Abstract
A total of 125 non-penicillin-susceptible Streptococcus pneumoniae isolates were received at the Norwegian Institute of Public Health in the period from 1995 to 2001. The strains were tested for antimicrobial susceptibility, serotyped, and genotyped by multilocus sequence typing (MLST); and their penicillin-binding proteins (PBPs) were typed by restriction fragment length polymorphism analysis of their pbp genes. Of the 125 strains, 48 (38%) were fully resistant to penicillin and 77 (62%) were intermediately resistant to penicillin. Most of the strains resistant to penicillin were also resistant to one or several additional antibiotics. The most frequent serotypes among the non-penicillin-susceptible strains were 14, 9V, 19F, 23F, and 6B. MLST analysis showed a high degree of genetic diversity among the 119 strains tested, with a total of 74 different sequence types. Six of the 26 internationally known resistant clones were present; the Spain9V-3 clone was the most frequent, with 19 isolates. A total of 74 (62%) of the isolates were related to 1 of the 26 international clones. Restriction enzyme analyses of the pbp1a, pbp2b, and pbp2x genes revealed 12, 12, and 19 different patterns, respectively; and a total of 43 different PBPs types were demonstrated. Our data indicate that the non-penicillin-susceptible strains in Norway are highly diverse genetically and that limited spread of the internationally known resistant strains occurred in the country in the period examined.
Streptococcus pneumoniae is a leading cause of community-acquired infections and causes a high burden of disease (5). Introduction of treatment with penicillin reduced the case fatality rate for systemic pneumococcal infections from 80% to 20% (17). Since the late 1970s, the proportion of pneumococcal isolates resistant to penicillin and other antimicrobial agents has increased at an alarming rate throughout the world (8). In some countries, like France and Spain, and in some states in the United States, the prevalence of penicillin resistance is over 50% (26). Epidemiological studies of pneumococcal resistance have revealed the worldwide dissemination of epidemic clones (21). In countries with a high occurrence of penicillin resistance, a limited number of clones are often responsible for the majority of cases (21, 27).
The major antibacterial action of penicillin is derived from its ability to bind to and inactivate penicillin-binding proteins (PBPs). PBPs are membrane-associated serine peptidases that catalyze the polymerization and transpeptidation of glycan strands during the final steps of peptidoglycan biosynthesis. Penicillin resistance in S. pneumoniae is caused by alterations in the PBPs that lead to organisms with reduced affinities for penicillin and other β-lactam antibiotics (9). S. pneumoniae contains six different PBPs; and changes in types 1A, 2B, and 2X have been reported to be the most important in the development of penicillin resistance (9). The pbp genes have a mosaic structure, and alterations are caused by the uptake and incorporation of DNA fragments from closely related species by genetic transformation (9, 19).
Norway is still in a favorable position regarding resistance to antimicrobial drugs, with less than 3% of pneumococcal strains causing systemic infections showing resistance to penicillin (24). Consequently, penicillin still remains the drug of choice for the treatment of pneumococcal infections.
In this study, we analyzed non-penicillin-susceptible strains isolated in Norway in the period from 1995 to 2001 to determine the nature of penicillin resistance in our pneumococcal population. The strains were serotyped and genotyped by multilocus sequence typing (MLST); and the genetic heterogeneity of their PBPs was analyzed by restriction fragment length polymorphism analysis of their pbp1a, pbp2b, and pbp2x genes.
MATERIALS AND METHODS
Bacterial strains.
In the period from 1995 to 2001, 4,850 pneumococcal isolates were submitted to the Norwegian Institute of Public Health (NIPH) from the different microbiological laboratories of the whole country; at the time, the population of Norway was about 4.5 million. The isolates included over 80% of the pneumococci recovered from cases of systemic disease in Norway and a selection of strains from cases of noninvasive infections. Strains from patients with systemic disease are sent routinely, while strains from patients with noninvasive infections may be sent if the strains are showing resistance or for other specific questions. Of the 4,850 strains submitted to NIPH, 226 were from patients with noninvasive infections, while the remaining 4,624 were from patients with systemic disease. The type of sample from which the strains were isolated, the age and gender of the patients, and the county of origin of the patients were recorded.
Antimicrobial susceptibility.
