Abstract
The usefulness of Candida ID 2 (CAID2) reformulated medium (bioMérieux, France) has been compared with that of the former Candida ID (CAID; bioMérieux), Albicans ID 2 (ALB2; bioMérieux), and CHROMagar Candida (CAC; Chromagar, France) chromogenic media for the isolation and presumptive identification of clinically relevant yeasts. Three hundred forty-five stock strains from culture collections, and 103 fresh isolates from different clinical specimens were evaluated. CAID2 permitted differentiation based on colony color between Candida albicans (cobalt blue; sensitivity, 91.7%; specificity, 97.2%) and Candida dubliniensis (turquoise blue; sensitivity, 97.9%; specificity, 96.6%). Candida tropicalis gave distinguishable pink-bluish colonies in 97.4% of the strains in CAID2 (sensitivity, 97.4%; specificity, 100%); the same proportion was reached in CAC, where colonies were blue-gray (sensitivity, 97.4%; specificity, 98.7%). CAC and CAID2 showed 100% sensitivity values for the identification of Candida krusei. However, with CAID2, experience is required to differentiate the downy aspect of the white colonies of C. krusei from other white-colony-forming species. The new CAID2 medium is a good candidate to replace CAID and ALB2, and it compares well to CAC for culture and presumptive identification of clinically relevant Candida species. CAID2 showed better results than CAC in some aspects, such as quicker growth and color development of colonies from clinical specimens, detection of mixed cultures, and presumptive differentiation between C. albicans and C. dubliniensis.
Candida albicans is the Candida species most frequently isolated from patients with candidiasis. However, other species with more reduced susceptibility to antifungal agents, such as Candida parapsilosis, Candida tropicalis, Candida glabrata, Candida krusei, and Candida guilliermondii, are steadily increasing their isolation frequency (16, 20). Identification of Candida isolates to the species level is required to aid the selection of the appropriate antifungal agent for treatment of invasive candidiasis and other severe Candida infections (22, 27). As identification of yeasts may take several days, the use of available chromogenic media may help to reduce the time for isolation and identification as well as to detect the presence of mixed cultures (11, 29).
There are several chromogenic media available for the isolation and presumptive identification of C. albicans based on the pigmentation of the developing colonies, which is due to different enzyme activities among Candida species (2, 17, 21). CHROMagar Candida (CAC) (CHROMagar Microbiology, Paris, France) shows different color colonies for C. albicans (green), C. tropicalis (dark blue, with a pink halo), and C. krusei (pink and downy appearance) (17, 24). Some authors have also indicated that CAC is useful for identification of C. glabrata (typical purple color colonies) (3, 19) and Candida dubliniensis (typical dark green colonies) (11, 26). However, other researchers did not agree with these findings (2, 13, 24).
Albicans ID 2 (ALB2) (bioMérieux, Marcy l'Étoile, France) allows the specific identification of C. albicans colonies on the basis of their blue color and smooth appearance (2, 4) but does not differentiate other species of Candida. Candida ID (CAID) (bioMérieux) differentiates three groups of yeasts based on the color of colonies: blue colonies for C. albicans, pink colonies for C. tropicalis, C. guilliermondii, Candida kefyr, and Candida lusitaniae, and white colonies for the rest of the species (14, 21, 28).
Candida ID 2 chromogenic medium (CAID2) (bioMérieux) is an improved version of CAID that is being marketed to replace the previous formulae, CAID and ALB2. CAID2 has been developed for the identification of C. albicans (blue colonies). Additionally, the isolates from the species C. tropicalis, C. lusitaniae, and C. kefyr grow like pink colonies on CAID2.
The present study has been designed to evaluate the performance of CAID2 in the isolation and presumptive identification of medically important yeasts and to compare the results to those obtained using CAID, ALB2, and CAC. For this purpose, we have studied 448 strains from our stock collection of yeasts and from fresh human clinical specimens.
MATERIALS AND METHODS
Strains.
