Abstract
During 2004 and 2005, cholera was recorded in 15 states of India, with 7 outbreaks. The newly emerged Vibrio cholerae O1 Inaba had a different antibiogram and ribotype, different pulsotypes, and different mutations in the wbeT gene. Due to the absence of serogroup O139, the Inaba serotype may have acquired the potential to affect the population at large.
Cholera continues to be a growing concern in most developing countries. Since the emergence of Vibrio cholerae O139, the incidence patterns of serogroup O1 have been constantly changing in the Indian subcontinent (12, 14). During 2004 and 2005, we investigated outbreaks of cholera in Delhi, Madhya Pradesh, and West Bengal. V. cholerae O1 isolates from outbreaks and those isolates received from four other states of India were included in this study (Table 1). From May 2004 to July 2005, sporadic cases of cholera were reported from 10 other areas (Table 1). To our knowledge, the emergence of the Inaba serotype and its spread in many parts of India, mostly in the form of outbreaks, were first detected during this time.
TABLE 1.
Vibrio cholerae isolates
| Place/state of isolation | Mo and yr of isolation | No. of V. cholerae isolates
|
|||
|---|---|---|---|---|---|
| Ogawa | Inaba | Total | |||
| Delhia/Delhi | May 2004 | 36 | 69 | 105 | |
| Ludhiana/Punjab | May 2004 | 23 | 25 | 48 | |
| Tripuraa/Assam | May 2004 | 6 | 6 | ||
| Bhinda/Madhya Pradesh | May 2004 | 2 | 2 | ||
| Chandigarh | July 2004 | 1 | 1 | 2 | |
| Madurai/Tamil Nadu | July 2004 | 25 | 1 | 26 | |
| Utter Pradesh | September 2004 | 6 | 6 | ||
| Haryana | September 2004 | 5 | 5 | ||
| Trivandrum/Kerala | October 2004 | 2 | 2 | ||
| Ahmedabad/Gujarat | October 2004, May 2005 | 29 | 27 | 56 | |
| Goa | February 2005 | 7 | 9 | 16 | |
| Kolkataa/West Bengal | April 2005 | 4 | 4 | ||
| Berhampura/Orissa | April 2005 | 4 | 4 | ||
| Chennai/Tamil Nadu | July 2005 | 44 | 3 | 47 | |
| Hyderabad/Andra Pradesh | July 2005 | 34 | 34 | ||
| Manipala/Karnataka | July 2005 | 9 | 18 | 27 | |
| Alleppya/Kerala | November 2005 | 12 | 12 | ||
| Total | 174 | 228 | 402 | ||
Cholera outbreak-affected area.
Stool specimens collected from diarrheal patients were processed for common enteric pathogens following standard methods. V. cholerae isolates were grown on thiosulfate citrate bile salts sucrose agar (Eiken Chemical Co. Ltd., Tokyo, Japan) at 37°C for 16 to 18 h and confirmed using polyvalent and monospecific antisera (Denka Seiken, Tokyo, Japan). The Clinical and Laboratory Standards Institute (formerly NCCLS) antimicrobial susceptibility test was adapted for V. cholerae O1 (3) with commercially available disks (Becton Dickinson Co., Sparks, MD). All of the V. cholerae O1 isolates were typed with El Tor phages (2).
The presence of the cholera toxin gene (ctxA) and tcpA variants (the major structural subunit gene of the toxin-coregulated pilus) of both biotypes were determined by a multiplex PCR assay (4). Uniplex PCR was performed for the amplification of wbeT, encoding the somatic antigen synthesis region (15), and rstR gene alleles, encoding regulation of the lysogeny of the CTX phage (12), in a standard PCR mixture. For ribotyping, BglI-digested chromosomal DNAs from the representative isolates were transferred to Hybond N+ membranes (Amersham International PLC, Buckinghamshire, England), and hybridization was done with the 7.5-kb BamHI fragment as a probe from plasmid pKK3535 (16) (ECL Nucleic Acid Detection System; Amersham). Pulsed-field gel electrophoresis (PFGE) was performed as described previously (20).
For wbeT mutation analysis, the 902-bp PCR products were purified (QIAGEN [Hilden, Germany] PCR purification kit), and sequencing was done with a Big Dye Terminator Cycle Sequencing kit (Applied Biosystems) using an ABI PRISM 3100 DNA sequencer (Applied Biosystems). The nucleotide and deduced protein sequences were analyzed with DNASIS (Hitachi, Yokohama, Japan), DNASTAR (DNA Star Inc., Madison, WI), and GenBank via the BLAST network.
