Abstract
A network of laboratories designated Centres for Molecular Diagnosis was funded in 2000 by Belgian National Health Insurance to provide clinically relevant molecular diagnostic tests. These included typing of nosocomial pathogens as a service to local hospital infection control programs. Two external quality assessment (EQA) surveys were performed in 2001 and 2003 to evaluate the proficiencies of the laboratories at Staphylococcus aureus typing. EQA panels included S. aureus isolates with either indistinguishable, clonally related, or unrelated pulsed-field gel electrophoresis (PFGE) patterns. A hypothetical hospital outbreak problem was also submitted for analysis. Typeability, reproducibility, discrimination (D) index, and epidemiological concordance were evaluated. Ten centers participated in each survey. Seven centers performed PFGE analysis, while others used repetitive-element or randomly amplified polymorphic DNA PCR, amplified fragment length polymorphism, or spa typing. Full typeability (100%) was achieved by all centers, and all but one showed 100% reproducibility. Discrimination was appropriate (D index, ≥96%) for centers performing PFGE analysis but not for all those using other methods (D index range, 72% to 97%). Correct answers to the epidemiological questions were provided by 7/10 and 10/10 centers in 2001 and 2003, respectively. Individual feedback of results was provided to each center together with specific technical recommendations for improving performance. Our findings indicate that surveys of lab proficiency are useful for validation and optimization of molecular typing services to local hospital infection control programs.
The use of molecular methods for epidemiological typing of nosocomial bacterial pathogens has become a standard service for hospital infection control programs (24). It is also an important tool for the surveillance of antibiotic-resistant pathogens, such as methicillin-resistant Staphylococcus aureus (MRSA) strains, which are causing epidemics in health care facilities. The use of in-house molecular typing methods and protocols developed by each laboratory raises the issues of standardization and quality control of these assays. The accuracy of typing methods can be checked by external quality assessment (EQA) programs. National and international EQA programs have been developed for antimicrobial susceptibility testing (3, 17, 29). Multicenter studies have examined the interlaboratory reproducibility of newly developed methods for genotyping of pathogens of public health importance, including S. aureus (1, 4, 5, 9, 22, 30, 32), Legionella pneumophila (14, 15), and Mycobacterium tuberculosis (2). However, EQA programs for molecular typing of nosocomial pathogens as a support to hospital surveillance and infection control policy have not been described.
In 2000, a network of 18 Centers for Molecular Diagnosis (CMDs) was established in Belgium by the National Disease and Disability Institute (INAMI) to provide clinically relevant molecular diagnostic tests to clinicians, including typing of nosocomial pathogens (http://webhost.ua.ac.be/cmd/). The CMD steering group agreed on indications for molecular typing. The first indication is outbreak investigation, typing of clinical isolates (and, when indicated, related environmental isolates) from presumed epidemic cases of nosocomial infection to delineate the extent of clonal spread, determine the sources, and test the hypothetical mode(s) of transmission. Typing is meant to complement the epidemiological outbreak investigation performed by the hospital infection control team to determine appropriate control measures. Bacterial isolates (<30) referred for typing must be documented by the infection control investigator by filling out a typing request form giving information about the outbreak and the origin of the isolates. The second indication is evaluation of policies for controlling the spread of multiresistant bacteria (e.g., contact isolation precautions, carrier decolonization). In the latter case, a sample of isolates from consecutive cases (maximum, 20/year) of colonization or infection by potentially transmissible antibiotic-resistant pathogens (e.g., methicillin-resistant S. aureus, glycopeptide-resistant enterococci, multiple-antibiotic-resistant Enterobacter aerogenes) could be submitted for typing to complement local epidemiologic surveillance of antibiotic resistance.
To evaluate the CMD laboratories' proficiency with bacterial typing methods, we organized and conducted two EQA surveys of typing of S. aureus isolates in support of local hospital infection programs. Performance criteria included typeability, reproducibility, discrimination (D) index, and appropriate epidemiological interpretation of results to assist with testing nosocomial transmission hypotheses.
MATERIALS AND METHODS
Participants.
The surveys were coordinated by the Reference Laboratory for Staphylococci, Université Libre de Bruxelles (ULB), Hôpital Erasme, Brussels, Belgium. The following 11 Belgian laboratories participated in one or more surveys: Cliniques Universitaires Saint Luc—Université Catholique de Louvain, Brussels (V. Avesani); Universitair Instelling Antwerpen, Edegem (M. Ieven); Heilig Hart Ziekenhuis, Roeselare (E. De Laere); Universitair Ziekenhuis Ghent, Gent (M. Vaneechoutte); Universitair Ziekenhuis—Katholieke Universiteit Leuven, Leuven (F. Vanstapel); Academisch Ziekenhuis van Vrije Universiteit Brussel, Brussels (O. Soetens); Openbaar Centrum voor Maatschappelijk Welzijn (M. Van De Vyvere), Algemeen Centrumziekenhuis Antwerpen, Antwerpen; Institut de Pathologie et de Génétique, Gerpinnes (M. Antoine); Centre Hospitalier Universitaire—Saint-Pierre, Brussels (R. Scheen); Algemeen Ziekenhuis Sint-Jan, Brugge (P. Descheemaeker); and Université Catholique de Louvain-Mont-Godinne, Yvoir (Y. Glupczynski). In 2003, these 11 centers together provided 1,496 (range, 5 to 465) typing tests for local or regional hospitals, with an average turnaround time of 10.1 days.
Proficiency test panels.