A total of 448 strains from the genera Candida, Cryptococcus, Geotrichum, Saccharomyces, and Trichosporon have been evaluated (Table 1). The 345 collection and stock strains were obtained from the National Collection of Pathogenic Fungi (NCPF, Bristol, United Kingdom), the American Type Culture Collection (ATCC, Manassas, Va.), and the collection of the Departamento de Inmunología, Microbiología y Parasitología of the Universidad del País Vasco (UPV, Bilbao, Spain).
TABLE 1.
Origin and distribution of yeast strains
| Species | No. of:
|
||
|---|---|---|---|
| Total strains | Stock strains | Clinical fresh isolatesh | |
| Candida albicansa | 168 | 96 | 72 |
| Candida dubliniensisb | 51 | 48 | 3 |
| Candida famata | 2 | 2 | 0 |
| Candida glabratac | 45 | 39 | 6 |
| Candida guilliermondiid | 22 | 20 | 2 |
| Candida haemulonii | 10 | 10 | 0 |
| Candida kefyr | 2 | 2 | 0 |
| Candida kruseie | 26 | 23 | 3 |
| Candida lipolytica | 3 | 3 | 0 |
| Candida lusitaniae | 11 | 10 | 1 |
| Candida parapsilosisf | 35 | 28 | 7 |
| Candida rugosa | 2 | 2 | 0 |
| Candida tropicalisg | 48 | 39 | 9 |
| Cryptococcus neoformans | 12 | 12 | 0 |
| Geotrichum capitatum | 2 | 2 | 0 |
| Saccharomyces cerevisiae | 7 | 7 | 0 |
| Trichosporon spp. | 2 | 2 | 0 |
| Total | 448 | 345 | 103 |
Including ATCC 20408, ATCC 64548, ATCC 90029, and NCPF 3153.
Including NCPF 3949.
Including ATCC 90030 and NCPF 3240.
Including NCPF 3099.
Including ATCC 6258, NCPF 3100, and NCPF 3321.
Including ATCC 22019.
Including NCPF 3111.
All were oral isolates except 2 vaginal C. albicans isolates.
Fresh oral or vaginal isolates were obtained from clinical specimens of patients attending Odontology and Gynecology clinics collaborating with the UPV. The identity of clinical isolates was confirmed by conventional mycological methods (7, 8), such as the germ tube induction test in serum, microscopic morphology, and chlamydospore formation in corn meal agar (Oxoid, Basingstoke, United Kingdom) with Tween 80 and carbon source assimilation by ID 32C (bioMérieux). If necessary, identification was confirmed by PCR with specific primers (12, 15).
Chromogenic media.
ALB2, CAID, and CAID2 agar plates were donated by bioMérieux Spain, while CAC plates were prepared in the laboratory according to the manufacturer's instructions. All plates were stored at 4°C and left to reach room temperature prior to inoculation.
Inoculation of media.
Stock collection strains were grown on Sabouraud glucose agar plates (Difco, St. Louis, Mo.) at 37°C for 24 to 48 h prior to inoculation onto chromogenic media. One hundred eighty oral and vaginal specimens were directly streaked onto chromogenic media agar plates and incubated for up to 10 days at 37°C, the temperature recommended by the manufacturers. Plates were checked daily for growth and read independently by the same three investigators. The following parameters of growth were recorded for each strain in a side-by-side comparison of the four chromogenic agar media at 24, 48, and 72 h (lecture recommended at 24 and 48 h for the three bioMérieux's chromogenic media and at 48 h for CHROMagar Candida): number of positive cultures, number and macroscopic features (size, color, morphology, and texture) of colonies, and the presence of mixed cultures by detecting phenotypically different colonies.
To check the influence of the incubation temperature in the growth of the different species of yeasts on the chromogenic media, 10 μl of a yeast suspension that contained between 30 and 300 viable cells grown on Sabouraud agar plates at 37°C was spread on the agar plates, and the number, size, and color of the colonies were registered at different times of incubation (18, 24, 48, and 72 h) at 37°C or room temperature (18 to 20°C). Growth on Sabouraud glucose agar plates was used as a reference of viability and colony features.
