for 1 h with occasional rocking, and Eagle's minimal essential medium containing pancreatin (5 μg/ml) was added. The cultures were incubated for 3 to 4 days at 37°C under a 5% CO2 atmosphere and were examined daily for any evidence of cytopathic effects (CPE). Isolated BCoVs were cloned by liquid-limiting dilution, and the highest dilution of the virus that caused any CPE was passaged an additional three times in the HRT-18G cells. The isolated BCoV was confirmed by direct immunofluorescence tests (31), ELISA, and RT-PCR, as described above.
HA, RDE activity, and HI tests.
HA tests were conducted using the microtiter method in V-bottom plates, as described elsewhere (33, 42). The HA titers were expressed as the reciprocal of the highest dilution of the virus showing complete HA of 0.4% and 0.2% suspensions of mouse and chicken erythrocytes, respectively, after 1 h of incubation at 4°C or 37°C. The plates incubated at 4°C were then incubated at 37°C for 2 h in order to determine the level of receptor inactivation reflected by the disaggregation of the BCoV-erythrocyte complexes mediated by the RDE activity. The hemagglutination inhibition (HI) test was performed using 96-well U-bottom plates (42). The MAb to the prototype strain, Mebus, was serially diluted 20-fold with veronal-buffered saline and mixed with the same volume of 8 HA units of purified BCoV, followed by incubation at 22°C for 1 h. After incubation, a 1% mouse erythrocyte suspension was added and incubated for a further 2 h at 22°C. The HI titers were expressed as the reciprocal of the highest dilution of the MAb that completely inhibited HA (pellet formation).
DNA sequencing.
The oligonucleotide primers used in the RT-PCR were designed from the published sequences of the S, HE, M, and E genes of strain Mebus (GenBank accession no. U00735). Table 2 shows the primer sequences and predicted product sizes. A one-step RT-PCR assay was performed as described above. The RT-PCR products were purified using a GeneClean II kit (Bio 101, Inc., La Jolla, CA) according to the manufacturer's instructions. DNA sequencing was carried out using an automated DNA sequencer (ABI system 3700; Applied Biosystems Inc., Foster City, CA). By using the DNA Basic module (DNAsis MAX, Alameda, CA), the S, HE, M, and E gene sequences of our BCoV isolates were compared with those of the other known BCoVs. The gene sequences compared are listed in Table 3. The deduced amino acid sequences were then assembled and analyzed using the Amino Acid Basic module (DNAsis MAX, Alameda, CA). A sequence similarity search was performed for the BCoV S, HE, M, and E proteins using the LALIGN Query program of the GENESTREAM network server at the Institut de Génétique Humaine, Montpellier, France (http://www.eng.uiowa.edu/∼tscheetz/sequence-analysis/examples/LALIGN/lalign-guess.html). Phylogenetic and bootstrap (1,000 replicates) analyses based on nucleotide and amino acid alignments were constructed by the neighbor-joining method and the unweighted-pair group method using average linkages of Molecular Evolutionary Genetics Analysis (MEGA, version 3.1) with pairwise distance (20).
TABLE 2.
Oligonucleotide primers designed from the S, HE, M, and E genes of BCoV strain Mebus and used for DNA sequencing
| Gene name | Primer namea | Sequence | Location | Product size (bp) |
|---|---|---|---|---|
| S | S1F | 5′-ATGTTTTTGATACTTTTAATTTCC-3′ | S gene 1-920 | 920 |
| S1R | 5′-ACACCAGTAGATGGTGCTAT-3′ | |||
| S2F | 5′-GGGTTACACCTCTCACTTCT-3′ | S gene 782-1550 | 769 | |
| S2R | 5′-GCAGGACAAGTGCCTATA CC-3′ | |||
| S3F | 5′-CTGTCCGTGTAAATTGGATG-3′ | S gene 1459-2286 | 828 | |
| S3R | 5′-TGTAGAGTA ATCCACACGT-3′ | |||
| S4F | 5′-TTC ACGACAGCTGCAACCTA-3′ | S gene 2151-3022 | 872 | |
| S4R | 5′-CCATGGTAACACCAATCC CA-3′ | |||
| S5F | 5′-CCCTGTATTAGGTTGTTTAG-3′ | S gene 2691-3606 | 916 | |
| S5R | 5′-ACCACTACCAGTGAACATCC-3′ | |||
| S6F | 5′-GTG CAGAATGCTCCATATGGT-3′ | S gene 3439-4092 | 653 | |
| S6R | 5′-TTAGTCGTCATGTGATGTTT-3′ | |||
| HE | HEAF | 5′-CAG TGA AGA AGA CTA AAC TCA GT-3′ | 32-kDa putative nonstructural protein 821 to HE 698 | 741 |
| HEAR | 5′-TAA ATA ACA CCA GTG TCT TTA TT-3′ | |||
| HEBF | 5′-TGA CGA GTA TAT CGT ACC ACT T-3′ | HE gene 591-1275 | 684 | |
| HEBR | 5′-CTA AGC ATC ATG CAG CCT AGT ACC-3′ | |||
| M | MF | 5′-CCA CCA GTT CTT GAT GTG GA-3′ | E gene 226 to N gene 59 | 817 |
| MR | 5′-CCA GAA CGA TTT CCA AAG GA-3′ | |||
| E | EF | 5′-CGK AGA CAG GAG TTA AAT GTT T′-3′ | 12.7-kDa nonstructural protein 301 to noncoding region 10 flanked by E and M genes | 280 |
| ER | 5′-TTT GGA TTA ACT AAA CGT CA-3′ |
F, upstream primer; R, downstream primer.
TABLE 3.
GenBank accession numbers of reference strains of genogroup 2 coronaviruses used in phylogenetic analysis
RESULTS
Detection of BCoV in fecal samples of adult cattle with diarrhea.