We analyzed 402 V. cholerae O1 isolates from 15 states collected during 2004 and 2005. Among these, 43.3 and 56.7% of the isolates were identified as Ogawa and Inaba serotypes, respectively (Table 1). Serotype Inaba was exclusively identified in five cholera outbreaks (Table 1). Even though V. cholerae O1 Inaba was found to coexist with the Ogawa serotype (10, 19), the prevalence of the latter was consistent in many regions where cholera is endemic (11, 17). Outbreaks of cholera exclusively caused by Inaba were reported in a few countries (7, 18). Cycles of serotype switching lasting between 2 and 8 years have been reported in Bangladesh (10). Periodic shifting between V. cholerae O1 Ogawa and O139 was observed in India from 1994 to 2000 (14). We assume that, due to the absence of O139 from 2000 to 2004, the O1 Inaba serotype perhaps reemerged in India in a high proportion.
Almost all of the Ogawa, as well as Inaba, isolates belonged to phage type T4 and type 27 with a new set of phages (2), and these types prevailed in India for many years (17). Compared to Ogawa, the Inaba isolates were resistant to chloramphenicol and streptomycin. The pattern of susceptibility of V. cholerae isolates to these two antimicrobials has changed over time (6). A majority (91%) of the Inaba isolates showed reduced susceptibility to ciprofloxacin, whereas 32% of the Ogawa isolates remained resistant to the drug. Increase in resistance to ciprofloxacin is a cause for concern, as this drug is extensively used in India for the treatment of diarrhea (5).
The rstR region is classified into rstRclass, rstRET, and rstRCalc for classical, El Tor, and O139 alleles, respectively (9). Interestingly, genetic hybrids of the El Tor and classical biotypes, which lack common phenotypic traits, have been detected in Mozambique and Bangladesh (1, 12). When they were screened for rstR alleles by PCR, we found that all the recent isolates were of the rstRET type. In addition, phage typing and polymyxin B susceptibility results confirmed our isolates as El Tor biotype.
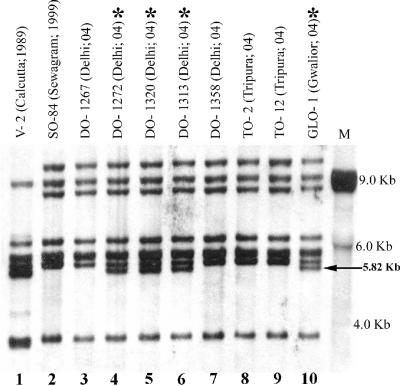
Up to 1993, three ribotypes (RI through RIII) were identified in the V. cholerae O1 serogroup (16). The RII and RIII ribotypes were identified after the emergence of V. cholerae O139 (16), and these two types were not recorded in the previous scheme (13). Most of the recent Inaba isolates from different states of India were identified as a new ribotype, RIV, and the Ogawa isolates during the same period were identified as RIII (Fig. 1). Prevalence of ribotype RIII was also detected in some of the 2004 Inaba isolates.
FIG. 1.
Ribotypes of the representative V. cholerae O1 Inaba isolates using BglI. The place and year of isolation are given in parentheses. The isolates indicated with asterisks belong to ribotype RIV. M, molecular size marker.
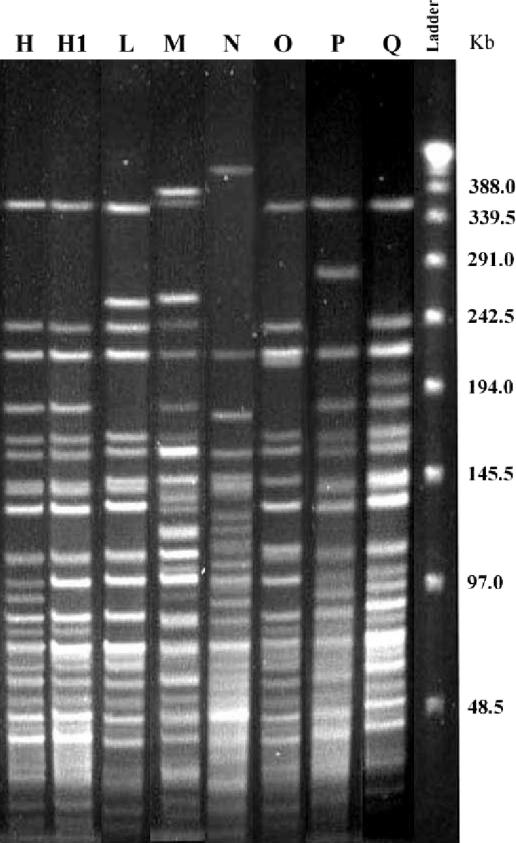
Twenty-seven V. cholerae isolates (20 Inaba and 7 Ogawa) were tested following the PFGE typing scheme (20), which consisted of 11 pulsotypes (A through K). The majority of the Inaba isolates encountered in this study belonged to the “H” type (12 isolates) or its subtype “H1” (Fig. 2). The H pulsotype was dominant among V. cholerae O1 isolates and was identified in India in July 1993. Six new pulsotypes (L through Q) were identified in this study (Fig. 2). Such an assortment of pulsotypes of V. cholerae O1 in a span of 2 years has not been reported before.