Two consecutive EQA surveys were conducted, in 2001 and 2003, respectively. Each survey used a panel of coded clinical S. aureus isolates selected at the Reference Laboratory for Staphylococci—ULB from a collection of strains collected as part of national surveillance studies and outbreak investigations (Table 1). Strains were selected based on their genotypes by pulsed-field gel electrophoresis (PFGE) analysis. Sets of isolates with indistinguishable, clonally related, and unrelated PFGE patterns were included in each panel. PFGE patterns were classified according to a previously described system (7). PFGE groups included profiles showing ≤6 DNA fragment differences and were designated by a capital letter (e.g., A). PFGE types included profiles showing ≤3 DNA fragment differences and were designated by the group letter followed by a roman numeral suffix (e.g., A1). A subtype included any variant PFGE profile within a type and was designated by a lowercase letter suffix (e.g., A1a). The first panel was composed of 15 S. aureus strains, including representative isolates of the epidemic MRSA strains that were most widespread in Belgium in 2001, designated BE-A1, BE-B2, and BE-C3 (8, 11), and the second panel included 20 S. aureus strains (Table 1). Duplicate isolates with distinct identifier codes were included in each panel for reproducibility assessment. Each panel was composed of strains subcultured in deep stab agar tubes, which were sent by the coordinating center to the 11 participating CMDs. The ULB standard operating procedure (SOP) for PFGE typing of S. aureus, as previously described (11, 22), was made available to participating centers. The S. aureus NCTC 8325 reference strain, used as a restriction fragment size marker in this SOP, was added to the panel for use as an internal standard. A fictitious hospital outbreak problem was submitted to each participant along with the panel. Several questions pertaining to outbreak control issues were asked, and a typing request was required in order to assess the ability of each center to make the appropriate epidemiological interpretation (Table 2).
TABLE 1.
Characteristics and origins of S. aureus panel strains for EQA surveys, 2001 and 2003
| Strain no. | Origina (reference) | Resistance(s)b | PCR result (mecA) | Typing resultc
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| PFGE | ST | SCCmec | spa repeats | spa type | agr group | ||||
| 2001-SA1 | Outbreak A, 2000 | OXA, CIP | + | B2a | 45 | IV | 8-39-34-34-13-17-34-16-34 | t038 | 1 |
| 2001-SA2 | Duplicate SA1 | OXA, CIP | + | B2a | 45 | IV | 8-39-34-34-13-17-34-16-34 | t038 | 1 |
| 2001-SA3 | Outbreak A, 2000 | OXA, ERY, CIP | + | B2e | 45 | IV | 8-39-34-34-13-17-34-16-34 | t038 | 1 |
| 2001-SA4 | Triplicate SA1 | OXA, CIP | + | B2a | 45 | IV | 8-39-34-34-13-17-34-16-34 | t038 | 1 |
| 2001-SA5 | EARSS, 1999 (8) | OXA, ERY, CIP | + | L1 | 22 | IV | 26-23-23-13-23-31-29-17-31-29-17-25-17-25-16-28 | t032 | 1 |
| 2001-SA6 | EARSS, 1999 (8) | − | J2 | 30 | 15-12-16-02-16-02-17-24 | t743 | 3 | ||
| 2001-SA7 | EARSS, 1999 (8) | OXA, AMK, CIP | + | F2 | 8 | IV | 11-19-21-17-34-24-34-22-25 | t121 | 1 |
| 2001-SA8 | EARSS, 1999 (8) | OXA | + | H | 88 | NT | 07-12-21-17-13-13-34-34-33-34 | t186 | 3 |
| 2001-SA9 | Surv., 1992 (11) | OXA, ERY, GEN, AMK, CIP | + | F1 | 247 | IV | 11-19-21-12-21-17-34-24-34-22-25 | t051 | 1 |
| 2001-SA10 | Outbreak A, 2000 | OXA, CIP | + | B2a | 45 | IV | 8-39-34-34-13-17-34-16-34 | t038 | 1 |
| 2001-SA11 | Surv., 1992 (11) | OXA, ERY, GEN, AMK, CLI | + | K1 | 247 | I | 11-19-21-21-12-21-17-34-24-34-22-25 | t075 | 1 |
| 2001-SA12 | Surv., 1992 (11) | ERY | − | W1 | 25 | 04-21-12-41-17-12-12-17 | t168 | 1 | |
| 2001-SA13 | BE-A1 (11) | OXA, ERY, GEN, AMK, CIP, CLI | + | A1a | 247 | I | 11-19-21-21-17-34-24-34-22-25 | t054 | 1 |
| 2001-SA14 | BE-B2 (11) | OXA, ERY, CIP | + | B2c | 45 | IV | 8-39-34-34-13-17-34-16-34 | t038 | 1 |
| 2001-SA15 | BE-C3 (11) | OXA, AMK, CIP | + | C3a | 5 | III | 26-17-20-17-12-17-16 | t045 | 2 |
| 2003-SA1 | EARSS, 2001 | OXA, ERY, GEN, AMK, CIP, CLI | + | D4c | 228 | I | 26-30-34-17-20-17-12-17-16 | t744 | 2 |
| 2003-SA2 | EARSS, 2001 | OXA, ERY, GEN, AMK, CIP, CLI | + | D4c | 228 | I | 26-30-34-17-20-17-12-17-16 | t744 | 2 |
| 2003-SA3 | EARSS, 2001 | OXA, ERY, GEN, AMK, CIP, CLI | + | D4c | 228 | I | 26-30-34-17-20-17-12-17-16 | t744 | 2 |
| 2003-SA4 | EARSS, 2001 | OXA, ERY, GEN, AMK, CIP, CLI | + | D4c | 228 | I | 26-30-34-17-20-17-12-17-16 | t744 | 2 |
| 2003-SA5 | Duplicate SA4 | OXA, ERY, GEN, AMK, CIP, CLI | + | D4c | 228 | I | 2 | ||
| 2003-SA6 | Surv., 2001 (7) | OXA, ERY, CIP, CLI | + | A20c | 8 | IV | 11-19-12-21-17-34-24-34-22-25 | t008 | 1 |
| 2003-SA7 | Outbreak A, 2000 | OXA, ERY, GEN, AMK, CIP, CLI | + | D4c | 228 | I | 26-30-34-17-20-17-12-17-16 | t744 | 2 |
| 2003-SA8 | Surv., 2001 (7) | OXA, CIP | + | B2b | 45 | IV | 8-39-34-34-34-13-17-34-16-34 | t739 | 1 |
| 2003-SA9 | Outbreak A, 2000 | OXA, ERY, GEN, AMK, CIP, CLI | + | D4c | 228 | I | 26-30-34-17-20-17-12-17-16 | t744 | 2 |
| 2003-SA10 | Surv., 2001 (7) | OXA, ERY, CIP, CLI | + | A20a | 8 | IV | 11-19-12-21-17-34-24-34-22-25 | t008 | 1 |
| 2003-SA11 | Surv., 2001 (7) | OXA, ERY, GEN, CLI | + | D4b | 228 | I | 26-30-34-17-20-17-12-17-16 | t744 | 2 |
| 2003-SA12 | Duplicate SA11 | OXA, ERY, GEN, AMK, CIP, CLI | + | D4b | 228 | I | 26-30-34-17-20-17-12-17-16 | t744 | 2 |
| 2003-SA13 | Surv., 2001 (7) | OXA, ERY, CIP | + | B2b | 45 | IV | 8-39-34-34-13-17-34-16-34 | t038 | 1 |
| 2003-SA14 | Surv., 2001 (7) | OXA, ERY, GEN, CIP, CLI, RIF | + | A1e | 247 | I | 11-21-12-21-17-34-24-34-22-25 | t052 | 1 |