Eleven out of 184 fecal samples (8/75 herds) tested BCoV positive by ELISA. A one-step RT-PCR assay, which targeted a 730-bp fragment of the N gene of BCoV, detected 13 positive fecal samples from 7 herds. A nested-PCR assay, which targeted a 407-bp fragment of the N gene, detected 83 positive fecal samples from 32 herds. The total numbers of fecal samples and herds that tested positive for BCoV were 107 and 47 (Table 4), respectively. Fecal samples and herds were determined to be positive if at least 1 sample tested positive by ELISA, RT-PCR, or nested PCR (data not shown). Seasonally, 25 out of 41 fecal samples (13/15 herds) in the spring, 21 out of 43 fecal samples (8/23 herds) in the summer, and 61 out of 100 fecal samples (26/37 herds) in the autumn tested positive for BCoV by ELISA, RT-PCR, or nested PCR (data not shown).
TABLE 4.
Summary of enteric pathogens found in fecal samples from adult cattle with diarrhea
| Enteric pathogen(s) present | No. (%) of positive farms | No. (%) of positive cattle |
|---|---|---|
| BCoV alone | 33 (44.0) | 90 (48.9) |
| BCoV, BRV A | 1 (1.3) | 1 (0.5) |
| BCoV, BRV C | 2 (2.7) | 3 (1.6) |
| BCoV, BEC-NBV | 3 (4.0) | 5 (2.7) |
| BCoV, BNoV | 1 (1.3) | 1 (0.5) |
| BCoV, BRV A, BToV | 1 (1.3) | 1 (0.5) |
| BCoV, Coccidium | 1 (1.3) | 1 (0.5) |
| BCoV, Salmonella | 1 (1.3) | 1 (0.5) |
| BCoV, BRV A, Coccidium | 1 (1.3) | 1 (0.5) |
| BCoV, BRV B, Coccidium | 1 (1.3) | 1 (0.5) |
| BCoV, BRV C, BEC-NBV, Coccidium | 1 (1.3) | 1 (0.5) |
| BCoV, Coccidium, Mycobacterium paratuberculosis | 1 (1.3) | 1 (0.5) |
| BNoV alone | 1 (1.3) | 1 (0.5) |
| BEC-NBV alone | 2 (2.7) | 3 (1.6) |
| BToV alone | 2 (2.7) | 2 (1.1) |
| BRV A alone | 2 (2.7) | 2 (1.1) |
| BRV C alone | 3 (4.0) | 4 (2.2) |
| BEC-NBV, BRV A | 1 (1.3) | 1 (0.5) |
| BEC-NBV, BRV C | 1 (1.3) | 1 (0.5) |
| BEC-NBV, Mycobacterium paratuberculosis | 1 (1.3) | 1 (0.5) |
| BNoV, Cryptosporidium | 1 (1.3) | 1 (0.5) |
| Cryptosporidium | 2 (2.7) | 3 (1.6) |
| Salmonella | 1 (1.3) | 2 (1.1) |
| Mycobacterium paratuberculosis | 1 (1.3) | 1 (0.5) |
| No. enteric pathogens detected | 10 (13.3) | 55 (29.9) |
| Total | 75 (100) | 184 (100) |
Other enteric pathogens.
Figure 1 shows representative RT-PCR and nested-PCR results for BCoV and other viral pathogens. Of the 107 BCoV-positive fecal specimens from the 47 BCoV-positive herds of adult cattle with diarrhea, 90 fecal samples from 33 herds tested positive for BCoV alone, while the other 17 BCoV-positive fecal samples from 14 BCoV-positive herds also tested positive for other enteric pathogens (Table 4). In addition, 16 fecal specimens from the 14 herds that tested negative for BCoV were positive for other enteric pathogens (Table 4). No enteric pathogens were detected in 55 fecal samples from 10 herds.
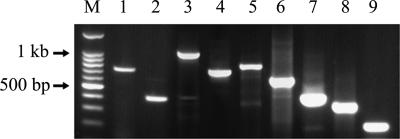
FIG. 1.
Representative RT-PCR and nested-PCR results for the detection of each BCoV, BRV A, BRV B, BRV C, BToV, BNoV, BEC-NBV, and BVDV in fecal samples from adult cattle with diarrhea. Lanes: M, 100-bp marker; 1, BCoV RT-PCR product; 2, BCoV nested-PCR product; 3, BRV A RT-PCR product; 4, BRV A nested-PCR product; 5, BRV B RT-PCR product; 6, BRV C RT-PCR product; 7, BToV nested-PCR product; 8, BNoV nested-PCR product; 9, BEC-NBV nested-PCR product.
Virus isolation in HRT-18G cells.
Of the 15 fecal samples that were found BCoV positive by ELISA, RT-PCR, or nested PCR, BCoV was isolated from 9 fecal samples from 9 herds. Of these isolates, one (KWD12) was isolated from a fecal specimen sampled in the spring, three (KWD14, -18, and -19) in the summer, and five (KWD11, -13, and -15 to -17) in the autumn. After the second or third passage, CPE, characterized by enlarged, rounded, and densely granular cells in clusters, was observed at postinoculation days 2 to 3 in the cultures inoculated with each fecal sample from cattle with diarrhea. No differences in CPE were observed among the isolates. CPE was not observed in the mock-infected HRT-18G cells. The direct immunofluorescence test detected BCoV-specific cytoplasmic fluorescence in the HRT-18G cells inoculated with each of the samples at the third or fourth passage. The number of fluorescing cells increased after further serial passages. ELISA detected the BCoV antigen in nine isolates. A specific band was detected after amplification with all nine isolates by using an RT-PCR assay targeting a 730-bp fragment of the nucleocapsid gene of BoCV. These nine isolates were designated strains KWD11 to KWD19, respectively.
HA, RDE, and HI titers of the KWD BCoV strains.