FIG. 2.
PFGE profiles generated with NotI-digested genomic DNAs of V. cholerae O1 isolates. The new pulsotypes H1, M, and P and L, N, and O were detected among Inaba and Ogawa isolates, respectively.
Mutations in the wbeT gene were responsible for serotype conversion in V. cholerae O1 (8). The DNA sequence analysis revealed that wbeT was homologous in Inaba isolates from Tripura, Madurai, Ludhiyana, Ahemedabad, and Kolkata. In all of these isolates, a novel mutation (substitution of C for T at position 538) was detected, which changed serine to proline. More molecular studies are warranted to confirm our findings in relation to the epidemiology of cholera with recent Inaba serotype isolates.
Nucleotide sequence accession number.
The nucleotide sequence of wbeT has been deposited in the NCBI database under accession number DQ401028.
Acknowledgments
We acknowledge Kasturba Medical College, Mannipal; Trivandrum Medical College, Trivandrum; T. D. Medical College, Alleppey; Sheth V. S. General Hospital, Ahmedabad; Sir Ronald Ross Institute of Tropical and Communicable Diseases, Hyderabad; M. K. C. G. Medical College Hospital, Berhampur; Madurai Medical College, Madurai; Christian Medical College, Ludhiana; Communicable Diseases Hospital, Chennai; IGM Hospital, Tripura; and Goa Medical College, Punjim, for sending the V. cholerae isolates.
The work was supported in part by the Indian Council of Medical Research, Japan International Cooperation Agency (JICA/NICED Project 054-1061-E-O), and the Ministry of Health, Labor and Family Welfare of Japan (Project H17-Shinkou-3).
REFERENCES
- 1.Ansaruzzaman, M., N. A. Bhuiyan, G. B. Nair, D. A. Sack, M. Lucas, J. L. Dean, J. Ampuero, C.-L. Chaignat, and The Mozambique Cholera Vaccine Demonstration Project Coordination Group. 2004. Cholera in Mozambique, variant of Vibrio cholerae. Emerg. Infect. Dis. 10:2057-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chattopadhyay, D. J., B. L. Sarkar, M. Q. Ansari, B. K. Chakrabarti, M. K. Roy, A. N. Ghosh, and S. C. Pal. 1993. New phage typing scheme for Vibrio cholerae O1 biotype El Tor strains. J. Clin. Microbiol. 31:1579-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial testing. 15th informational supplement M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 4.De, K., T. Ramamurthy, A. C. Ghose, M. S. Islam, Y. Takeda, G. B. Nair, and R. K. Nandy. 2001. Modification of the multiplex PCR for ambiguous difference of the El Tor and classical biotypes of Vibrio cholerae O1. Indian J. Med. Res. 114:77-82. [PubMed] [Google Scholar]
- 5.Garg, P., S. Sinha, R. Chakraborty, S. K. Bhattacharya, G. B. Nair, T. Ramamurthy, and Y. Takeda. 2001. Emergence of fluoroquinolone-resistance strains of Vibrio cholerae O1 biotype El Tor among hospitalized patients with cholera in Calcutta, India. Antimicrob. Agents Chemother. 45:1605-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garg, P., S. Chakraborty, I. Basu, S. Datta, K. Ranjendran, T. Bhattacharya, S. Yamasaki, S. K. Bhattacharya, Y. Takeda, G. B., Nair, and T. Ramamurthy. 2000. Expanding multiple antibiotic resistance among clinical strains of Vibrio cholerae isolated from 1992-7 in Calcutta, India. Epidemiol. Infect. 