RESULTS
After 24 h at 37°C, 94.8% of stock collection strains grew on CAID2, 94.2% on CAID, 92.8% on both, Albicans ID 2, and CAC. However, only CAID2 medium permitted the growth of all the strains (100%) after 48 h of incubation. Moreover, color and other characteristics of colonies were easier to read after 48 h of incubation at 37°C on CAID2. The slowest colony growth was observed on ALB2. Colonies grew earlier with a faster and stronger color on CAID2.
Table 2 shows the main characteristics of yeast colonies grown on the four chromogenic media studied. Nearly all isolates grew as colonies of the expected color on each chromogenic media. Only one isolate of C. albicans showed white colonies at 24 h on CAID or ALB2, and 2 strains did not grow on ALB2. On CAC, one strain gave white colonies and 3 strains did not grow at 24 h. Moreover, CAID2 allowed a clear differentiation between C. albicans, growing as cobalt blue colonies (91.7% of isolates), and C. dubliniensis, growing as turquoise blue colonies (97.9% of isolates). This colony differentiation between C. albicans and C. dubliniensis was clearer after 48 h of incubation (Fig. 1A and C). In this medium, the turquoise blue color of colonies had a sensitivity of 97.9% and a specificity of 96.6% for the identification of C. dubliniensis. This differentiation was also observed on CAID (sensitivity, 85.4%; specificity, 97%), but 4 C. dubliniensis isolates grew as white colonies at 48 h in this medium. No differences were observed between both species on CAC, where colonies were of different green tones. On ALB2, we could not find any differentiating characteristics between C. albicans and C. dubliniensis colonies, some strains of C. tropicalis and Candida rugosa also gave blue colonies. The latter species grew also as blue colonies on CAID and CAID2 (Table 2).
TABLE 2.
Identity and colony characteristics of 345 yeast isolates grown 24 to 48 h at 37°C on the chromogenic media Candida ID 2, Candida ID, Albicans ID 2, and CHROMagar Candida
| Species (no. of strains) | No. of strains with colony color after time (h) on medium:
|
|||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Candida ID 2
|
Candida ID
|
Albicans ID 2
|
CHROMagar
|
|||||||||||||||||||||||||||
| Blue (cobalt)
|
Blue (turquoise)
|
Pink
|
White
|
Blue (cobalt)
|
Blue (turquoise)
|
Pink
|
White
|
Blue
|
White
|
Green
|
Blue
|
Pink
|
Violet
|
Other
|
||||||||||||||||
| 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | |
| Candida albicans (96) | 96 | 88 | 0 | 8 | 0 | 0 | 0 | 0 | 95 | 89 | 0 | 7 | 0 | 0 | 1 | 0 | 93 | 96 | 1 | 0 | 92 | 96 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Candida dubliniensis (48) | 37 | 1 | 9 | 47 | 0 | 0 | 2 | 0 | 34 | 3 | 7 | 41 | 0 | 0 | 7 | 4 | 47 | 47 | 1 | 1 | 45 | 48 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Candida famata (2) | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 |
| Candida glabrata (39) | 0 | 0 | 0 | 0 | 0 | 0 | 39 | 39 | 0 | 0 | 0 | 0 | 0 | 0 | 38 | 39 | 0 | 0 | 39 | 39 | 0 | 0 | 0 | 0 | 0 | 0 | 38 | 38 | 1 | 1 |
| Candida guilliermondii (20) | 0 | 0 | 0 | 0 | 13 | 19 | 6 | 1 | 0 | 0 | 0 | 0 | 15 | 19 | 5 | 1 | 0 | 0 | 19 | 20 | 0 | 0 | 0 | 0 | 3 | 0 | 12 | 20 | 2 | 0 |