Table 5 summarizes the HA, RDE, and HI titers of the purified KWD BCoV strains. All the KWD BCoV strains that were isolated year round agglutinated mouse and chicken erythrocytes at 4°C, with HA titers ranging from 4 to 256 for mouse erythrocytes and from 2 to 64 for chicken erythrocytes. This diversity influenced the diversity in the M/C ratio (the ratio of HA titers for mouse erythrocytes to HA titers for chicken erythrocytes) at 4°C: higher M/C ratios were observed for some of the KWD BCoVs isolated in the autumn and summer. At 37°C, the KWD BCoVs agglutinated mouse erythrocytes with almost the same HA titers as those observed at 4°C. However, some of the KWD BCoVs (KWD12 to -16) did not agglutinate chicken erythrocytes. Remarkably, no KWD BCoV strains isolated in the autumn, summer, or spring appeared to have RDE activity against mouse erythrocytes; there was no reduction in their HA titers when the incubation temperature was shifted to 37°C. In contrast, the 10 WD-BCoVs isolated in winter showed higher RDE activity against mouse erythrocytes. All KWD BCoVs, regardless of the season, had low HI titers against the hyperimmune guinea pig serum raised against strain Mebus. On the other hand, relatively lower HI titers were detected in the KWD BCoVs isolated in autumn, summer, and spring, except for KWD11 and -13 in autumn.
TABLE 5.
Comparison of HA, RDE activity, and HI titers between BCoV strains isolated in warmer seasons and WD-BCoV strains
| Season | BCoV straina | HA titerb at 4°C
|
M/C ratio | HA titer at 37°C
|
RDE titerc
|
HI titerd | |||
|---|---|---|---|---|---|---|---|---|---|
| Mouse | Chicken | Mouse | Chicken | Mouse | Chicken | ||||
| Winter | KWD1 | 64 | 32 | 2 | 64 | 32 | +2 | 1 | 16 |
| KWD2 | 64 | 32 | 2 | 64 | 16 | +8 | 1 | 8 | |
| KWD3 | 32 | 32 | 1 | 32 | 32 | +128 | +2 | 8 | |
| KWD4 | 64 | 32 | 2 | 64 | 16 | +32 | 1 | 16 | |
| KWD5 | 128 | 64 | 2 | 128 | 16 | +16 | +2 | 8 | |
| KWD6 | 32 | 32 | 1 | 64 | 16 | +16 | +4 | 8 | |
| KWD7 | 32 | 32 | 1 | 32 | 16 | +64 | +2 | 8 | |
| KWD8 | 128 | 64 | 2 | 64 | 16 | +8 | 1 | 16 | |
| KWD9 | 32 | 16 | 2 | 32 | 8 | +128 | 1 | 16 | |
| KWD10 | 32 | 32 | 1 | 32 | 16 | +128 | −4 | 16 | |
| Autumn | KWD11 | 256 | 16 | 16 | 256 | 16 | 1 | 1 | 16 |
| KWD13 | 64 | 4 | 26 | 64 | 2 | 1 | +32 | 16 | |
| KWD15 | 16 | 2 | 8 | 16 | 2 | 1 | +64 | 2 | |
| KWD16 | 4 | 4 | 1 | 4 | 2 | 1 | +2 | 2 | |
| KWD17 | 16 | 16 | 1 | 16 | 16 | 1 | 1 | 2 | |
| Summer | KWD14 | 8 | 2 | 4 | 8 | 2 | 1 | 1 | 4 |
| KWD18 | 8 | 2 | 4 | 8 | 2 | 1 | +8 | 2 | |
| KWD19 | 8 | 2 | 4 | 8 | 2 | 1 | 1 | 4 | |
| Spring | KWD12 | 8 | 4 | 2 | 8 | 2 | 1 | +2 | 4 |
KWD1 to KWD10, Korean WD-BCoV strains (19); KWD11 to KWD19, BCoV strains isolated in the autumn, summer, and spring.
Expressed as the reciprocal of the highest dilution of virus showing complete HA of 0.4 and 0.2% suspensions of mouse and chicken erythrocytes, respectively, after a 1-h incubation at 4°C or 37°C.
Expressed as the reciprocal of the highest dilution of virus causing complete disappearance of HA patterns at 4°C after a 2-h incubation at 37°C.
Expressed as the reciprocal of the highest dilution of serum inhibiting HA activity against hyperimmune guinea pig serum raised against BCoV strain Mebus.
Molecular analysis of S, HE, M, and E genes of BCoV strains.
All the S, HE, M, and E genes of the nine KWD strains contained open reading frames of 4,092, 1,272, 690, and 252 nucleotides, respectively. These nucleotide sequences encoded predicted proteins of 1,363, 424, 230, and 84 amino acid (aa) residues, respectively. The spike protein was further divided into the S1 and S2 segments, of approximately 86 and 65 kDa, respectively, at the cleavage site, amino acid 768. In comparison with strain Mebus, a total of 99 and 84 polymorphic nucleotides were identified in the S1 and S2 subunits of the BCoV strains isolated in this study, respectively. These polymorphisms led to 31 and 16 amino acid changes at the randomly distributed sites, respectively. Nucleotide substitutions in the HE, M, and E genes of the BCoVs isolated in this study in comparison with the reference strain Mebus were detected in 34, 14, and 4 sites, respectively, leading to 11, 2, and 1 amino acid change at different sites, respectively. No frameshift, deletion, insertion, or nonsense mutations were observed in the nucleotide sequences of the S, HE, M, and E genes of our strains compared with those of the prototype strain Mebus. By drawing comparisons of the deduced amino acids of the S and HE genes between our strains and the other known BCoVs, the two most similar sequences were those of strain KWD13 and strain RBCV OK (99.26%) for the S gene and those of strain KWD16 and the CD strain BCQ701 (99.54%) for the HE gene. In addition, the most distant were those of strain KWD16 and the CD strain BCQ20 (94.76%) for the S gene and those of strain KWD17 and enteric BCoV (EBCoV) LY-138 (97.16%) for the HE gene. Generally, the amino acid sequence homologies of the M and E genes were relatively more conserved between the strains isolated in this study and the other known BCoVs than those of the S and HE genes. All the virulent KWD strains tended to be distant from the ancestral enteric strain, Mebus.
