Abstract
We identified three encapsulated Bacillus cereus strains, isolated from patients with severe pneumonia, in a collection of B. cereus isolates associated with human illness. We found that the extent of capsule expression was influenced by culturing conditions. Our findings highlight consequent clinical and laboratory diagnostic challenges posed by such isolates.
Bacillus cereus is a gram-positive, spore-forming, opportunistic pathogen (5, 13). Although it has been implicated in severe respiratory infections (9, 12), the majority of reported illnesses involving B. cereus are food-borne intoxications (5, 6, 11). Phylogenetically, B. cereus is closely related to both Bacillus anthracis and Bacillus thuringiensis, and these species are commonly referred to as the B. cereus group (1, 3, 10). Classically these species are differentiated by multiple phenotypic tests (2). One recognized phenotype used to distinguish B. anthracis from B. cereus and B. thuringiensis is the presence of a poly-γ-d-glutamic acid capsule in B. anthracis (7); B. cereus and B. thuringiensis typically are not encapsulated (2).
Recently, Hoffmaster et al. reported a unique strain of encapsulated B. cereus (G9241) associated with severe pneumonia (9). The encapsulated cells of G9241 did not react with antibodies specific for the B. anthracis poly-γ-d-glutamic acid capsule and did not contain the capBCA operon, which encodes the enzymes necessary for the production of the B. anthracis capsule. However, a plasmid-encoded (pBC218), putative capsule biosynthesis operon was identified in G9241 that was hypothesized to encode genes necessary for capsule biosynthesis in this strain.
To examine the prevalence of encapsulated B. cereus strains associated with human disease, B. cereus isolates from the CDC's Special Bacteriology Reference Laboratory culture collection were screened using India ink staining to demonstrate the presence of capsule and PCR to detect putative capsule biosynthesis genes, which were identified in G9241 (9). A total of 45 B. cereus strains from the culture collection, which were isolated in the United States from 1954 to 2000, were included in this study. This convenient sample represents a spectrum of human illness associated with B. cereus: 21 isolates from 21 sporadic severe systemic human infections (atypical, unusual presentations) of the blood or lungs and 24 isolates from cases of gastrointestinal illness or from food remnants associated with food-borne outbreaks. B. cereus strains G9241 and ATCC 14579 were used throughout as positive and negative controls, respectively.
Isolates were grown on Trypticase soy agar plates containing 5% (vol/vol) sheep blood (SBA) (Becton Dickinson Microbiology Systems, Cockeysville, MD) overnight at 37°C in 5% CO2 and stained using India ink (Remel, Lenexa, KS) (9). Of 45 B. cereus isolates tested, only 1 B. cereus strain, G9898 (12), expressed capsule (Table 1).
TABLE 1.
Laboratory and epidemiologic findings associated with reported encapsulated B. cereus isolates
| Strain | State where isolated | Yr isolated | Occupation of patient | Capsule expression | PCR for pBC218 genesa | Reference |
|---|---|---|---|---|---|---|
| G9241 | LA | 1994 | Welder | + | + | 9 |
| G9898 | LA | 1996 | Welder | + | + | 12 |
| 03BB87 | TX | 2003 | Muller | + | + | 8 |
| 03BB102 | TX | 2003 | Welder | + | − | 8 |
PCR was performed to detect the putative capsule biosynthesis genes, polysaccharide polymerase and polysaccharide translocase.
The same 45 isolates were screened by PCR to detect the putative polysaccharide polymerase and translocase genes, hypothesized to be required for capsule biosynthesis in G9241 (9). DNA was isolated using the High Pure PCR template preparation kit (Roche Diagnostics, Indianapolis, IN), with lysozyme (100 mg/ml) to lyse the cells. Forward and reverse primers were designed to amplify 607 nucleotides of the putative polysaccharide polymerase (forward, 5′-TTAATAAAGTGGCCGTTACAT-3′; reverse, 5′-GTACAACAAATCCTGCGAATA-3′) and 432 nucleotides of the putative polysaccharide translocase (forward, 5′-AATTTGGGATGTATTCGATGT-3′; reverse, 5′-ATGAACTTACTTCCGCCAACA-3′) genes. PCR amplification was performed using Platinum Taq polymerase (Invitrogen, Foster City, CA). Amplicons were analyzed using E-gel agarose (2%) gel electrophoresis (Invitrogen). G9898 was the only B. cereus strain that contained the putative capsule biosynthesis genes.
Two additional B. cereus strains (03BB87 and 03BB102), identified after our study began (8), were also analyzed. B. cereus strains 03BB87 and 03BB102 were isolated from a welder and a muller, respectively, from two unrelated fatal pneumonia cases in Texas in 2003. A detailed report of these cases and the epidemiologic investigation is in preparation (S. B. Avashia, submitted for publication). Capsule was observed in both strains, using India ink staining (Table 1). When these two B. cereus strains were tested by PCR, the putative capsule biosynthesis genes were detected in strain 03BB87 only (Table 1). The failure to detect these capsule genes in 03BB102 may be due to slightly divergent sequences in primer binding regions, or capsule genes unrelated to these may be responsible for the capsule in this isolate.