| Candida haemulonii (10) | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 4 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 10 | 0 | 0 |
| Candida kefyr (2) | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 1 |
| Candida krusei (23) | 0 | 0 | 0 | 0 | 0 | 0 | 22 | 23a | 0 | 0 | 0 | 0 | 0 | 0 | 22 | 23 | 0 | 0 | 22 | 23 | 0 | 0 | 0 | 0 | 23 | 23a | 0 | 0 | 0 | 0 |
| Candida lipolytica (3) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| Candida lusitaniae (10) | 0 | 0 | 0 | 0 | 9 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 9 | 0 | 1 | 0 | 0 | 10 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 8 | 0 | 2 |
| Candida parapsilosis (28) | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 28 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 28 | 0 | 0 | 28 | 28 | 0 | 0 | 0 | 0 | 1 | 0 | 18 | 22 | 9 | 6 |
| Candida rugosa (2) | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Candida tropicalis (39)b | 0 | 0 | 0 | 0 | 36 (21) | 38 (37) | 2 | 1 | 0 | 0 | 0 | 0 | 36 (2) | 38 (12) | 2 | 1 | 13 | 27 | 24 | 11 | 0 | 0 | 35 | 37 | 0 | 0 | 3 | 1 | 0 | 0 |
| Cryptococcus neoformans (12) | 3 | 3 | 0 | 0 | 2 | 5 | 6 | 4 | 0 | 2 | 0 | 0 | 0 | 1 | 10 | 9 | 0 | 1 | 8 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 8 | 2 | 4 |
| Geotrichum capitatum (2) | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 0 |
| Saccharomyces cerevisiae (7) | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 7 | 0 | 0 | 6 | 7 | 0 | 0 | 0 | 0 | 1 | 1 | 6 | 6 | 0 | 0 |
| Trichosporon spp. (2) | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 0 |
Downy aspect of colonies.
Number in parentheses are numbers of strains with pink-bluish colonies.
FIG. 1.
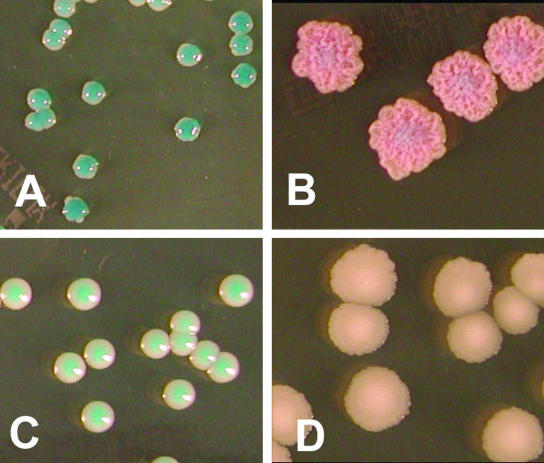
Colony aspect of Candida spp. grown on CAID2 at 37°C for 48 h. (A) Candida albicans; (B) Candida tropicalis; (C) Candida dubliniensis; (D) Candida krusei.
On CAID and CAID2, isolates of C. tropicalis (94.7%), C. guilliermondii (95%), Candida haemulonii (60%), C. kefyr (100%), C. lusitaniae (90% and 100%, respectively), and Cryptococcus neoformans (10% and 41.7%, respectively) grew as colonies with different pink shades. Pink-bluish colonies on CAID2 allowed a presumptive identification of 97.4% of C. tropicalis isolates at 48 h (Fig. 1B). In comparison, 94.9% of C. tropicalis isolates and 100% of C. krusei isolates grew on CAC, showing their characteristic blue or pink colonies, respectively. A downy white texture of C. krusei colonies was a differential feature for this species on CAID2 (Fig. 1D).
A total of 85 of 180 clinical specimens were positive for yeast. The recovery rates were equivalent on the four media. In 47 specimens, there were 5 or more colonies from a single yeast species distributed in the following manner: 47 were positive on CAID2, 47 on CAID, 47 on ALB2, and 45 on CAC (Table 3). Thirteen specimens were positive for two or more species (Table 4). However, only CAID2 medium revealed all cultures of mixed species (13 of 13, 100% efficiency). CAID detected 9 of 13, and ALB2 and CAC showed less than 50% efficiency (6 of 13). Detection of mixed populations was more difficult when a heavy presence of yeast was present in the specimen. Bacterial contamination was not a problem on any media tested.