| 2003-SA15 | Surv., 2001 (7) | OXA, GEN, SXT | + | C3b | 5 | IV | 26-23-17-34-17-20-17-12-17-16 | t002 | 2 |
| 2003-SA16 | Surv., 2001 (7) | OXA, CIP | + | B2a | 45 | IV | 8-39-34-13-17-34-16-34 | t740 | 1 |
| 2003-SA17 | Surv., 2001 (7) | OXA, ERY, CIP, CLI, MIN | + | G10a | 5 | II | 26-23-17-34-17-20-17-12-17-16 | t002 | 2 |
| 2003-SA18 | Surv., 2001 (7) | − | E1 | 182 | 04-34-17-66-32-17-23-24 | t493 | 1 | ||
| 2003-SA19 | Surv., 2001 (7) | OXA, CIP | + | Q1 | 5 | IV | 26-23-16 | t769 | 2 |
| 2003-SA20 | Surv., 2001 (7) | − | F3 | 8 | 11-19-12-21-17-34-24-34-22-25 | t008 | 1 | ||
EARSS, European Antimicrobial Resistance Surveillance System (studies on bloodstream infections, conducted in 1999 and 2001); Surv., surveillance studies conducted in Belgian Hospitals in 1992 and 2004.
Antibiotics tested: OXA, oxacillin; ERY, erythromycin; CLI, clindamycin; CIP, ciprofloxacin; GEN, gentamicin; AMK, amikacin; MIN, minocycline; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; FUS, fusidic acid; MUP, mupirocin.
TABLE 2.
Fictitious hospital outbreak problems submitted with questions and answers about interpretation of typing results in the EQAs performed in 2001 and 2003
| EQA yr | Problem statement | Infection control question | Correct answer |
|---|---|---|---|
| 2001 | The set of S. aureus strains (labeled SA1 to SA15) | Q1: Is this a clonal outbreak? | A1: Yes. |
| consists of 15 strains submitted for typing. They originated from clinical cultures and health care staff screening cultures carried out during an | Q2: If so, is the epidemic strain related to one of the reference Belgian epidemic strains? Which one? | A2: Yes, related to BE-B2 type. | |
| outbreak investigation in hospital B. In summary, this outbreak of S. aureus surgical-site | Q3: How many patients are part of this clonal outbreak? Who are they? | A3: Four, patients 1, 2, 3, and 4 (SA1 to SA4). | |
| infections occurred over a 2-mo period in an orthopedic surgery department. Nine patients (patients 1 to 9) were colonized or infected with S. aureus. Patients 1 to 5 were operated on by the same surgical team (surgeons X and Y; nurses W and Z). Three of these four staff members (X, Y, and Z) were found to carry S. aureus. For type comparison, three reference Belgian epidemic MRSA strains are included (SA13 to SA15). | Q4: Are any of the S. aureus carriers on the health care staff harboring the outbreak strain? | A4: Yes, surgeon X. | |
| 2003 | The avg incidence rate of MRSA at the intensive-care unit of hospital A was about 1.5 cases per 100 admissions for the past year. During the | Q1: Was the July 2002 cluster of MRSA colonization due to a clonal outbreak? | A1: Yes. |
| first 2 wk of July 2002, the infection control team observed a cluster of eight cases | Q2: If so, which isolates/patients were part of the outbreak? | A2: SA1 to SA5 and SA7. | |
| associated with an increase in the incidence rate to 3%. Control measures involving reinforcement of contact isolation precautions and decontamination of colonized patients were undertaken on 17 July. One month later, the incidence had dropped back to the baseline rate. To verify that the cluster of cases was due to cross-infection and to verify if transmission was interrupted by control measures, 20 clinical isolates were typed. Isolates SA1 to SA8 were collected during the outbreak period, isolates SA9 to SA10 in May and June 2002, and isolates SA11 to SA20 during the period from 20 July to 15 September. | Q3: Do you confirm that the control measures were effective? If so, please comment on these results in the light of your typing results. | A3: Yes, isolates recovered after control measures were different from the epidemic type. |
Panel validation.
Isolates were further genetically characterized by PCR analysis of the staphylococcal cassette chromosomal mec (SCCmec) element (23), multilocus sequence typing (MLST) (13), determination of accessory gene regulator (agr) type (16), and spa typing using Ridom Staph Type software (18). Isolates in both panels were classified into 10 sequence types (ST), 18 spa types, 4 SCCmec types, and 3 agr groups (Table 1). Concordant classification into PFGE types, and classification obtained by the combination of MLST, SCCmec, spa, and agr analysis, was performed for 91% of MRSA isolates (data not shown). Discordances were observed for three S. aureus isolates: (i) one isolate classified as PFGE type K1 that presented an ST/SCCmec/spa/agr type (247/I/t075/1) similar to that of PFGE type A1 isolates, differing only by one deletion of two consecutive repeats in its spa allelic profile; (ii) one PFGE type F2 isolate that showed a combined type (8/IV/t121/1) similar to that of PFGE type A20, differing by one repeat deletion in its spa profile; and (iii) one PFGE type F3 isolate (susceptible to methicillin) that presented an ST/spa/agr profile (8/t008/1) identical to that observed for MRSA PFGE A20 isolates (Table 1).
Panel testing.
Participants were asked to analyze one panel per survey by the methods that were routinely used in their laboratory for epidemiological typing of S. aureus. Patterns were classified into types by each participant, who completed a table giving the type determined for each isolate. Participants also filled out a questionnaire about the testing procedure(s) (e.g., in-house, commercial, or the ULB PFGE SOP) and interpretation criteria used. Classification was verified by the coordinator based on gel pictures referred by participants. In addition, each participant completed the questionnaire about the hospital outbreak problem (Table 2).