The drug susceptibility patterns of the strains were initially tested by the disc diffusion method for penicillin G (by applying 1-μg oxacillin depots), lincomycin, erythromycin, tetracycline, and trimethoprim-sulfamethoxazole (Neo-Sensitabs; Rosco Diagnostics, Taastrup, Denmark). Isolates showing reduced zone sizes for any one of these antibiotics were further tested by the Etest method (AB Biodisk, Solna, Sweden) to determine the MICs of penicillin, erythromycin, chloramphenicol, doxycycline, cefotaxime, and clindamycin. For both methods, the Biodisk PDM medium with 5% horse blood was used and the plates were incubated in 5% CO2. For control purposes we used S. pneumoniae TIGR4 (NIPH 7/87), ATCC 49619, and NIPH 76/00; these strains are wild-type, intermediately penicillin-sensitive, and penicillin-resistant isolates, respectively (24). Strains were defined as antibiotic susceptible, intermediately resistant, or resistant, in accordance with the Clinical and Laboratory Standards Institute guidelines (6).
When several isolates were obtained from a patient during the same disease episode, only one of them was included in the analysis. A total of 125 isolates showed reduced susceptibility to penicillin (MICs > 0.064 μg/ml); 50 of these isolates were from patients with systemic disease and 75 were from patients with localized infections. All these strains were included in the study.
Serotyping.
Serotyping was performed by the Quellung reaction with antisera produced by the Statens Seruminstitut, Copenhagen, Denmark. All strains were serogrouped and serotyped by using factor-specific sera to identify types within groups.
MLST.
The strains were analyzed by MLST. The sequences of the seven housekeeping loci included in the pneumococcal MLST scheme were determined for each isolate, as described elsewhere (12). The sequence types (STs) were obtained by using the MLST website (http://www.mlst.net). New alleles and new STs were submitted to the curator of the database and assigned designations.
To view the genetic relationships among the different strains, a dendrogram was generated from the distance matrix between STs by using the unweighted pair group method with arithmetic averages. The relationship of our strains to the 26 international clones was analyzed with the clustering algorithm BURST by using the programs on the website http://pubmlst.org/.
Analysis of PBPs.
PCR fragments of the pbp1a, pbp2b, and pbp2x genes were amplified by using the following primers: Pbp 1A-F (dCGG CAT TCG ATT TGA TTC GCT TCT), Pbp 1A-R (dTCG TAC TAT TAT TTG TGC TTG GAG T) (11), Pbp 2B-F (dGAT CCT CTA AAT GAT TCT CAG GTG G), Pbp 2B-R (dCAA TTA GCT TAG CAA TAG GTG TTG G) (10), Pbp 2x-F (dTGC CAA TTC ACA CGA TTT GC), and Pbp 2X-R (dTCA CAA TTC CAG CAC TGA TG) (13). The PCRs were run for 30 cycles with an annealing temperature of 54°C for 2 min. The PCR products were cleaned by using QIAquick purification columns (QIAGEN, Oslo, Norway), and the products were eluted in 30 μl distilled water.
The purified PCR products were digested separately with 10 U HinfI (Invitrogen, Carlsbad, CA), according to the manufacturer's instructions, as described by Coffey et al. (7). The digested products were separated on a 1.8% (wt/vol) agarose gel at 100 V for 4 h. DNA was visualized following ethidium bromide staining. The restriction fragment length patterns for each gene were grouped by visual judgment. Gels were also scanned, and the different patterns were compared by using BioNumerics software (Applied Maths BVBA, Sint-Martens-Latem, Belgium) by using the Dice coefficient of similarity and cluster analysis with the unweighted pair group method with arithmetic averages. The position tolerance and the optimization were set equal to 0.6 and 1.0, respectively.
RESULTS
Nonsusceptible strains.
Of the 4,624 pneumococcal isolates from patients with systemic disease received at NIPH in the years from 1995 to 2001, 50 strains showed reduced susceptibility to penicillin. Of these, 48 were from cultures of blood samples and 2 were from cerebrospinal fluid samples. Of the 226 isolates received from patients with localized infections, 75 showed reduced susceptibility to penicillin. The most frequent sources of isolation were the nose and sputum. All these 125 isolates with reduced susceptibility to penicillin were included in the study. Males and females were equally represented among the cases infected with non-penicillin-susceptible isolates: 61 were male and 64 were female.