TABLE 3.
Results for 180 clinical specimens on each chromogenic mediuma
| Fungal species | Total no. of isolates | No. of isolates with expected colony color after time (h) on medium:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Candida ID 2
|
Candida ID
|
Albicans ID 2
|
CHROMagar
|
||||||
| 24 | 48 | 24 | 48 | 24 | 48 | 24 | 48 | ||
| Candida albicans | 42 | 39b | 42 | 40c | 42 | 40c | 42 | 26d | 41 |
| Candida guilliermondii | 1 | 0c | 1 | 0c | 1 | 1 | 1 | 1 | 1 |
| Candida krusei | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 1 | 1 |
| Candida parapsilosis | 1 | 1 | 1 | 1 | 1 | ||||
| Candida tropicalis | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Total | 47 | 45 | 47 | 45 | 47 | 44 | 47 | 43 | 45 |
Only specimens yielding more than five colonies are shown.
Two specimens yielded white colonies.
One specimen yielded white colonies.
Fourteen specimens yielded white colonies.
TABLE 4.
Detection of multiple yeast species in fresh clinical specimens when inoculated on Candida ID 2, Candida ID, Albicans ID 2, or CHROMagar after 48 h of incubation at 37°C
| Species (no. of occurrences) | No. of mixed cultures detected on medium:
|
|||
|---|---|---|---|---|
| Candida ID 2 | Candida ID | Albicans ID 2 | CHROMagar | |
| C. albicans + C. dubliniensis (1) | 1 | 0 | 0 | 0 |
| C. albicans + C. glabrata (2) | 2 | 2 | 2 | 2 |
| C. albicans + C. tropicalis (4) | 4 | 4 | 4 | 4 |
| C. lusitaniae + C. parapsilosis (1) | 1 | 1 | 0 | 0 |
| C. albicans + C. glabrata + C. tropicalis (2) | 2 | 1 | 0 | 0a |
| C. albicans + C. krusei + C. tropicalis (1) | 1 | 1 | 0 | 0 |
| C. dubliniensis + C. glabrata + C. tropicalis (1) | 1 | 0 | 0 | 0a |
| C. dubliniensis + C. glabrata + C. parapsilosis (1) | 1b | 0 | 0 | 0c |
In mixed infections where growth was very abundant, C. tropicalis showed purple colonies on CAC.
C. glabrata and C. parapsilosis showed white colonies with different morphologies on CAID2.
In CAC, only two colony colors were appreciated.
One hundred three isolates were recovered from 180 fresh oral and vaginal specimens. C. albicans was the most frequently isolated species (72 isolates, 69.9%), followed by C. tropicalis (9 isolates, 8.74%), C. parapsilosis (7 isolates, 6.8%), C. glabrata (6 isolates, 4.83%), C. krusei and C. dubliniensis (3 isolates each, 2.91%), C. guilliermondii (2 isolates, 1.94%), and C. lusitaniae (1 isolate, 0.97%). All three of the C. dubliniensis isolates were obtained from oral specimens of patients with human immunodeficiency virus (HIV) infection in mixed populations with C. albicans, with both C. glabrata and C. parapsilosis, or with both C. glabrata and C. tropicalis (Table 4).
All chromogenic media had excellent sensitivity (97.8 and 100% at 24 h and 48 h, respectively), and most of them showed high specificity values (96.1% at 48 h) for the identification of germ tube-positive species (C. albicans or C. dubliniensis). However, ALB2 had lower specificity values (84.7%). Few isolates were registered as false positive or false negative, with the exception of C. tropicalis isolates: 27 of 39 C. tropicalis showed blue colonies on ALB2 medium, but this blue color was lighter and could be clearly distinguished from C. albicans isolates. On CAID medium, some C. rugosa and Trichosporon spp. strains showed the same blue color as strains of C. albicans or C. dubliniensis.