Data analysis.
Data were returned electronically by individual laboratories to the coordinating center. The data included (i) type classification of all isolates, (ii) typing method and classification criteria used, and (iii) answers to the infection control questions. Performance was evaluated according to typeability, intralaboratory reproducibility in type assignment of identical isolates (duplicate strains), and discrimination of unrelated strains as recommended (26). The discrimination index was determined by calculation of the Simpson index of diversity, D (19, 26), for epidemiologically unrelated strains (SA5 to SA8 and SA11 to SA15 in the 2001 EQA; SA1, SA6 and SA15 to SA20 in the 2003 EQA). The concordance of type classification by local typing with the reference PFGE analysis was examined. Epidemiological concordance was evaluated by correct clonal grouping of isolates and ability to give the appropriate answers to the infection control questions (Table 2).
RESULTS
Participation and typing methods used.
A total of 11 centers participated in the EQA program, with 10 centers participating in each survey (Table 3). Centers returned results within a median of 18 days (range, 2 to 66 days) of receipt of the proficiency panels. The majority of centers (7/10) performed PFGE analysis in both surveys, but they used different separation protocols and classification criteria. PFGE patterns were analyzed visually or/and by computer (Table 3). A few centers (3/10 and 1/10 in 2001 and 2003, respectively) performed randomly amplified polymorphic DNA (RAPD) PCR or inter-IS256 PCR analysis. Centers that used PCR-based assays variously defined a distinct type based on either ≥1 or ≥3 DNA fragment differences. In the second survey, one center used amplified fragment length polymorphism (AFLP) analysis by resolution on a capillary sequencer and another center used sequencing of the spa gene (25), analyzed by visual comparison of sequences (Table 3).
TABLE 3.
Methods, classification criteria, and pattern analysis used by participating centers in the two EQA surveys on S. aureus typing
| Center | First EQA, 2001
|
Second EQA, 2003
|
||||||
|---|---|---|---|---|---|---|---|---|
| Method(s) | Protocol | Classification criterion | Pattern analysis | Method(s) | Protocol | Classification criterion | Pattern analysis | |
| 1 | IS256 PCR | Deplano et al. (10) and Ready-To-Go beadsa | Distinct type if ≥1-fragment difference | Visual | AFLP | Koeleman et al. (21) with a capillary sequencer | Distinct type if ≥3-fragment difference | Visual |
| 2 | SmaI PFGE | GenePath kitb | Tenover et al. (28) | Computer | SmaI PFGE | GenePath kitb | Tenover et al. (28) | Computer |
| 3 | SmaI PFGE | In-house for 20 h and ULB SOP, but run of 22 h | Tenover et al. (28) | Visual | SmaI PFGE | ULB SOP, but run shortened to 22 h | Denis et al. (7) | Visual |
| 4 | SmaI PFGE | GenePath kit | Denis et al. (7) | Visual | SmaI PFGE | ULB SOP, but run shortened to 23 h | Denis et al. (7) | Visual |
| 5 | SmaI PFGE | In-house, run of 22 h | Tenover et al. (28) | Computer | SmaI PFGE | In-house, run of 22 h | Tenover et al. (28) | Computer |
| 6 | SmaI PFGE | In-house, run of 22 h | Tenover et al. (28) | Computer | SmaI PFGE | In-house, run of 22 h | Tenover et al. (28) | Computer |
| 7 | RAPD primer 4 | In-house | Distinct type if ≥3-fragment difference | Visual | Not done | |||
| 8 | RAPD ERIC2 | In-house | Distinct type if ≥1-fragment difference | Visual | RAPD ERIC2 | In-house | Distinct type if ≥3-fragment difference | Visual |
| 9 | SmaI PFGE | Descheemaeker et al. (12) | Tenover et al. (28) | Visual + computer | SmaI PFGE | Descheemaeker et al. (12) | Tenover et al. (28) | Visual + computer |
| 10 | Not done | Sequencing of spa gene | Shopsin et al. (25) | Distinct type if ≥3-base difference in sequence | Visual | |||
| 11 | SmaI PFGE | In-house, run of 12 h | Tenover et al. (28) | Visual | SmaI PFGE | In house, run of 12 h | Denis et al. (7) | Visual |
| Coordinator | SmaI PFGE | ULB SOP (11) | Denis et al. (7) | Visual + computer | SmaI PFGE | ULB SOP (11) | Denis et al. (7) | Visual + computer |
From Amersham-Biosciences (Roosendaal, The Netherlands).
From Bio-Rad (Nazareth, Belgium).
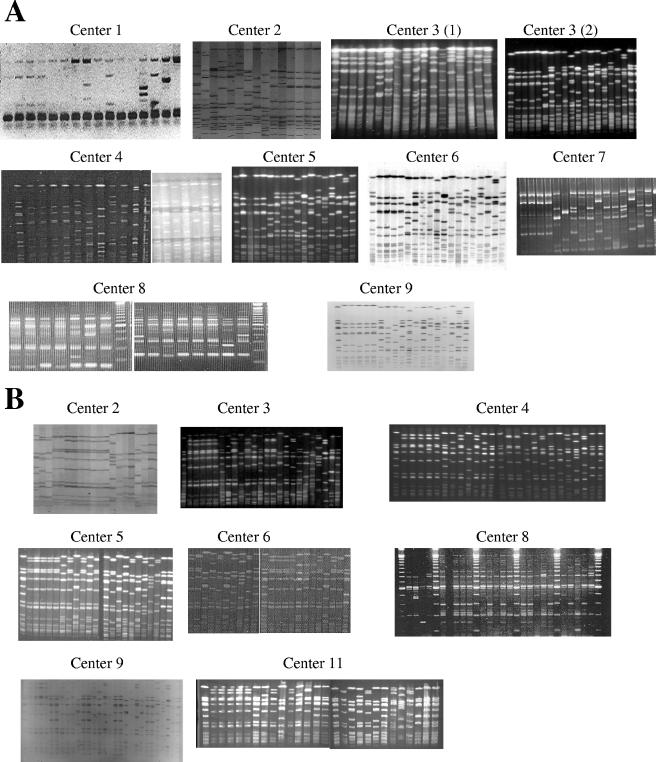
Quality of patterns.