The N-terminal region of the S1 subunit (aa 1 to 330) (22), which was shown to function as a receptor-binding domain in mouse hepatitis virus, had a total of 21 amino acid changes compared with strain Mebus. Within this region, a unique amino acid substitution was observed at aa 149 in all the KWD strains (data not shown). The S1A and S1B immunoreactive domains identified within aa 351 to 403 and aa 517 to 621 had a total of three amino acid changes in comparison with strain Mebus (data not shown). Compared with all the known BCoVs, the sequence KRRSRR at the predicted proteolytic cleavage site was conserved in all KWD strains. The amino acid sequences of the S2 subunit of the present KWD strains were comparatively well conserved in comparison with the S1 subunit of strain Mebus. The only remarkable finding for the S2 subunit was found in the first hydrophobic region, in which all known virulent BCoVs, including the respiratory BCoV (RBCoV) strains LSU and OK, EBCoV strains LY and F15, and all KWD strains, had markedly higher hydrophobicity due to the amino acid substitution at aa 965 than the avirulent strains Vacc, L9, and Mebus (data not shown). All KWD strains showed several conserved features of the HE, M, and E genes with all the BCoVs, such as a hydrophobic putative signal sequence of 18 aa at the N terminus of HE, 9 potential N-linked glycosylation sites of HE, and the putative active site for neuraminate-O-acetylesterase activity, FGDS, at the N terminus of HE.
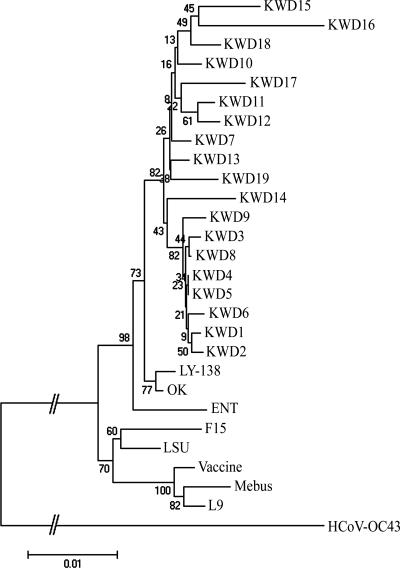
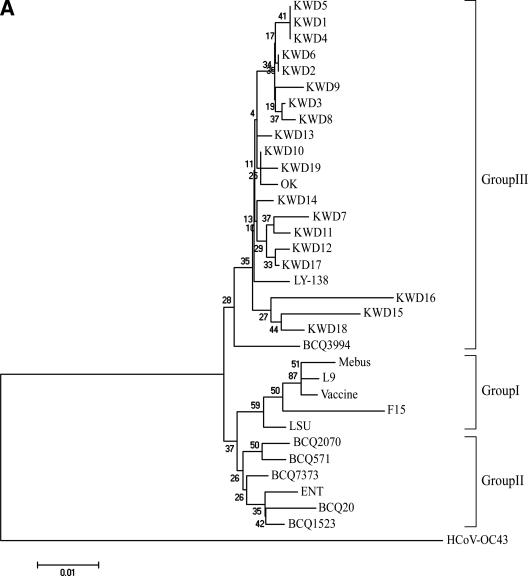
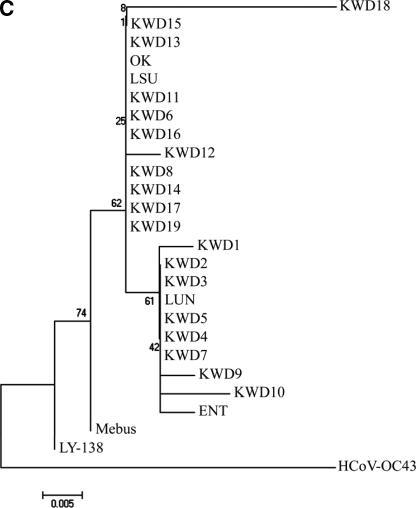
Based on the total number of amino acid substitutions, a phylogenetic comparison of the entire S gene sequence was constructed using the neighbor-joining method of Molecular Evolutionary Genetics Analysis (20) with the CD, RBCoV, EBCoV, and WD strains or isolates. The alignment indicated that the amino acid sequences of the nine KWD strains isolated in the warmer seasons were more homologous to each other and were closely related to the Korean WD strains isolated in winter as well as to the virulent RBCoV strain OK and EBCoV strain LY-138 among the other known BCoVs examined in the paired comparisons (Fig. 2). Moreover, a phylogenetic tree of the hypervariable region of the S1 subunit showed that all Korean WD strains isolated in the warmer seasons were also clustered in groups with the Korean WD strains isolated in the winter season as well as with the virulent RBCoV strains OK and BCQ3994 and EBCoV strain LY-138 (Fig. 3A). The Canadian CD and WD BCQ strains, the American RBCoV strain LSU, the French EBCoV strain F15, and the avirulent strains Vacc, L9, and Mebus are clustered on a separate major branch (Fig. 3A).
FIG. 2.
The phylogenetic tree of the S genes of RBCoV, EBCoV, a CD strain, WD strains, the BCoV strains isolated in this study, avirulent strains or isolates, and human antigenic II coronavirus strain HCoV-OC43 was made using the neighbor-joining method of Molecular Evolutionary Genetics Analysis (20). KWD1 to KWD10, Korean WD strains; KWD11 to KWD19, Korean BCoV isolates recovered from adult cattle with diarrhea in the spring, summer, and autumn; ENT, F15, and LY-138, EBCoV strains; LSU and OK, RBCoV strains; L9 and Vaccine, avirulent vaccine strains; Mebus, prototype CD strain; HCoV-OC43, human antigenic II coronavirus strain.