We performed direct fluorescent-antibody staining for the detection of B. anthracis-specific capsule antigens to determine if the capsule of these B. cereus isolates contained poly-γ-d-glutamic acid (4). B. anthracis Pasteur (ATCC 4229) and B. cereus (ATCC 14579) were used as positive and negative controls, respectively, for capsule direct fluorescent-antibody assays. The three encapsulated B. cereus strains did not react with the poly-γ-d-glutamic acid capsule antibody following incubation at either 30°C or 37°C.
To determine if culturing conditions affected capsule expression, we evaluated capsule production for the three B. cereus strains G9898, 03BB87, and 03BB102 and the positive control, G9241, using India ink staining following growth on SBA plates incubated overnight at 30°C or 37°C in 5% or 12% CO2 atmosphere (four combinations) or in broth culture. Approximately 107 cells grown on SBA at 30°C in 5% CO2 were used to inoculate broth cultures, which contained the following: (i) 450 μl of heart infusion (HI) broth alone, (ii) HI broth containing 50% (vol/vol) heat-inactivated horse serum (Sigma, St. Louis, MO), (iii) HI broth containing 0.8% (wt/vol) sodium bicarbonate (ICN Biomedicals, Aurora, MO), or (iv) HI broth containing both 50% heat-inactivated horse serum and 0.8% sodium bicarbonate. Cultures were incubated without shaking at 30°C or 37°C in 5% CO2 for 3 h, and pelleted cells were stained by India ink (9). Tests were performed in triplicate. The level of capsule production varied for the three strains according to culturing conditions and the media used (Table 2; Fig. 1). After observing variation in capsule expression from these three strains, we retested 10 of the nonencapsulated strains from the original collection in HI plus horse serum and HI plus sodium bicarbonate broths at 30°C and 37°C. None of these strains formed a capsule under these conditions.
TABLE 2.
Capsule production by B. cereus strains grown under various culturing conditions
| Growth conditionsa | Growth of B. cereus strainb
|
||||
|---|---|---|---|---|---|
| G9898 | 03BB87 | 03BB102 | G9241 (+ ctrl) | 14579 (− ctrl) | |
| SBA plates | |||||
| 30°C (5% CO2) | V | V | − | + | − |
| 30°C (12% CO2) | − | − | − | + | − |
| 37°C (5% CO2) | V | V | V | + | − |
| 37°C (12% CO2) | − | − | + | + | − |
| Broth media | |||||
| 30°C (HI broth) | V | − | V | + | − |
| 30°C (HI + H. serum) | + | + | + | + | − |
| 30°C (HI + NaHCO3) | + | + | + | + | − |
| 30°C (HI + H. serum+ NaHCO3) | + | + | + | + | − |
| 37°C (HI broth) | − | − | V | + | − |
| 37°C (HI + H. serum) | − | − | + | + | − |
| 37°C (HI + NaHCO3) | − | − | + | + | − |
| 37°C (HI + H. serum + NaHCO3) | − | − | + | + | − |
H. serum, horse serum.
Each symbol represents three independent experiments, where each of the three test strains and two control strains was cultured under the designated growth conditions on three separate occasions and then observed by light microscopy at magnification ×100 after staining. +, capsule visualized; −, capsule not visualized; V, capsule visualized on less than 5% of cells observed; + ctrl, positive control; − ctrl, negative control.
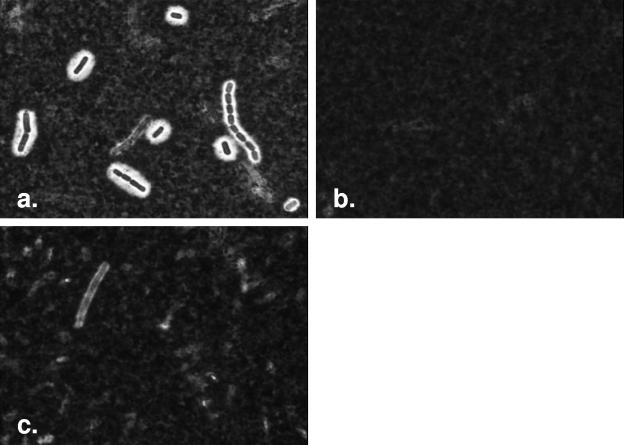
FIG. 1.
Culture conditions affecting capsule production in rare, encapsulated B. cereus strains. Microscopic images captured (at ×100) of B. cereus 03BB87 following incubation (a) at 30°C in HI plus horse serum plus sodium bicarbonate, (b) at 30°C in HI broth, and (c) at 30°C on SBA in 5% CO2. These images represent the degree of capsule formation observed with India ink staining of cells, noted in Table 1 as +, −, and V, respectively. Each represents three independent experiments, where each of the three test strains and two control strains was cultured under the various growth conditions.