Gel pictures or tagged-image format files obtained with DNA fingerprint methods were reviewed by the coordinator for scoring of quality and resolution. The ULB SOP and “in-house” separation protocols showed well-resolved PFGE analysis patterns (Fig. 1). In contrast, center 2, which used the GenePath kit (Bio-Rad) for PFGE analysis, showed poorly resolved patterns due to an underload of DNA and faintly staining bands (Fig. 1). Center 4 improved PFGE resolution and pattern quality in the second survey over that in the first survey after substituting the ULB SOP separation protocol (Fig. 1). In the second survey, two centers obtained poorly resolved PFGE patterns due to suboptimal separation conditions (center 11) or showed faintly stained DNA bands (center 9). Centers using RAPD PCR analysis showed well-resolved amplified DNA patterns (Fig. 1). One center that performed AFLP analysis showed well-resolved densitometric curves (data not shown). Sequence data from spa gene analysis performed in one center could not be scored for quality, because ABI sequence files were not sent to the coordinator.
FIG. 1.
Gel images sent by CMDs in EQA surveys conducted in 2001 (A) and 2003 (B). In 2001, center 1 used IS256 PCR; centers 2 to 6 and 9 performed PFGE, with center 3 testing two protocols, an in-house protocol [center 3 (1)] and the ULB SOP [center 3 (2)]; centers 7 and 8 performed RAPD PCR; and center 11 sent no picture. In 2003, centers 2 to 6, 9, and 11 performed PFGE, and center 8 used RAPD PCR.
Typeability and reproducibility.
All centers obtained complete typeability for both strain panels with all methods (Table 5). Excellent reproducibility with replicate strains (100%) was observed for all centers, except for one center that found a single DNA fragment difference in the PFGE profiles of a duplicate strain (Fig. 1B, center 4, strain SA4). After repeat testing, the “additional” band was shown to be artifactual, resulting from incomplete DNA fragment digestion.
TABLE 5.
Evaluation of typing method performance of participating centers by EQAs in 2001 and 2003
| Yr and center | Method | Typeability (%) | Reproducibility (%) | D index (%) | Concordancea (%) | Correct answersb |
|---|---|---|---|---|---|---|
| 2001 | ||||||
| 1 | IS256 PCR | 100 | 100 | 72 | 67 | No |
| 2 | PFGE | 100 | 100 | 100 | 100 | Yes |
| 3 | PFGE | 100 | 100 | 100 | 100 | Yes |
| 4 | PFGE | 100 | 100 | 100 | 100 | Yes |
| 5 | PFGE | 100 | 100 | 97 | 93 | No |
| 6 | PFGE | 100 | 100 | 97 | 93 | Yes |
| 7 | RAPD PCR | 100 | 100 | 97 | 87 | Yes |
| 8 | RAPD PCR | 100 | 100 | 80 | 47 | No |
| 9 | PFGE | 100 | 100 | 100 | 100 | Yes |
| 10 | ||||||
| 11 | PFGE | 100 | 100 | 97 | 93 | Yes |
| 2003 | ||||||
| 1 | AFLP | 100 | 100 | 86 | 70 | Yes |
| 2 | PFGE | 100 | 100 | 100 | 100 | Yes |
| 3 | PFGE | 100 | 100 | 100 | 90 | Yes |
| 4 | PFGE | 100 | 75 | 100 | 95 | Yes |
| 5 | PFGE | 100 | 100 | 100 | 100 | Yes |
| 6 | PFGE | 100 | 100 | 100 | 100 | Yes |
| 7 | ||||||
| 8 | RAPD PCR | 100 | 100 | 79 | 85 | Yes |
| 9 | PFGE | 100 | 100 | 100 | 100 | Yes |
| 10 | Sequencing of spa | 100 | 100 | 93 | 80 | Yes |
| 11 | PFGE | 100 | 100 | 96 | 85 | Yes |
With PFGE analysis performed by the coordinating center.
To all infection control questions about the fictitious hospital outbreak (see Table 2).
Discrimination.
In both surveys, PFGE analysis showed a discrimination index of ≥96%, with the majority of centers displaying a D index of 100% (Table 5). Only three strain type misclassifications were noted for centers using PFGE typing, due to suboptimal separation conditions in one (center 11) and mislabeling of strain SA7 in two centers (centers 5 and 6). In contrast, RAPD PCR and inter-IS256 PCR typing showed lower discrimination index values, ranging between 72% and 80%, except for center 7, which achieved a D index of 97%. AFLP analysis also showed limited discrimination (D index, 86%). Sequencing of the spa gene, performed by center 10 as described by Shopsin et al. (25), showed a D index of 93%.
Concordance of type classification with reference typing data.
In both surveys, excellent concordance of type classification with the reference PFGE analysis, ranging from 85 to 100%, was obtained for centers using PFGE (Table 5). Center 11, which used suboptimal separation conditions (electrophoresis run time, 12 h, compared to 22 to 25 h for other centers), achieved concordance for 93% and 85% of isolates tested in 2001 and 2003, respectively (Table 5). Center 3 misclassified two isolates (SA13 and SA16) as distinct PFGE types (Table 5, 2003). Review of gel pictures by the coordinator indicated that discordant classification by this center was due to misinterpretation of similar PFGE patterns showing 2-fragment differences by visual pattern comparison.
Because of the lower discrimination of RAPD and inter-IS256 PCR analysis, centers using these methods showed concordant type assignment with reference PFGE analysis for 47 and 87% of isolates, respectively. Discrepancies of these PCR typing results with PFGE analysis were also confirmed by MLST and spa analysis data. Center 1, using AFLP analysis, classified test isolates with 70% concordance with PFGE analysis and 85% concordance with MLST data. By using spa typing, center 10 obtained 80% concordance with PFGE analysis, 85% concordance with MLST data, and 89% concordance with spa typing data generated by using the Ridom Staph Type protocol and software.
Interpretation of typing results.