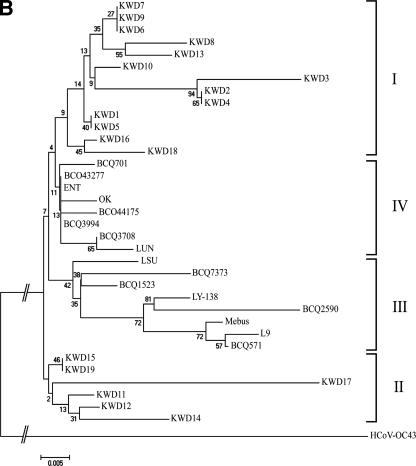
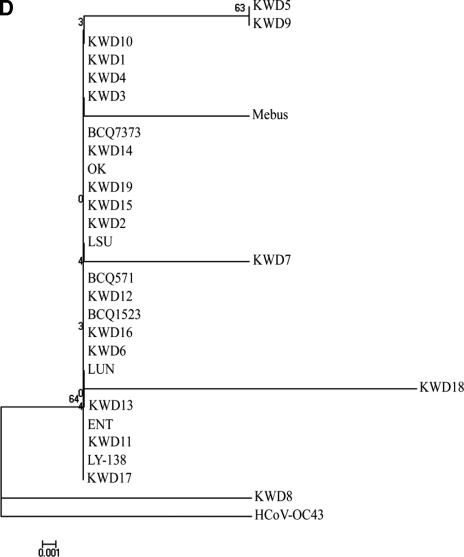
FIG.3.
The phylogenetic trees of the hypervariable region of the S protein (A) and of the HE (B), M (C), and E (D) proteins of RBCoV, EBCoV, CD strains, WD strains, and avirulent strains or isolates were made using the neighbor-joining method of Molecular Evolutionary Genetics Analysis (20). KWD1 to KWD10, Korean WD strains; KWD11 to KWD19, Korean BCoV isolates recovered from adult cattle with diarrhea in the spring, summer, and autumn; BCQ7373 and BCQ2590, WD strains; Mebus, BCQ20, BCQ571, BCQ701, BCQ1523, BCQ2070, and BCQ3708, CD strains; ENT, F15, and LY-138, EBCoV strains; BCQ3994, LSU, OK, LUN, BCO43277, and BCO44175, RBCoV strains; L9, avirulent strain.
Phylogenetically, the amino acid sequence of the HE gene also showed distinct patterns. The alignment indicated that the HE genes among the BCoVs could be divided into four groups (Fig. 3B). The first group included the Korean WD-BCoVs, which consisted mainly of Korean WD strains isolated in winter, with three strains (KWD13, -16, and -18) isolated in the warmer seasons, while the second group contained only Korean WD strains sampled in the warmer seasons. The third group contained mainly CD strains, with WD, EBCoV, and RBCoV strains, while the fourth group contained mainly the RBCoVs, with CD and EBCoV strains. However, phylogenetic analysis of the amino acid sequences of the M and E genes revealed no characteristic pattern of relatedness between the strains isolated in this study and the other known BCoVs (Fig. 3C and D).
FIG. 3—
Continued.
FIG. 3—
Continued.
DISCUSSION
Bovine coronavirus is now recognized as an important cause of WD in adult cattle because it is always identified in sick animals (19, 30, 31). It most often occurs in the winter, and it has been reported in North America, Europe, Australia, and East Asia (30). Although BCoV has been detected, isolated, and characterized in adult cattle with diarrhea in the winter from different regions of South Korea (19), the exact epidemiology and the causative agent of diarrhea in adult cattle in the warmer seasons have never been studied. This represents the first large-scale study of BCoV infection in adult cattle with diarrhea in the warmer seasons in South Korea. The high prevalence and widespread geographical distribution of BCoV suggest that it is endemic in adult cattle with diarrhea in the warmer seasons in South Korea.
Bovine coronavirus infections of adult cattle with epizootic diarrhea occur in the winter as a result of cold stress, e.g., chilling due to low temperatures, with predisposing events including drinking cold water or wide temperature fluctuations, which is an important risk factor for winter dysentery (30, 46). Like other enveloped viruses, BCoV is moderately sensitive to heat (8, 31). Therefore, the occurrence of epizootic diarrhea in adult cattle in the warmer seasons has not been studied in detail. To our knowledge, there is only one report of detection of BCoV in fecal samples from adult cows during an outbreak of epizootic diarrhea in Japan in August (12). In the present study, there were no predisposing events such as chilling or wide fluctuations in temperature (data not shown). Therefore, cold stress might not always be required to initiate the adult cow diarrhea associated with BCoV. This is also compatible with findings showing that experimental WD could be successfully reproduced in adult cattle by inoculating them with WD-BCoV in the warm seasons (41). Although BCoV might play a key role in the occurrence of epidemic diarrhea in herds of adult cattle in the warmer seasons in South Korea, its precise role in the epizootic diarrhea of adult cattle is unclear. One possible explanation is the high prevalence of BCoV infections in adult cattle with WD in South Korea (19), which could precipitate continued exposure to BCoV during the warmer seasons. A similar seasonal incidence pattern has been also reported in the epidemic occurrence of porcine epidemic diarrhea (PED) caused by the group 1 coronavirus, which causes PED all year round in the countries with high prevalences of PED (17, 40).