124:393-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garg, P., R. K. Nandy, P. Chaudhury, N. R. Chowdhury, K. De, T. Ramamurthy, S. Yamasaki, S. K. Bhattacharya, Y. Takeda, and G. B. Nair. 2000. Emergence of Vibrio cholerae O1 biotype El Tor serotype Inaba from the prevailing O1 Ogawa serotype strains in India. J. Clin. Microbiol. 38:4249-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito, T., K. Hiramatsu, Y. Ohshita, and T. Yokota. 1993. Mutations in the rfbT gene are responsible for the Ogawa to Inaba serotype conversion in Vibrio cholerae O1. Microbiol. Immunol. 37:281-288. [DOI] [PubMed] [Google Scholar]
- 9.Kimsey, H. H., G. B. Nair, A. Ghose, and M. K. Waldor. 1998. Diverse CTXφ and evolution of new pathogenic Vibrio cholerae. Lancet 352:457-458. [DOI] [PubMed] [Google Scholar]
- 10.Longini, I. M., Jr., M. Yunus, K. Zaman, A. K. Siddique, R. B. Sack, and A. Nizam. 2002. Epidemic and endemic cholera trends over a 33-year period in Bangladesh. J. Infect. Dis. 186:246-251. [DOI] [PubMed] [Google Scholar]
- 11.Mishra, M., F. Mohammed, S. L. Akulwar, V. J. Katkar, N. S. Tankhiwale, and R. M. Powar. 2004. Re-emergence of El Tor Vibrio in outbreak of cholera in and around Nagpur. Indian J. Med. Res. 120:478-480. [PubMed] [Google Scholar]
- 12.Nair, G. B., S. M. Faruque, N. A. Bhuiyan, M. Kamruzzaman, A. K. Siddique, and D. A. Sack. 2002. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J. Clin. Microbiol. 40:3296-3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popovic, T., C. Bopp, O. Olsvik, and K. Wachsmuth. 1993. Epidemiologic application of a standardized ribotyping scheme of Vibrio cholerae O1. J. Clin. Microbiol. 31:2474-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramamurthy, T., S. Yamasaki, Y. Takeda, and G. B. Nair. 2003. Vibrio cholerae O139 Bengal: odyssey of a fortuitous variant. Microbes Infect. 5:329-344. [DOI] [PubMed] [Google Scholar]
- 15.Rijpkema, S. G., Z. Durrani, T. Ramamurthy, and G. B. Nair. 2004. Assessing clonality of Vibrio cholerae Inaba isolates by characterization of nonsense mutations in wbeT. J. Med. Microbiol. 53:1105-1107. [DOI] [PubMed] [Google Scholar]
- 16.Sharma, C., G. B. Nair, A. K. Mukhopadhyay, S. K. Bhattacharya, R. K. Ghosh, and A. Ghosh. 1997. Molecular characterization of Vibrio cholerae O1 biotype El Tor strains isolated between 1992 and 1995 in Calcutta, India: evidence for the emergence of a new clone of El Tor biotype. J. Infect. Dis. 175:1134-1141. [DOI] [PubMed] [Google Scholar]
- 17.Sundaram, S. P., J. Revathi, B. L. Sarkar, and S. K. Bhattacharya. 2002. Bacteriological profile of cholera in Tamil Nadu. Indian J. Med. Res. 116:258-263. [PubMed] [Google Scholar]
- 18.Swerdlow, D. L., G. Malenga, G. Begkoyian, D. Nyangulu, M. Toole, R. J. Waldman, D. N. Puhr, and R. V. Tauxe. 1997. Epidemic cholera among refugees in Malawi, Africa: treatment and transmission. Epidemiol. Infect. 118:207-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verenkar, M., R. Savio, N. Venkatesh, M. J. Pinto, and I. Singh. 1994. Cholera in Goa. Indian J. Pathol. Microbiol. 37:289-292. [PubMed] [Google Scholar]
- 20.Yamasaki, S., G. B. Nair, S. K. Bhattacharya, S. Yamamoto, H. Kurazono, and Y. Takeda. 1997. Cryptic appearance of a new clone of Vibrio cholerae O1 biotype El Tor in Calcutta, India. Microbiol. Immunol. 41:1-6. [DOI] [PubMed] [Google Scholar]