Epidemiological interpretation was evaluated according to answers given to the questions about the hospital infection control problem (Table 2). In the 2001 survey, all centers correctly recognized the existence of a clonal outbreak (Q1) and all but one center (center 8) identified the outbreak strain as the epidemic Belgian MRSA strain BE-B2 (Q2). This error was probably due to inversion between reference Belgian MRSA strains BE-A1 and BE-B2 by center 8 during testing. Due to the lower discrimination by IS256 and RAPD PCR typing, centers 1 and 8 incorrectly included unrelated strains as part of the outbreak and therefore provided inaccurate answers to questions related to the extent and potential source of the outbreak (Q3 and Q4) (Tables 2 and 4). In contrast, in the 2003 survey, in spite of different discrimination levels and occasional discrepancies between typing methods, all centers interpreted their results appropriately and answered all infection control questions correctly.
TABLE 4.
Type classification of the S. aureus collection by participating centers, EQA surveys, 2001 and 2003
| Strain no. | Type designationa reported by the following center (method):
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coordinator (SmaI PFGE) | 1 (IS256 PCR or AFLPb) | 2 (SmaI PFGEc) | 3 (SmaI PFGE) | 4 (SmaI PFGEc) | 5 (SmaI PFGE) | 6 (SmaI PFGE) | 7 (RAPD PCR) | 8 (RAPD PCR) | 9 (SmaI PFGE) | 10 (sequencing of spa) | 11 (SmaI PFGE) | |
| 2001:SA1 | B2a | 2 | 2a | 2a | 2a | 2a | 2a | 2 | 2 | 2a | 2a | |
| 2001:SA2d | B2a | 2 | 2a | 2a | 2a | 2a | 2a | 2 | 2 | 2a | 2a | |
| 2001:SA3 | B2b | 2 | 2a | 2b | 2a | 2b | 2b | 2 | 2 | 2b | 2a | |
| 2001:SA4d | B2a | 2 | 2a | 2a | 2a | 2a | 2a | 2 | 2 | 2a | 2a | |
| 2001:SA5 | L1 | 4 | 4 | 4 | 4 | 4a | 4 | 4 | 4 | 4 | 4 | |
| 2001:SA6 | J2 | 4 | 5 | 5 | 5 | 5a | 5 | 5 | 5 | 5 | 5 | |
| 2001:SA7 | F2 | 4 | 6 | 6 | 6 | 5a | 5 | 1 | 2 | 6 | 6 | |
| 2001:SA8 | H1 | 5 | 7 | 7 | 7 | 6a | 6 | 6 | 3 | 7 | 7 | |
| 2001:SA9 | F1 | 4 | 8 | 8 | 8 | 7a | 7 | 7 | 2 | 8 | 8 | |
| 2001:SA10 | B2a | 2 | 2b | 2a | 2a | 2c | 2c | 2 | 2 | 2a | 2a | |
| 2001:SA11 | K1 | 4 | 9 | 9 | 9 | 8a | 8 | 7 | 2 | 9 | 1b | |
| 2001:SA12 | W1 | 4 | 10 | 10 | 10 | 9a | 9 | 8 | 2 | 10 | 9 | |
| 2001:SA13 | A1a | 1 | 1 | 1 | 1 | 1a | 1 | 1 | 2 | 1 | 1a | |
| 2001:SA14 | B2c | 2 | 2a | 2a | 2b | 2a | 2a | 2 | 1 | 2c | 2a | |
| 2001:SA15 | C3a | 3 | 3 | 3 | 3 | 3a | 3 | 3 | 3 | 3 | 3 | |
| 2003:SA1 | D4c | A | 1a | A1a | A1a | A1 | 1a | 5a | A1 | A | A1a | |
| 2003:SA2 | D4c | A | 1a | A1a | A1a | A1 | 1a | 5a | A1 | A | A1a | |
| 2003:SA3 | D4c | A | 1a | A1a | A1a | A1 | 1a | 5a | A1 | A | A1a | |
| 2003:SA4 | D4c | A | 1a | A1a | A1b | A1 | 1a | 5a | A1 | A | A1a | |
| 2003:SA5e | D4c | A | 1a | A1a | A1a | A1 | 1a | 5a | A1 | A | A1a | |
| 2003:SA6 | A20c | B | 7a | B1a | B1a | B1 | 2a | 1a | B1 | B1 | A2 | |
| 2003:SA7 | D4c | A | 1a | A1a | A1a | A1 | 1a | 5a | A1 | A | A1a | |
| 2003:SA8 | B2b | C | 2 | C | C1a | C1 | 3a | 6 | C1 | C | B12 | |
| 2003:SA9 | D4c | A | 1a | A1a | A1a | A1 | 1a | 5a | A1 | A | A1a | |
| 2003:SA10 | A20a | B | 7a | B1b | B1a | B1 | 2a | 1a | B2 | B1 | A2 | |
| 2003:SA11 | D4b | D1 | 1b | A2a | A2a | A2 | 1b | 5a | A2 | A | A1b | |
| 2003:SA12f | D4b | D1 | 1b | A2a | A2a | A2 | 1b | 5a | A2 | A | A1b | |
| 2003:SA13 | B2b | C | 2 | D | C1a | C1 | 3a | 6 | C1 | D1 | B12 | |
| 2003:SA14 | A1e | D2 | 7b | B1c | B1b | B2 | 2b | 1b | B2 | B2 | A2 | |
| 2003:SA15 | C3b | D3 | 4 | E | D | H1 | 4a | 5a | D1 | E | C12 | |
| 2003:SA16 | B2a | C | 2 | F | C1b | C2 | 3b | 6 | C2 | F1 | B12 | |
| 2003:SA17 | G10a | D4 | 5 | G | E | D1 | 5a | 5b | E1 | E | D12 | |
| 2003:SA18 | E1 | E | 9 | H | F | E1 | 6a | 7 | F1 | F2 | E12 | |
| 2003:SA19 | Q1 | D4 | 3 | I | G | F1 | 7a | 5a | G1 | D2 | C1b | |
| 2003:SA20 | F3 | B | 8 | J | H | G1 | 8a | 8 | H1 | B1 | F12 | |
Results showing major discrepancies in group and/or type classification with the PFGE classification performed by the coordinator are boldfaced.
IS256 PCR was performed in 2001 using Ready-To-Go Beads (Amersham-Biosciences). AFLP was performed in 2003.
PFGE of SmaI was performed using the GenePath kit (Bio-Rad).
SA2 is a duplicate, and SA4 is a triplicate, of 2001:SA1.
Duplicate of 2003:SA4.
Duplicate of 2003:SA11.
Report and recommendations.