Our findings demonstrate that capsule expression in B. cereus is rare; we observed capsules in only 3 of the 47 total B. cereus isolates we examined. Therefore, the total number of reported encapsulated B. cereus strains is four, including G9241. The clinical sources of these four strains are notably similar (9, 12). Each of the encapsulated B. cereus strains was implicated in either fatal (three strains) or near-fatal (one strain) pneumonia in individuals characterized as immunocompetent (9, 12). Furthermore, each of these individuals was a metal worker and resided in either Texas or Louisiana (9, 12).
Discovery of these encapsulated B. cereus strains shows that the distinctions classically used to define B. cereus group organisms may not always be clear-cut, highlighting the danger of relying exclusively upon a single phenotypic trait to characterize an isolate and emphasizing the importance of using multiple tests when identifying isolates associated with human disease.
Our findings demonstrate the challenges for clinical and public health laboratories charged with the identification and differentiation of B. cereus and B. anthracis. With an increased awareness of the importance of rapid and accurate identification of B. anthracis in today's world, the discovery of several capsule-producing B. cereus strains, all isolated from patients with severe pneumonia, also emphasizes the need for a better understanding of the prevalence and risk factors associated with the diseases caused by these organisms.
Acknowledgments
We acknowledge Leta Helsel of the Special Bacteriology Reference Laboratory, CDC, for her help in providing the isolates used in this study.
This research was supported in part by an appointment to the Emerging Infectious Diseases Fellowship Program administered by the Association for Public Health Laboratories and funded by the Centers for Disease Control and Prevention (CDC), awarded to D.S.
REFERENCES
- 1.Ash, C., J. A. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 2.Bergey, D. H., J. G. Holt, and N. R. Krieg. 1984. Bergey's manual of systematic bacteriology, 1st ed. Williams & Wilkins, Baltimore, Md.
- 3.Daffonchio, D., A. Cherif, and S. Borin. 2000. Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S-23S internal transcribed spacers describe genetic relationships in the “Bacillus cereus group." Appl. Environ. Microbiol. 66:5460-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De, B. K., S. L. Bragg, G. N. Sanden, K. E. Wilson, L. A. Diem, C. K. Marston, A. R. Hoffmaster, G. A. Barnett, R. S. Weyant, T. G. Abshire, J. W. Ezzell, and T. Popovic. 2002. A two-component direct fluorescent-antibody assay for rapid identification of Bacillus anthracis. Emerg. Infect. Dis. 8:1060-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granum, P. E., and T. Lund. 1997. Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Lett. 157:223-228. [DOI] [PubMed] [Google Scholar]
- 7.Green, B. D., L. Battisti, T. M. Koehler, C. B. Thorne, and B. E. Ivins. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmaster, A. R., K. K. Hill, J. E. Gee, C. K. Marston, B. K. De, T. Popovic, D. Sue, P. P. Wilkins, S. B. Avashia, R. Drumgoole, C. H. Helma, L. O. Ticknor, R. T. Okinaka, and P. J. Jackson. 2006. Characterization of Bacillus cereus isolates associated with fatal pneumonias: isolates are closely related to Bacillus anthracis and harbor B. anthracis virulence genes. J. Clin. Microbiol. 44:3352-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmaster, A. R., J. Ravel, D. A. Rasko, G. D. Chapman, M. D. Chute, C. K. Marston, B. K. De, C. T. Sacchi, C. Fitzgerald, L. W. Mayer, M. C. Maiden, F. G. Priest, M. Barker, L. Jiang, R. Z. Cer, J. Rilstone, S. N. Peterson, R. S. Weyant, D. R. Galloway, T. D. Read, T. Popovic, and C. M. Fraser. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. USA 101:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaneko, T., R. Nozaki, and K. Aizawa. 1978. Deoxyribonucleic acid relatedness between Bacillus anthracis, Bacillus cereus and Bacillus thuringiensis. Microbiol. Immunol. 22:639-641. [DOI] [PubMed] [Google Scholar]
- 11.Logan, N. A., and P. C. Turnbull. 2003. Bacillus and related genera, p. 445-460. In P. R. Murray and E. J. Baron (ed.), Manual of clinical microbiology, 8th ed., vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 12.Miller, J. M., J. G. Hair, M. Hebert, L. Hebert, F. J. Roberts, Jr., and R. S. Weyant. 1997. Fulminating bacteremia and pneumonia due to Bacillus cereus. J. Clin. Microbiol. 35:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbull, P. C., K. Jorgensen, J. M. Kramer, R. J. Gilbert, and J. M. Parry. 1979. Severe clinical conditions associated with Bacillus cereus and the apparent involvement of exotoxins. J. Clin. Pathol. 32:289-293. [DOI] [PMC free article] [PubMed] [Google Scholar]