On CAID2, 97.4% of C. tropicalis strains developed pink-bluish colonies, while CAC had a similar sensitivity (94.9%) for the blue-gray color developed by this species. The specificity for C. tropicalis identification ranged between 98.7% for CAC and 100% for CAID2. Infrequent Candida spp. studied showed a whitish color, but the aspect of C. krusei colonies was rough, not brilliant; however, the latter was easier to distinguish in CAC, where colonies showed a rough pink aspect. Concerning C. krusei, CAC and CAID2 showed 100% sensitivity values for its identification; however, CAID2 required an expert observer to differentiate the downy aspect of the white colonies developed by C. krusei in this medium. For C. neoformans, different colony colors (blue, pink, or whitish, etc.) were observed in CAID2; however, the mucous texture was a clear characteristic that guided to their presumptive identification. In CAC, C. neoformans colonies had an aspect similar to that in CAID2 and colors varied from whitish to violet.
To determine if it would be practical for the physician to inoculate and grow samples at room temperature, we studied the effect of incubation temperature (37°C or 18 to 20°C) on the growth and color development of yeast isolated from fresh specimens. The growth was very poor at room temperature on the 4 chromogenic media, and consequently, the small size and weak color development of colonies did not permit the correct identification of the different species tested (data not shown).
DISCUSSION
An essential prerequisite for the laboratory detection of mixed fungal populations on clinical specimens is a suitable primary culture medium which facilitates the recovery and differentiation of phenotypically similar colonies. New chromogenic media can complement traditional identification methods for identifying clinical yeast isolates. Although many other chromogenic media can differentiate mixed cultures and identify C. albicans, CAC, due to its wide identification performance, has become a gold standard for comparison and evaluation of new commercialized chromogenic media (10, 11, 13). Few false-positive and -negative results have been described, but in a recent report (25), 4 isolates of C. albicans from 4 different patients were misidentified because their colonies were pink and were initially classified as non-C. albicans.
Many authors have also reported that C. albicans and C. dubliniensis can be distinguished by a different green color intensity of colonies when growing on CAC medium. Jabra-Rizk et al. (11), using a reformulated CAC medium (Becton Dickinson, BBL, Cockeysville, Md.), observed that C. albicans isolates gave a yellowish shade of green colonies, while C. dubliniensis isolates gave a typical dark green color. This color divergence reinforced the different shade of green described by Schoofs et al. (26) for C. dubliniensis. We have not been able to confirm this observation in a consistent way when using the CAC medium provided by CHROMagar (France), with results that are in agreement with recent publications (6, 23). In the present study, we have evaluated many stock culture strains, and it has been reported that the dark green color of the colonies may be lost upon repeated subculture or storage (11, 25, 26). The reason for our results may be due to the use of stock culture strains or to the possibility that other brands of CHROMagar could yield different results in the presumptive identification of C. dubliniensis based on the color of the colonies. The identification of this species can be facilitated using CAC supplemented with Pal's agar, as has been recently reported (23). We have observed that CAID2 allowed a differentiation at 48 h between C. albicans, growing as cobalt blue colonies, and C. dubliniensis, growing as turquoise blue colonies. This diagnostic accuracy is reduced because 8 of 96 C. albicans isolates grew as turquoise blue colonies. However, considering the percentage (8.3%) of C. albicans growing as C. dubliniensis does and the low prevalence of the latter species in most clinical specimens, except oral ones from HIV-infected patients, a reasonable approach could be to use another identification test only to confirm the identity of those isolates growing in this medium as C. dubliniensis. This approach could save personnel time and reduce laboratory costs.