An individual report summarizing the results of each survey was sent to each participating center. A global report on all results was made available on the CMD website (http://webhost.ua.ac.be/cmd/). Laboratories were each labeled with a confidential code. Reports scored the quality of the results and offered specific suggestions to improve methods or interpretation of results. These recommendations included (i) separation conditions for adequate resolution of PFGE patterns as validated in a previous multicenter study (22), (ii) use of computer-assisted analysis verified by visual inspection to prevent misclassification of PFGE patterns, and (iii) caution against the use of rep-PCR and RAPD PCR as stand-alone methods for typing of S. aureus due to their limited discrimination.
DISCUSSION
We report on the first EQA surveys conducted with laboratories performing routine molecular typing of nosocomial pathogens with a variety of in-house methods, which were evaluated against reference typing based on PFGE typing. Typeability and within-run reproducibility were generally excellent. All centers that used PFGE analysis achieved excellent discrimination between outbreak-related and unrelated isolates based on validated criteria for PFGE profile interpretation (7, 27, 28). Laboratories that used PFGE typing achieved better performance if they applied the ULB SOP or similar in-house protocols than if they used the GenePath reagent kit and protocol, as previously described in another multicenter study (31). These findings underline the importance of using validated separation conditions for optimal resolution of DNA fragments. Use of computer-assisted pattern analysis verified by visual inspection helped to prevent misclassification (22). This is crucial when a large number of S. aureus PFGE profiles are compared across multiple gels.
In contrast with the good performance obtained with PFGE typing, centers using single-primer PCR-based typing methods generally showed insufficient discrimination. This led some participants to perform incorrect classification of outbreak-related and unrelated isolates. Among these PCR-based methods, the previously validated inter-IS256 PCR typing method (10) also presented low discrimination with the panel of isolates provided in the present study. This unexpectedly low discrimination may be due to genetic variation over time at the level of insertion element-like sequences in S. aureus isolates. Because of limited discrimination, isolates grouped together by indistinguishable RAPD/rep-PCR profiles should be confirmed as genotypically related either by further PCR testing with an additional primer(s) or, preferably, by another genotyping technique. Despite previous studies showing the good performance of AFLP analysis for typing L. pneumophila (14, 15), Acinetobacter baumannii (6, 21), and S. aureus (20), the performance of AFLP as used by a single center also showed suboptimal discrimination for S. aureus typing in this survey.
DNA sequence-based typing approaches have become more frequently used because data are easily exchangeable between laboratories via the Internet. MLST is a discriminating method that scores the genetic variation accumulated relatively slowly in house keeping genes. MLST has been successfully used for global epidemiology and population genetic studies of S. aureus. It is less suitable for routine typing in outbreak investigations or local surveillance studies because of its high cost and workload. Single-locus DNA sequence analysis of hypervariable regions such as the repeat region of the spa gene (protein A) is a suitable method for S. aureus typing in a hospital setting (18, 25). In the present study, one center that used spa typing achieved good performance. Commercial software (Ridom Staph Type) is available for spa typing analysis and interpretation according to an international spa nomenclature (1, 18).
This study had an obvious limitation relating to its design: the “gold standard” criteria for typing results were defined on the basis of a particular method, PFGE typing with SmaI macrorestriction analysis. The better performance of PFGE typing over other methods was therefore to be expected based on the selection of isolate panels by PFGE classification into indistinguishable, clonally related, and unrelated patterns. Nevertheless, the strains used in the present study were characterized by extended genotypic analysis. These data confirmed that the clonal classification by PFGE patterns was robust. These variations illustrate the difficulty of defining the “gold standard” typing method, because each method explores different specific genomic regions (highly polymorphic or more conserved). The choice of typing method depends on the question asked (26). Outbreak investigations require the use of methods with high discriminatory power, such as PFGE or spa typing. In the EQA surveys reported here, the goal was to assess the laboratories' abilities to delineate an outbreak of cross-infection and answer the infection control questions for implementation of appropriate control measures.
Our findings underline the importance of using validated typing protocols irrespective of the typing strategy. All centers recognized the clonal outbreak, and the proportion capable of correctly answering the infection control questions increased in the second survey. We suggest that similar EQA surveys should be conducted to (i) evaluate the performance of typing laboratories, (ii) compare the effectiveness of routinely used methods, and (iii) provide recommendations to optimize method resolution and appropriate interpretation of results. These EQA exercises should be organized regularly and should include a range of nosocomial pathogens causing infection control problems.
Acknowledgments
This study was supported a financial grant from the National Disease and Disability Institute (INAMI), Belgium.
We thank all participating CMDs for their participation and feedback.