Of the 125 strains, 77 were intermediately resistant (MICs ≥ 0.064 to 1 μg/ml) and 48 were fully resistant to penicillin (MICs ≥ 2 μg/ml). Resistant strains were recovered from patients in all age groups (Table 1). Isolates from children between 0 and 4 years old represented nearly 25% of the resistant isolates, and this age group was especially overrepresented among the patients with nonsystemic cases of infection due to resistant isolates (35.7%). Most of the strains resistant to penicillin showed additional resistance to one or several other drugs, especially erythromycin, doxycycline, clindamycin, and, to a lesser extent, chloramphenicol (Table 2).
TABLE 1.
Non-penicillin-susceptible S. pneumoniae isolates in relation to age of the patients, Norway, 1995 to 2001
| Age (yr) | No. (%) of nonsusceptible isolates from the following cases:
|
|
|---|---|---|
| Systemic | Nonsystemic | |
| 0-4 | 3 (6.0) | 26 (35.7) |
| 5-9 | 0 | 6 (8.0) |
| 10-19 | 2 (4.0) | 1 (1.3) |
| 20-39 | 7 (14.0) | 10 (13.3) |
| 40-59 | 9 (18.0) | 10 (13.3) |
| 60-79 | 19 (38.0) | 18 (24.0) |
| ≥80 | 10 (20.0) | 4 (5.3) |
TABLE 2.
Numbers of S. pneumoniae isolates resistant or intermediately resistant to penicillin and simultaneously resistant to other antibiotics, Norway, 1995 to 2001
| Antibiotic | No. (%) of isolates
|
|||
|---|---|---|---|---|
| Penicillin resistant
|
Intermediately penicillin resistant
|
|||
| Additional resistance | Additional intermediate resistance | Additional resistance | Additional intermediate resistance | |
| Cefotaxime | 1 (2) | 37 (77) | 3 (4) | 14 (18) |
| Erythromycin | 31 (65) | 0 | 24 (31) | 0 |
| Clindamycin | 20 (42) | 1 (2) | 14 (18) | 0 |
| Chloramphenicol | 13 (27) | 0 | 10 (13) | 0 |
| Doxycycline | 30 (63) | 1 (2) | 37 (48) | 2 (3) |
The number of strains showing intermediate susceptibility to penicillin slightly increased from 1995 to 2001, from 7 to 15 strains. The number of strains resistant to penicillin fluctuated somewhat during the years. Nonsusceptible strains were recovered from the whole country but most frequently from the most populated, southern part.
All but one patient had only one disease episode. The exceptional patient had five blood cultures positive for non-penicillin-susceptible S. pneumoniae in the period from April 1999 to November 2000. The five isolates were included in the analysis and were serotypes 23F (two isolates), 19F, 21, and 9V and ST-277 (two isolates), ST-336, ST-375, and ST-156, respectively. This patient, a refugee from Kosovo, was a 30-year-old man with myelomatosis, a type of cancer that begins in plasma cells.
Serotypes.
Seven serotypes (serotypes 14, 9V, 23F, 19F, 6B, 19A, and 3) were present among the fully resistant strains, while one strain was nontypeable. The intermediately resistant strains belonged to 20 different serotypes, and four strains were nontypeable. The most frequent serotypes among the intermediately resistant strains were 19F, 9V, 14, 23F, and 6B (Table 3).
TABLE 3.
Serotypes among penicillin-resistant and intermediately penicillin resistant isolates of S. pneumoniae, Norway, 1995 to 2001
| Serotypes | No. (%) of isolates
|
|
|---|---|---|
| Penicillin resistant | Intermediately penicillin resistant | |
| 14 | 12 (25.0) | 10 (13.0) |
| 9V | 11 (22.9) | 10 (13.0) |
| 23F | 10 (20.8) | 8 (10.4) |
| 19F | 7 (14.6) | 12 (15.6) |
| 6B | 5 (10.5) | 7 (9.1) |
| 19A | 1 (2.1) | 4 (5.2) |
| 6A | 0 | 4 (5.2) |
| Othersa | 3 (4.2) | 26 (28.6) |
Others include the following: penicillin resistant, one serotype 3 isolate, one serotype 19A isolate, and one nontypeable isolate; intermediately penicillin resistant, two isolates each of serotypes 1, 3, 5, 15C, and 23A; one isolate each of serotypes 4, 7B, 7F, 21, 17F, 22F, 33F, and 35B; and four nontypeable isolates.