Another factor is alteration of the biological properties of the BCoV, which might improve the survival of the virus in the warmer seasons and cause diarrhea in adult cattle. From the present biological comparisons among the BCoV strains, the most striking observation was that the BCoV strains isolated in the warmer seasons had no RDE activity against mouse erythrocytes. In contrast, the BCoV isolated in the winter season had higher RDE activity against mouse erythrocytes. HE is known to have acetylesterase (AE) activity, which cleaves the acetyl groups from 9-O-acetylated neuraminic acid, thereby preventing or reversing the hemagglutination or hemadsorption induced by S or HE (26, 35, 45). In addition, RDE activity could enhance the virulence of the BCoV by facilitating the elution of mature virions from the surfaces of infected cells. This might create a higher potential for rapid dissemination of the virus (higher shedding) in herds (9). Although the lack of RDE activity against mouse erythrocytes was conspicuous for our BCoV strains isolated in the warmer seasons, it is still not clear whether the lack of RDE activity affects the survival rate of the BCoV in the warmer seasons.
From the present phylogenetic analysis of the S gene, the amino acid sequences of the Korean BCoV strains, irrespective of season, were closer to each other than to those of the other known BCoVs. Moreover, the most notable finding for the sequence of the hypervariable region of the S1 subunit was the distribution of the BCoVs in highly related distinct clusters: the BCoV strains isolated in this study, regardless of season, were clustered together, whereas the Canadian and the American BCoVs were clustered on a separate major branch. Considering the results of the phylogenetic analysis of the S gene, the BCoV strains isolated in this study had genetic similarities based on their geographical origin. Because the BCoV strains isolated in this study were more closely related to the virulent RBCoV strain OK and the virulent EBCoV strain LY-138, these results support and extend the previous hypothesis that recently isolated BCoVs may be diverging over time from an enteric tropism to a dual respiratory and enteric tropism via intermediates (15, 18). Also, the BCoV strains isolated in this study tended to be distant from the ancestral enteric strain, Mebus, lending further support to the hypothesis that the genetic differences may be more in accordance with the time of the appearance of an outbreak (15, 18).
Phylogenetic data of amino acid sequences of the HE gene revealed that all Korean BCoV strains, regardless of season, were more homologous to each other and were distinct from the other known BCoVs. This supports our hypothesis that the Korean BCoV has a distinct evolutionary pathway. However, the relative conservation of the M and E proteins of all BCoVs, including the BCoV strains isolated in this study, suggests that the structural constraints on these proteins are rigid, resulting in more limited evolution of these proteins than of the S proteins. In addition, the putative site (FGDS) for the AE domain was well conserved in all BCoVs, including the BCoV strains isolated in this study, as well as in mouse hepatitis virus and influenza virus C. It has previously been suggested that an alteration of the RDE activity of WD-BCoV strains could be attributed in part to conformational changes in the HE protein due to a proline substitution near the putative AE domain of the HE protein (47). However, a previous report (13) and the present data do not support this hypothesis, because a proline substitution was identified in all the virulent BCoV strains studied, including those characterized by other investigators (47).
Coronaviruses are believed to mutate at a high frequency, like most RNA viruses, because of the high error frequencies of RNA polymerases (21). The most striking example of the biological importance of coronavirus mutants is the emergence of porcine respiratory coronavirus from transmissible gastroenteritis virus (23, 24, 28). Therefore, an attempt was made to identify genetic differences, responsible for the clinical occurrence of the BCoV strains isolated in this study in warmer seasons, between these strains and other known BCoV strains (including the BCoVs isolated in this study in the winter). Although there appear to be differences in RDE activity between the BCoV strains isolated in this study, sequence analyses failed to find any distinct divergences in the HE, S, M, and E genes, which are common to all field isolates. The factors involved in the occurrence of adult cattle diarrhea in the warmer seasons need to be established.
Although BCoV is believed to be the main causative agent of diarrhea in adult cattle in the warmer seasons, other bovine enteric pathogens might play a role in the clinical and pathological presentation of this disease, because many other enteric viruses were found to be associated with diarrhea in cattle and to cause lesions of villous atrophy (Breda virus, rotaviruses, enteric calicivirus, etc.). Increases in the severity of clinical and pathological findings after natural coinfection with BCoV and other enteric pathogens have been reported (1, 4, 11, 16, 29). Therefore, an etiologic diagnosis is needed for BCoV and other enteric pathogens (1, 4, 11, 16, 29). In accordance with these findings, 18.2% of the BCoV-positive farms (61.6%) included in this investigation tested positive for other pathogens, including rotavirus groups A, B, and C, sapovirus, norovirus, torovirus, Salmonella, M. paratuberculosis, and Coccidium. Although a coinfection with BCoV might play a role in the increased severity of the clinical and pathological presentation of diarrhea in adult cattle, its influence on the development of diarrhea in adult cattle cannot be estimated.
FIG. 3—
Continued.
Acknowledgments
This work was supported by the Regional Technology Innovation Program of the Ministry of Commerce, Industry, and Energy (MOCIE), Republic of Korea. We acknowledge a graduate fellowship provided by the Korean Ministry of Education and Human Resources Development through the Brain Korea 21 project.