REFERENCES
- 1.Aires-de-Sousa, M., K. Boye, H. de Lencastre, A. Deplano, M. C. Enright, J. Etienne, A. Friedrich, D. Harmsen, A. Holmes, X. W. Huijsdens, A. M. Kearns, A. Mellmann, H. Meugnier, J. K. Rasheed, C. Spalburg, B. Strommenger, M. J. Struelens, F. Tenover, J. Thomas, U. Vogel, H. Westh, J. Xu, and W. Witte. 2006. High interlaboratory reproducibility of DNA sequence-based typing of bacteria in a multicenter study. J. Clin. Microbiol. 44:619-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braden, C. R., J. T. Crawford, and B. A. Schable. 2002. Quality assessment of Mycobacterium tuberculosis genotyping in a large laboratory network. Emerg. Infect. Dis. 8:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaitram, J. M., L. A. Jevitt, S. Lary, and F. C. Tenover. 2003. The World Health Organization's External Quality Assurance System Proficiency Testing Program has improved the accuracy of antimicrobial susceptibility testing and reporting among participating laboratories using NCCLS methods. J. Clin. Microbiol. 41:2372-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sa-Leao, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Cookson, B. D., P. Aparicio, A. Deplano, M. Struelens, R. Goering, and R. Marples. 1996. Inter-centre comparison of pulsed-field gel electrophoresis for the typing of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 44:179-184. [DOI] [PubMed] [Google Scholar]
- 6.D'Agata, E. M., M. M. Gerrits, Y. W. Tang, M. Samore, and J. G. Kusters. 2001. Comparison of pulsed-field gel electrophoresis and amplified fragment-length polymorphism for epidemiological investigations of common nosocomial pathogens. Infect. Control Hosp. Epidemiol. 22:550-554. [DOI] [PubMed] [Google Scholar]
- 7.Denis, O., A. Deplano, C. Nonhoff, R. De Ryck, R. de Mendonca, S. Rottiers, R. Vanhoff, and M. J. Struelens. 2004. National surveillance of methicillin-resistant Staphylococcus aureus in Belgian hospitals indicates rapid diversification of epidemic clones. Antimicrob. Agents Chemother. 48:3625-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denis, O., J. Magdalena, A. Deplano, C. Nonhoff, E. Hendrickx, and M. J. Struelens. 2002. Molecular epidemiology of resistance to macrolides-lincosamides-streptogramins in methicillin-resistant Staphylococcus aureus (MRSA) causing bloodstream infections in patients admitted to Belgian hospitals. J. Antimicrob. Chemother. 50:755-757. [DOI] [PubMed] [Google Scholar]
- 9.Deplano, A., A. Schuermans, J. Van Eldere, W. Witte, H. Meugnier, J. Etienne, H. Grundmann, D. Jonas, G. T. Noordhoek, J. Dijkstra, A. van Belkum, W. van Leeuwen, P. T. Tassios, N. J. Legakis, A. van der Zee, A. Bergmans, D. S. Blanc, F. C. Tenover, B. C. Cookson, G. O'Neil, M. J. Struelens, and The European Study Group on Epidemiological Markers of the ESCMID. 2000. Multicenter evaluation of epidemiological typing of methicillin-resistant Staphylococcus aureus strains by repetitive-element PCR analysis. J. Clin. Microbiol. 38:3527-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deplano, A., M. Vaneechoutte, G. Verschraegen, and M. J. Struelens. 1997. Typing of Staphylococcus aureus and Staphylococcus epidermidis strains by PCR analysis of inter-IS256 spacer length polymorphisms. J. Clin. Microbiol. 35:2580-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deplano, A., W. Witte, W. J. van Leeuwen, Y. Brun, and M. J. Struelens. 2000. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighboring countries. Clin. Microbiol. Infect. 6:239-245. [DOI] [PubMed] [Google Scholar]
- 12.Descheemaeker, P., C. Lammens, B. Pot, P. Vandamme, and H. Goossens. 1997. Evaluation of arbitrarily primed PCR analysis and pulsed-field gel electrophoresis of large genomic DNA fragments for identification of enterococci important in human medicine. Int. J. Syst. Bacteriol. 47:555-561. [DOI] [PubMed] [Google Scholar]
- 13.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fry, N. K., B. Afshar, P. Visca, D. Jonas, J. Duncan, E. Nebuloso, A. Underwood, and T. G. Harrison. 2005. Assessment of fluorescent amplified fragment length polymorphism analysis for epidemiological genotyping of Legionella pneumophila serogroup 1. Clin. Microbiol. Infect. 11:704-712. [DOI] [PubMed] [Google Scholar]
- 15.Fry, N. K., J. M. Bangsborg, A. Bergmans, S. Bernander, J. Etienne, L. Franzin, V. Gaia, P. Hasenberger, J. B. Baladron, D. Jonas, D. Lindsay, S. Mentula, A. Papoutsi, M. Struelens, S. A. Uldum, P. Visca, W. Wannet, and T. G. Harrison. 2002. Designation of the European Working Group on Legionella Infection (EWGLI) amplified fragment length polymorphism types of Legionella pneumophila serogroup 1 and results of intercentre proficiency testing using a standard protocol. Eur. J. Clin. Microbiol. Infect. Dis. 21:722-728. [DOI] [PubMed] [Google Scholar]
- 16.Gilot, P., G. Lina, T. Cochard, and B. Poutrel. 2002. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hageman, J. C., S. K. Fridkin, J. M. Mohammed, C. D. Steward, R. P. Gaynes, and F. C. Tenover. 2003. Antimicrobial proficiency testing of National Nosocomial Infections Surveillance System hospital laboratories. Infect. Control Hosp. Epidemiol. 24:356-361. [DOI] [PubMed] [Google Scholar]
- 18.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter, P. R. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koeleman, J. G., J. Stoof, D. J. Biesmans, P. H. Savelkoul, and C. M. Vandenbroucke-Grauls. 1998. Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J. Clin. Microbiol. 36:2522-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murchan, S., M. E. Kaufmann, A. Deplano, R. De Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson, L. R., and G. A. Noskin. 2001. New technology for detecting multidrug-resistant pathogens in the clinical microbiology laboratory. Emerg. Infect. Dis. 7:306-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Struelens, M. J. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, et al.. 1997. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect. Control Hosp. Epidemiol. 18:426-439. [DOI] [PubMed] [Google Scholar]
- 28.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenover, F. C., M. J. Mohammed, J. Stelling, T. O'Brien, and R. Williams. 2001. Ability of laboratories to detect emerging antimicrobial resistance: proficiency testing and quality control results from the World Health Organization's external quality assurance system for antimicrobial susceptibility testing. J. Clin. Microbiol. 39:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Belkum, A., J. Kluytmans, W. van Leeuwen, R. Bax, W. Quint, E. Peters, A. Fluit, C. Vandenbroucke-Grauls, A. van den Brule, and H. Koeleman. 1995. Multicenter evaluation of arbitrarily primed PCR for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 33:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Forey, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O'Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. El Solh, R. De Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Leeuwen, W. B., S. Snoeijers, C. Werken-Libregts, A. Tuip, A. van der Zee, D. Egberink, M. de Proost, E. Bik, B. Lunter, J. Kluytmans, E. Gits, I. van Duyn, M. Heck, K. van der Zwaluw, W. Wannet, G. T. Noordhoek, S. Mulder, N. Renders, M. Boers, S. Zaat, D. van der Riet, M. Kooistra, A. Talens, L. Dijkshoorn, T. van der Reyden, D. Veenendaal, N. Bakker, B. Cookson, A. Lynch, W. Witte, C. Cuny, D. Blanc, I. Vernez, W. Hryniewicz, J. Fiett, M. Struelens, A. Deplano, J. Landegent, H. A. Verbrugh, and A. van Belkum. 2002. Intercenter reproducibility of binary typing for Staphylococcus aureus. J. Microbiol. Methods 51:19-28. [DOI] [PubMed] [Google Scholar]