MLST.
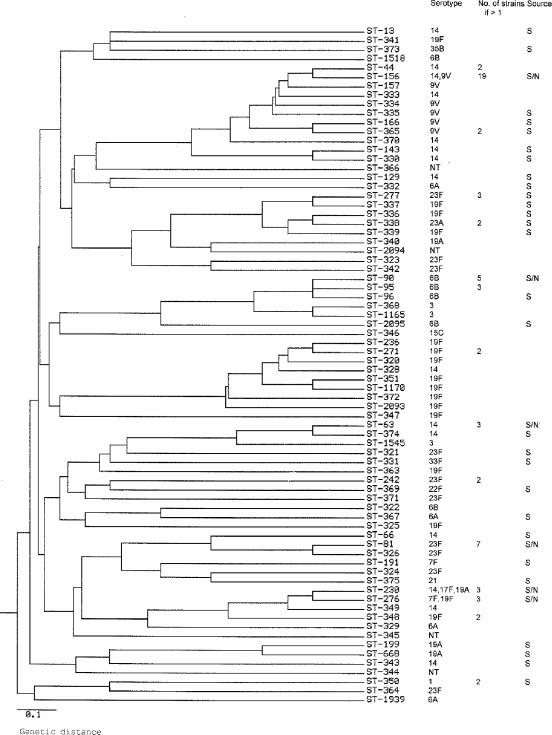
To examine the genetic relationship of the strains in our collection and to identify internationally circulating resistant clones, 119 of the 125 isolates were genotyped by MLST. A total of 74 different STs were detected. New STs were found for 48 of the strains, with each ST represented by a single isolate; 5 of them were single-locus variants (SLVs) of internationally known resistant clones. Of these 48 isolates with new STs, 20 (42%) were fully resistant to penicillin. The genetic relationships among the 74 STs are shown in the dendrogram in Fig. 1. The penicillin-resistant and intermediately resistant strains are equally distributed within the dendrogram. Except for one cluster of eight STs, isolates from patients with nonsystemic disease and systemic disease are also spread throughout the dendrogram; the isolates in that one cluster of eight STs (which included serotype 14 and 19F strains) were isolated only from patients with nonsystemic disease.
FIG. 1.
Genetic relationships among the 74 STs of 119 non-penicillin-susceptible S. pneumoniae isolates from Norway, 1995 to 2001. The dendrogram was generated from the distance matrix between STs by using the unweighted pair group method with arithmetic averages. The serotypes of the strains, the number of isolates, and the STs isolated from patients with systemic disease are indicated. S, isolated from patients with systemic disease; S/N, isolated from both patients with systemic and patients with nonsystemic disease.
Only 15 STs were represented by multiple isolates. The predominant ST, ST-156, known as the Spain9V-3 clone, was represented by 14 serotype 9V strains and, additionally, by 5 serotype 14 strains. The Spain23F-1 clone (ST-81) was represented by seven strains, and the Spain6B-2 clone (ST-90) was represented by five strains (Fig. 1).
BURST analysis of the 74 STs, together with the 26 international clones, identified 11 groups of STs and 33 singletons. Nine of the groups were related to 1 of the 26 international clones: (i) 1 serotype 14 isolate was an SLV of the England14-9 clone; (ii) 30 serotype 14 or 9V isolates were related to the Spain9V-3 clone; (iii) 5 isolates represented the DCC-Pn1476 clone, an SLV (ST-374), or a double-locus variant (DLV) of ST-374 (ST-1545); (iv) ST-66 was an SLV of the Tenessee14-18 clone; (v) ST-340 was an SLV of the Hungary19A-6 clone (vi) in addition to the 7 strains of the Spain23F-1 clone, a novel ST, ST-326, an SLV of this clone that also expressed serotype 23, was represented by 1 strain; (vii) 11 serotype 6B or serotype 3 isolates were related to the Spain6B-2 clone (ST-90); (viii) the Taiwan19F-14 clone (ST-236) was represented by 1 strain, and an SLV of it, ST-271, a known multiresistant clone from Korea, was represented by 2 strains; (ix) finally, the Colombia23F-26 clone (ST-338) was represented by 2 strains, and 6 additional isolates represented SLVs or DLVs of this clone. Thus, in our strain collection, 37 isolates belonged to 1 of 6 of the international clones, and a total of 74 isolates (62%) were SLVs or DLVs of the 26 international clones.