REFERENCES
- 1.Athanassious, R., G. Marsolais, R. Assaf, S. Dea, J. P. Descôteaux, S. Dulude, and C. Montpetit. 1994. Detection of bovine coronavirus and type A rotavirus in neonatal calf diarrhea and winter dysentery of cattle in Quebec: evaluation of three diagnostic methods. Can. Vet. J. 35:163-169. [PMC free article] [PubMed] [Google Scholar]
- 2.Barman, P., S. Ghosh, S. Das, V. Varghese, S. Chaudhuri, S. Sarkar, T. Krishnan, S. K. Bhattacharya, A. Chakrabarti, N. Kobayashi, and T. N. Naik. 2004. Sequencing and sequence analysis of VP7 and NSP5 genes reveal emergence of a new genotype of bovine group B rotavirus in India. J. Clin. Microbiol. 42:2816-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benefield, D. A., and L. J. Saif. 1990. Cell culture propagation of a coronavirus isolated from cows with winter dysentery. J. Clin. Microbiol. 28:1454-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulgin, M. S., A. C. S. Ward, D. P. Barrett, and V. M. Lane. 1989. Detection of rotavirus and coronavirus shedding in two beef cow herds in Idaho. Can. Vet. J. 30:236-239. [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, K. O., A. V. Parawani, and L. J. Saif. 1996. The characterization of VP7 (G type) and VP4 (P type) genes of bovine group A rotavirus from field samples using RT-PCR and RFLP analysis. Arch. Virol. 141:1727-1739. [DOI] [PubMed] [Google Scholar]
- 6.Cho, K. O., M. Hasoksuz, P. R. Nielsen, K. O. Chang, S. Lathrop, and L. J. Saif. 2001. Cross-protection studies between respiratory and calf diarrhea and winter dysentery coronavirus strains in calves and RT-PCR and nested PCR for their detection. Arch. Virol. 146:2401-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, K. O., P. G. Halbur, J. D. Bruna, S. D. Sorden, K. J. Yoon, B. H. Janke, K. O. Chang, and L. J. Saif. 2000. Detection and isolation of coronavirus from feces of three herds of feedlot cattle during outbreaks of winter dysentery-like disease. J. Am. Vet. Med. Assoc. 217:1191-1194. [DOI] [PubMed] [Google Scholar]
- 8.Clark, M. A. 1993. Bovine coronavirus. Br. Vet. J. 149:51-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dea, S., L. Michaud, and G. Milane. 1995. Comparison of bovine coronavirus isolates associated with neonatal calf diarrhea and winter dysentery in adult dairy cattle in Quebec. J. Gen. Virol. 76:1263-1270. [DOI] [PubMed] [Google Scholar]
- 10.Dea, S., A. J. Verbeek, and P. Tijssen. 1990. Antigenic and genomic relationships among turkey and bovine enteric coronaviruses. J. Virol. 64:3112-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durham, P. J. K., B. J. Stevenson, and B. C. Farquharson. 1979. Rotavirus and coronavirus associated diarrhea in domestic animals. N. Z. Vet. J. 27:30-32. [DOI] [PubMed] [Google Scholar]
- 12.Fukutomi, T., H. Tsunemitsu, and H. Akashi. 1999. Detection of bovine coronaviruses from adult cows with epizootic diarrhea and their antigenic and biological diversities. Arch. Virol. 144:997-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelinas, A. M., M. Boutin, A. M. J. Sasseville, and S. Dea. 2001. Bovine coronaviruses associated with enteric and respiratory diseases in Canadian dairy cattle display different reactivities to anti-HE monoclonal antibodies and distinct amino acid changes in their HE, S and ns4.9 protein. Virus Res. 76:43-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Givens, M. D., A. M. Heath, R. L. Carson, K. V. Brock, M. S. Edens, J. G. Wenzel, and D. A. Stringfellow. 2003. Analytical sensitivity of assays used for detection of bovine viral diarrhea virus in semen samples from the Southeastern United States. Vet. Microbiol. 96:145-155. [DOI] [PubMed] [Google Scholar]
- 15.Hasoksuz, M., S. L. Lathrop, K. L. Gadfield, and L. J. Saif. 1999. Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biological and antigenic properties with bovine enteric coronaviruses. Am. J. Vet. Res. 60:1227-1233. [PubMed] [Google Scholar]
- 16.Hoet, A. E., P. R. Nielsen, M. Hasoksuz, C. Thomas, T. E. Wittum, and L. J. Saif. 2003. Detection of bovine torovirus and other enteric pathogens in feces from diarrhea cases in cattle. J. Vet. Diagn. Investig. 15:205-212. [DOI] [PubMed] [Google Scholar]
- 17.Hwang, E. K., J. H. Kim, Y. H. Jean, Y. C. Bae, S. S. Yoon, C. K. Park, C. H. Kweon, Y. D. Yoon, and M. Ackermann. 1994. Current occurrence of porcine epidemic diarrhea in Korea. RDA J. Agric. Sci. 36:587-596. [Google Scholar]
- 18.Jeong, J. H., G. Y. Kim, S. S. Yoon, S. J. Park, Y. J. Kim, C. M. Sung, S. S. Shin, B. J. Lee, M. I. Kang, N. Y. Park, H. B. Koh, and K. O. Cho. 2005. Molecular analysis of S gene of spike glycoprotein of winter dysentery bovine coronavirus circulated in Korea during 2002-2003. Virus Res. 108:207-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeong, J. H., G. Y. Kim, S. S. Yoon, S. J. Park, Y. J. Kim, C. M. Sung, S. S. Shin, H. B. Koh, B. J. Lee, C. Y. Lee, M. I. Kang, H. J. Kim, N. Y. Park, and K. O. Cho. 2005. Detection and isolation of winter dysentery bovine coronavirus circulated in Korea during 2002-2004. J. Vet. Med. Sci. 67:187-189. [DOI] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, and N. Masatoshi. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 21.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae: the viruses and their replication, p. 1163-1185. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., Lippincott Williams & Wilkins, Philadephia, Pa.