PBPs.
To study the diversity of PBPs 1A, 2B, and 2X of the Norwegian non-penicillin-susceptible strains of S. pneumoniae, the PCR products of their respective genes from 79 of the isolates were digested with the enzyme HinfI. PCR fragments showed 12, 12, and 19 different patterns for pbp1a, pbp2b, and pbp2x, respectively. A total of 43 different pbp profiles were distinguished among the 79 isolates by the combination of the restriction patterns of the three pbp genes. Seven profiles were identified more than once, and these were designated by roman numerals I to VII (Table 4). The largest group (profile I) comprised 27 strains of nine different STs belonging to serotypes 9V, 23F, and 14; and the penicillin MICs ranged from 1 to 4 μg/ml. Profile II included six serotype 6B strains and three strains of three genetically closely related STs. Profile III contained one serotype 14 strain and one nontypeable strain; these two strains had different STs. Profile IV included two strains of ST-230 and of serotypes 17F and 19A. Profile V included two strains of ST-277, serotype 23F; profile VI included two serotype 6B strains of ST-90, and profile VII contained two strains with two different STs (ST-668 and ST-1545) and were serotypes 19A and 3, respectively.
TABLE 4.
pbp profiles represented by multiple isolates of non-penicillin-susceptible S. pneumoniae, Norway, 1995 to 2001
| pbp profile | Allele at:
|
No. of isolates | MIC range (mg/liter) | ||
|---|---|---|---|---|---|
| pbp2b | pbp1a | pbp2x | |||
| I | 1 | 1 | 1 | 27 | 1-4 |
| II | 5 | 1 | 7 | 6 | 1-2 |
| III | 2 | 5 | 8 | 2 | 0.125-0.5 |
| IV | 2 | 11 | 11 | 2 | 0.5 |
| V | 4 | 3 | 5 | 2 | 0.25-0.5 |
| VI | 7 | 1 | 7 | 2 | 0.5 |
| VII | 2 | 5 | 10 | 2 | 0.125 |
Strains of the same ST often harbored more than one pbp profile: the 14 ST-156 strains analyzed showed two different profiles, and the 6 strains of ST-90 showed four different profiles (Table 5). Strains of the same ST with the same pbp profile often had similar MICs.
TABLE 5.
pbp profiles of STs of non-penicillin-susceptible S. pneumoniae represented by multiple isolates, Norway, 1995 to 2001
| ST | Relationship of the ST to international clones | pbp profile(s) | Serotype(s) | MIC range (mg/liter) | No. of isolates |
|---|---|---|---|---|---|
| 156 | Spain9V-3 | I, XIV | 9V, 14 | 0.25-4 | 14 |
| 81 | Spain23F-1 | I | 23F | 1-2 | 6 |
| 90 | Spain6B-2 | II, VII, XXXIV, XLI | 6B | 1-2 | 5 |
| 95 | SLV of Spain6B-2 | XXXIV | 6B | 1-2 | 3 |
| 230 | None | V, XXVIII | 14, 17F, 19A | 0.5-1 | 3 |
| 44 | SLV of Spain9V-3 | I | 14 | 2-4 | 2 |
| 277 | DLV of Colombia23F-26 | VI | 23F | 0.25-0.5 | 2 |
| 330 | None | I | 14 | 4 | 2 |
| 338 | Colombia23F-26 | XVII, XLII | 23F | 0.125-0.25 | 2 |
| 348 | None | XI, XXVII | 19F | 1-4 | 2 |
DISCUSSION
Norway is in a favorable position regarding penicillin-resistant pneumococcal strains. In the 7-year period from 1995 to 2001, only 1% of the isolates from patients with systemic disease received at NIPH showed reduced susceptibility to penicillin. This is a low proportion, and data from the surveillance program for antimicrobial resistance in Norway show no signs of an increasing trend regarding penicillin resistance among S. pneumoniae isolates (23). Most other European countries are reporting much higher rates of resistance, especially Spain and France, where the rates of resistance among pneumococcal isolates have reached 62% and 48%, respectively (26).
To get a wider perspective on the non-penicillin-susceptible pneumococci present in Norway, a further 75 nonsusceptible isolates from patients with localized infections were included in the analysis. The fully resistant strains were recovered from patients of all age groups but were more often found in children 0 to 4 years of age. This is in accordance with the findings of studies that show that children, especially those attending day care centers, are at increased risk of carrying resistant strains (15, 25, 30). There was no difference in the proportion of cases of pneumococcal disease caused by resistant strains by gender. This is in accordance with the data on the occurrence of pneumococcal disease in general in Norway (24).
One patient presented with five blood cultures positive for non-penicillin-susceptible S. pneumoniae within a 20-month period. These five isolates showed four different STs and four different serotypes. This indicates that, despite the general level of low resistance, there are niches in our society as well where the prevalence of resistant strains is high. This particular patient was a refugee. In the beginning of their stay in our country, refugees live in centers where resistant strains are overrepresented due to continual import from various countries, and crowded conditions facilitate spread between individuals.
Many of the strains resistant to penicillin showed additional resistance to erythromycin, clindamycin, doxycycline, and, sometimes, chloramphenicol. The reasons why pneumococci develop simultaneous resistance to several antimicrobial classes are not quite clear, but some resistance determinants have been shown to be transferred together on the same transposon (1). Thus, the emergence of penicillin resistance has been associated with the emergence of multidrug resistance. Most of the known internationally resistant clones are multidrug resistant (21).
Serotypes 6B, 9V,14, 19A, 19F, and 23F have been associated with a high burden of resistance (26). Of the Norwegian isolates with reduced susceptibilities, serotypes 9V, 14, 19F, 23F, and 6B accounted for 73.6% of the cases. These serotypes are known to have the ability to colonize the upper respiratory tracts of healthy carriers and persist there for a long time (18, 28, 29), making them prone to the development of antibiotic resistance selection and gene transfer (22). The seven-valent pneumococcal vaccine could cover 64% of the invasive cases caused by nonsusceptible strains in Norway.
MLST analyses showed that the non-penicillin-susceptible strains in Norway were a very heterogeneous group with a high degree of genetic diversity; 42% of the strains had genotypes not previously identified elsewhere. However, several of the internationally known resistant clones were also identified, and 62% of the isolates were related to 1 of the 26 international clones. International resistant clones are an important cause of the increase of antibiotic resistance among pneumococci. In several countries most of the resistance observed is caused by a relatively few clones circulating in the community (3, 4, 16, 20, 27). The international Spain9V-3 clone, ST-156, was the most frequent resistant clone present in Norway, but it appeared only in solitary cases; only one to four cases occurred each year, and these were in different geographic regions. The Spain23F-1 clone, the Taiwan19F-14 clone, a cluster containing the Spain6B-2 clone, a variant of the Sweden15A-25 clone, and the Colombia23F-26 clone were also all present; but fortunately, none of them have yet had any success in spreading in the Norwegian population. The reason why these resistant strains do not establish themselves in Norway might be the relatively low selective pressure for resistant strains (2). Norway has a relatively strict routine for the prescription of antibiotics, and the consumption of penicillin was reported to be 7 defined daily doses/1,000 inhabitants/day in the period from 1995 to 2001 (23); in comparison, for example, the consumption of penicillin in France during that period was 16 defined daily doses/1,000 inhabitants/day (14). In addition, broad-spectrum penicillins are little used in Norway (14).
The majority of the STs identified by MLST were represented by only one strain. The high degree of diversity shown among the non-penicillin-susceptible strains suggests that many of the resistant cases in Norway result from imported strains rather than from the spread of resistant clones within the Norwegian community. This hypothesis is further supported by the analysis of the PBP patterns, which also revealed a high degree of genetic diversity in the pbp genes among the non-penicillin-susceptible strains. This high degree of diversity indicated that resistance has developed by genetic transformation of the pbp genes on many occasions rather than from the interclone transfer of modified genes. The fact that strains of the same serotype and strains of the same ST showed several different PBP patterns strengthens this point of view.