- 22.Lathrop, S. L., T. E. Wittum, K. V. Brock, S. C. Loerch, L. J. Perino, H. R. Bingham, F. T. McCollum, and L. J. Saif. 2000. Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am. J. Vet. Res. 61:1062-1066. [DOI] [PubMed] [Google Scholar]
- 23.Laude, H., M. Godet, S. Bernard, J. Gelfi, M. Duarte, and B. Delmas. 1995. Functional domains in the spike protein of transmissible gastroenteritis virus. Adv. Exp. Med. Biol. 380:299-304. [DOI] [PubMed] [Google Scholar]
- 24.Laude, H., K. Van Reeth, and M. Pensaert. 1993. Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet. Res. 24:125-150. [PubMed] [Google Scholar]
- 25.Millar, D. S., S. J. Withey, M. L. V. Tizard, J. G. Ford, and J. Hermon-Taylor. 1995. Solid-phase hybridization capture of low-abundance target DNA sequences: application to the polymerase chain reaction detection of Mycobacterium paratuberculosis and Mycobacterium avium subsp. sylvaticum. Anal. Biochem. 226:325-330. [DOI] [PubMed] [Google Scholar]
- 26.Parker, M. D., D. Yoo, and L. A. Babiuk. 1990. Expression and secretion of the bovine coronavirus hemagglutinin esterase glycoprotein by insect cells infected with recombinant baculoviruses. J. Virol. 64:1625-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pensaert, M., P. Callebaut, and E. Cox. 1994. Enteric coronaviruses of animals, p. 627-696. In A. Z. Kapikian (ed.), Viral infections of the gastrointestinal tract, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 28.Rasschaert, D., M. Duarte, and H. Laude. 1990. Porcine respiratory coronavirus differs from transmissible gastroenteritis virus by a few genomic deletions. J. Gen. Virol. 71:2599-2607. [DOI] [PubMed] [Google Scholar]
- 29.Saif, L. J., K. V. Brock, D. R. Redman, and E. M. Kohler. 1991. Winter dysentery in dairy herds: electron microscopic and serological evidence for an association with coronavirus infection. Vet. Rec. 128:447-449. [DOI] [PubMed] [Google Scholar]
- 30.Saif, L. J. 1990. A review of evidence implicating bovine coronavirus in the etiology of winter dysentery in cows: an enigma resolved? Cornell Vet. 80:303-311. [PubMed] [Google Scholar]
- 31.Saif, L. J., and R. Heckert. 1990. Enteropathogenic coronaviruses, p. 185-252. In L. J. Saif and K. W. Theil. (ed.), Viral diarrheas of man and animals. CRC Press, Boca Raton, Fla.
- 32.Saif, L. J., D. R. Redman, K. V. Brock, E. M. Kohler, and R. A. Heckert. 1988. Winter dysentery in adult dairy cattle: detection of coronavirus in the faeces. Vet. Rec. 123:300-301. [DOI] [PubMed] [Google Scholar]
- 33.Sato, K., Y. Inaba, H. Kurogi, E. Takahashi, K. Satado, T. Omori, and M. Matsumoto. 1977. Hemagglutination by calf diarrhea coronavirus. Vet. Microbiol. 2:83-87. [Google Scholar]
- 34.Schultze, B., K. Wahn, H. D. Klenk, and G. Herrier. 1991. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-binding activity. Virology 180:221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schultze, B., H. J. Gross, R. Brossmer, and G. Herrler. 1991. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J. Virol. 65:6232-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smiley, J. R., A. E. Hoet, M. Tråvén, H. Tsunemitsu, and L. J. Saif. 2003. Reverse transcription-PCR assays for detection of bovine enteric calicivirus (BEC) and analysis of the genetic relationships among BEC and human caliciviruses. J. Clin. Microbiol. 41:3089-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, D. R., H. Tsunemitsu, R. A. Heckert, and L. J. Saif. 1996. Evaluation of two antigen-capture ELISAs using polyclonal or monoclonal antibodies for the detection of bovine coronavirus. J. Vet. Diagn. Investig. 8:99-105. [DOI] [PubMed] [Google Scholar]
- 38.Spaan, W., D. Cavanagh, and M. C. Horzinek. 1988. Coronaviruses: structure and genome expression. J. Gen. Virol. 69:2939-2952. [DOI] [PubMed] [Google Scholar]
- 39.Storz, J., W. Purdy, X. Lin, M. Burrell, R. E. Truax, R. E. Briggs, G. H. Frank, and R. W. Loan. 2000. Isolation of respiratory bovine coronavirus, other cytocidal viruses, and Pasteurella spp. from cattle involved in two natural outbreaks of shipping fever. J. Am. Vet. Med. Assoc. 216:1539-1604. [DOI] [PubMed] [Google Scholar]
- 40.Sueyoshi, M., T. Tsuda, K. Yamazaki, K. Yoshida, M. Nakazawa, K. Sato, T. Minami, K. Iwashita, M. Watanabe, Y. Suzuki, and M. Mori. 1995. An immunohistochemical investigation of porcine epidemic diarrhea. J. Comp. Pathol. 113:59-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tråvén, M., K. Näslund, N. Linde, B. Linde, A. Silván, C. Fossum, K. O. Hedlund, and B. Larsson. 2001. Experimental reproduction of winter dysentery in lactating cows using BCV—comparison with BCV infection in milk-fed calves. Vet. Microbiol. 81:127-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsunemitsu, H., and L. J. Saif. 1995. Antigenic and biological comparisons of bovine coronaviruses derived from neonatal calf diarrhea and winter dysentery of adult cattle. Arch. Virol. 140:1303-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsunemitsu, H., H. Yonemichi, T. Hirai, T. Kudo, S. Onoe, K. Mori, and M. Shimizu. 1991. Isolation of bovine coronavirus from feces and nasal swabs of calves with diarrhea. J. Vet. Med. Sci. 53:433-437. [DOI] [PubMed] [Google Scholar]
- 44.Vautherot, J. F., M. F. Madelanine, P. Boireau, and J. Laporte. 1992. Bovine coronavirus peplomer glycoproteins: detailed antigenic analysis of S1, S2 and HE. J. Gen. Virol. 73:1725-1737. [DOI] [PubMed] [Google Scholar]
- 45.Vlask, R., W. Luytjes, J. Leider, W. Spann, and P. Palese. 1988. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J. Virol. 62:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White, M. E., Y. H. Schukken, and B. Tanksley. 1989. Space-time clustering of, and risk factors for, farmer-diagnosed winter dysentery in dairy cattle. Can. Vet. J. 30:948-951. [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, X., K. G. Kousoulas, and J. Storz. 1991. The hemagglutinin/esterase glycoprotein of bovine coronaviruses: sequence and functional comparisons between virulent and avirulent strains. Virology 185:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]