The PBP analyses revealed seven different profiles represented by more than one isolate. Isolates with profile I showed a MIC range of 1 to 4 mg/liter. This difference might indicate that mechanisms other than the PBPs are influencing penicillin resistance. Alterations in PBP 2X have been recognized as the most important factor in high-level penicillin resistance. Twenty of the strains showing the type 1 pattern of digested pbp2x were fully resistant to penicillin, with the MIC range being from 2 to 4 mg/liter, while seven strains had an MIC of 1 mg/liter.
So far, the spread of non-penicillin-susceptible pneumococcal strains seems to be limited in Norway. The level of resistance shown in our material seems to be a result of multiple, independent imports of resistant strains. As long as the occurrence of resistant pneumococci in the world around us is increasing, it is probable that we will have to live with the continual import of resistant strains at a level revealed in this study. Continuous monitoring for resistance and evaluation of the characteristics of the resistant strains in Norway are important so that we may be able to detect an eventual change in this favorable situation and rapidly try to control a possible clonal spread within the country.
Acknowledgments
We thank Torill Alvestad, Anne-Marie Klem, Anne Ramstad Alme, Inger-Lise Bergstrøm, and Gro Lermark for skillful technical assistance.
Funding was provided by the Norwegian Research Council (grant 143253/310 to D.A.C.).
REFERENCES
- 1.Appelbaum, P. C. 2002. Resistance among Streptococcus pneumoniae: implications for drug selection. Clin. Infect. Dis. 34:1613-1620. [DOI] [PubMed] [Google Scholar]
- 2.Austin, D. J., and R. M. Anderson. 1999. Studies of antibiotic resistance within the patient, hospitals and the community using simple mathematical models. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:721-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, D., B. Lennon, H. Humphreys, and M. Cafferkey. 2003. Penicillin susceptibility and epidemiological typing of invasive pneumococcal isolates in the Republic of Ireland. J. Clin. Microbiol. 41:3641-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogaert, D., G. A. Syrogiannopoulos, I. N. Grivea, R. de Groot, N. G. Beratis, and P. W. Hermans. 2000. Molecular epidemiology of penicillin-nonsusceptible Streptococcus pneumoniae among children in Greece. J. Clin. Microbiol. 38:4361-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartwright, K. 2002. Pneumococcal disease in western Europe: burden of disease, antibiotic resistance and management. Eur. J. Pediatr. 161:188-195. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing. M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 7.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. J. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 8.Dowson, C. G., T. J. Coffey, and B. G. Spratt. 1994. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to beta-lactam antibiotics. Trends Microbiol. 2:361-366. [DOI] [PubMed] [Google Scholar]
- 9.Dowson, C. G., A. Hutchison, J. A. Brannigan, R. C. George, D. Hansman, J. Linares, A. Tomasz, J. M. Smith, and B. G. Spratt. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 86:8842-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowson, C. G., A. Hutchison, N. Woodford, A. P. Johnson, R. C. George, and B. G. Spratt. 1990. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 87:5858-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.du Plessis, M., E. Bingen, and K. P. Klugman. 2002. Analysis of penicillin-binding protein genes of clinical isolates of Streptococcus pneumoniae with reduced susceptibility to amoxicillin. Antimicrob. Agents Chemother. 46:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 13.Ferroni, A., and P. Berche. 2001. Alterations to penicillin-binding proteins 1A, 2B and 2X amongst penicillin-resistant clinical isolates of Streptococcus pneumoniae serotype 23F from the nasopharyngeal flora of children. J. Med. Microbiol. 50:828-832. [DOI] [PubMed] [Google Scholar]
- 14.Goosens, H., M. Ferech, R. Vander Stichele, and M. Elseviers. 2005. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579-587. [DOI] [PubMed] [Google Scholar]
- 15.Henderson, F. W., P. H. Gilligan, K. Wait, and D. A. Goff. 1988. Nasopharyngeal carriage of antibiotic-resistant pneumococci by children in group day care. J. Infect. Dis. 157:256-263. [DOI] [PubMed] [Google Scholar]
- 16.Hermans, P. W., M. Sluijter, K. Elzenaar, A. van Veen, J. J. Schonkeren, F. M. Nooren, W. J. van Leeuwen, A. J. de Neeling, B. van Klingeren, H. A. Verbrugh, and R. de Groot. 1997. Penicillin-resistant Streptococcus pneumoniae in The Netherlands: results of a 1-year molecular epidemiologic survey. J. Infect. Dis. 175:1413-1422. [DOI] [PubMed] [Google Scholar]
- 17.Holm, A. M., D. Berild, S. H. Ringertz, L. L. Haheim, and E. A. Høiby. 2002. Occurrence and clinical presentation of systemic pneumococcal infections in an unselected population in Oslo, Norway, between 1993 and 1997. Eur. J. Clin. Microbiol. Infect. Dis. 21:465-467. [DOI] [PubMed] [Google Scholar]
- 18.Hussain, M., A. Melegaro, R. G. Pebody, R. George, W. J. Edmunds, R. Talukdar, S. A. Martin, A. Efstratiou, and E. Miller. 2005. A longitudinal household study of Streptococcus pneumoniae nasopharyngeal carriage in a UK setting. Epidemiol. Infect. 133:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laible, G., B. G. Spratt, and R. Hakenbeck. 1991. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 5:1993-2002. [DOI] [PubMed] [Google Scholar]
- 20.Marchese, A., M. Ramirez, G. C. Schito, and A. Tomasz. 1998. Molecular epidemiology of penicillin-resistant Streptococcus pneumoniae isolates recovered in Italy from 1993 to 1996. J. Clin. Microbiol. 36:2944-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the pneumococcal molecular epidemiology network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Normark, B. H., and S. Normark. 2002. Evolution and spread of antibiotic resistance. J. Intern. Med. 252:91-106. [DOI] [PubMed] [Google Scholar]
- 23.NORM/NORM-VET 2004. 2005. Usage of antimicrobial agents and occurrence of antimicrobial resistance in Norway. Microbiological Department, University of North Norway, Tromsø, Norway.
- 24.Pedersen, M. K., E. A. Høiby, L. O. Frøholm, V. Hasseltvedt, G. Lermark, and D. A. Caugant. 2004. Systemic pneumococcal disease in Norway 1995-2001: capsular serotypes and antimicrobial resistance. Epidemiol. Infect. 132:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Principi, N., P. Marchisio, G. C. Schito, S. Mannelli, et al. 1999. Risk factors for carriage of respiratory pathogens in the nasopharynx of healthy children. Pediatr. Infect. Dis. J. 18:517-523. [DOI] [PubMed] [Google Scholar]
- 26.Reinert, R. R., S. Reinert, M. van der Linden, M. Y. Cil, A. Al Lahham, and P. Appelbaum. 2005. Antimicrobial susceptibility of Streptococcus pneumoniae in eight European countries from 2001 to 2003. Antimicrob. Agents Chemother. 49:2903-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richter, S. S., K. P. Heilmann, S. L. Coffman, H. K. Huynh, A. B. Brueggemann, M. A. Pfaller, and G. V. Doern. 2002. The molecular epidemiology of penicillin-resistant Streptococcus pneumoniae in the United States, 1994-2000. Clin. Infect. Dis. 34:330-339. [DOI] [PubMed] [Google Scholar]
- 28.Sandgren, A., K. Sjöström, B. Olsson-Liljequist, B. Christensson, A. Samuelsson, G. Kronvall, and N. B. Henriques. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189:785-796. [DOI] [PubMed] [Google Scholar]
- 29.Syrjanen, R. K., T. M. Kilpi, T. H. Kaijalainen, E. E. Herva, and A. K. Takala. 2001. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J. Infect. Dis. 184:451-459. [DOI] [PubMed] [Google Scholar]
- 30.Yagupsky, P., N. Porat, D. Fraser, F. Prajgrod, M. Merires, L. McGee, K. P. Klugman, and R. Dagan. 1998. Acquisition, carriage, and transmission of pneumococci with decreased antibiotic susceptibility in young children attending a day care facility in southern Israel. J. Infect. Dis. 177:1003-1012. [DOI] [PubMed] [Google Scholar]