In our hands, on CAID2, C. tropicalis strains developed pink-bluish colonies, which implied a sensitivity of 95% for its presumptive identification, while CAC sensitivity was 97.4% for the blue-gray color developed by this species. The growth of many colonies may influence the color of the colonies and alter the development of neighboring colonies, changing the expected color of the colonies for a determined species. This has been the case for oral specimens from patients with HIV infection, where C. tropicalis colonies gave a violet color on CAC instead of the expected blue-gray color. According to Fricker-Hidalgo et al. (9) pink isolates on CAID required further conventional tests for definitive identification because 4 frequently isolated non-C. albicans Candida species (C. tropicalis, C. kefyr, C. lusitaniae, and C. guilliermondii) and other less common species can produce such colored colonies. Conversely, CAID2 permits the differentiation of C. tropicalis as far as it develops pink-bluish colonies. C. tropicalis shows blue colonies on CAC, usually with a halo of diffusible pigment that cannot be observed in other species, such as Candida catenulata (2), C. guilliermondii, Cryptococcus humicola (3), and Saccharomyces cerevisiae (29). However, in our study, C. rugosa and some strains of Candida famata and Trichosporon spp. also showed bluish color on CAC, but colonies of this latter species were readily differentiated from those of C. tropicalis by their folded-lace appearance.
Pfaller et al. (19) and other authors (3, 10) also considered CAC to be reliable for the presumptive identification of C. glabrata, although other researchers (2, 6, 24) did not agree. Pink or violet colonies on CAC should be interpreted with caution as presumptive C. glabrata isolates, since other yeasts also grow as pink, dark pink, or violet colonies. In our study, C. glabrata grew as violet colonies, a color similar to that of the colonies of an additional 8 different yeast species. Infrequent Candida spp. studied showed a whitish color on these chromogenic agars, but the aspect of C. krusei colonies was rough, not brilliant. However, C. krusei was easier to distinguish in CAC, where colonies showed a rough pink aspect. Concerning C. krusei, CAC and CAID2 showed 100% sensitivity values for its identification. Nevertheless, CAID2 required an expert observer to differentiate the downy aspect of the white colonies developed by C. krusei in this medium.
Candida rugosa colonies can be mistaken with those of Candida dubliniensis in CAID2 or in CAID. This finding does not decrease significantly the usefulness of these chromogenic media, as Candida rugosa has been rarely described as a human pathogen. However, some outbreaks have been reported in Brazil with a very high crude mortality rate, despite therapy with intravenous amphotericin B (5).
Growth was very poor at room temperature (18 to 20°C) on the four chromogenic media, and consequently, the small size and weak color development of colonies did not permit the correct identification of the different species tested. This behavior was initially reported for CAC by Odds and Davidson (18), who determined that growth at temperatures below 30°C cannot be recommended for reliable presumptive identification of Candida spp. in this medium. A similar recommendation could be proposed for the use of the rest of the chromogenic media evaluated in the present study. We also recommend incubating the agar plates at 37°C to obtain faster growth and color development of colonies, so that identification may be accomplished as soon as possible.
The potential impact of a faster identification on patient care could be important in the selection of the therapeutic choice. This choice between antifungal agents depends primarily on the physician's knowledge of the etiologic agent and patient's prior exposure to antifungal therapy. Many patients are initially treated with caspofungin or voriconazole instead of fluconazole as first-line treatment when yeast growth is reported in blood cultures. This approach covers the possibility of fluconazole-resistant non-C. albicans species until the isolate is identified. However, caspofungin and voriconazole are much more expensive than fluconazole. A faster identification of isolated species could reduce the cost of treatment (1). In our center of reference (Hospital de Cruces, Barakaldo, Spain), fluconazole-resistant non-C. albicans species, such as C. krusei and C. glabrata, accounted for less than 5% of Candida isolates in the last 10-year period (unpublished data). Nearly 85% of blood isolates were C. albicans and C. parapsilosis, and an additional 10% included other fluconazole-susceptible species such as C. tropicalis or C. guilliermondii. Chromogenic media, such as CAID2 or CAC, with a sensitivity of >90% and >95% specificity for the identification of C. albicans and other fluconazole-susceptible species of Candida, would reduce laboratory and antifungal treatment costs because empirical use of caspofungin or voriconazole could be avoided.
In summary, yeast colonies grown on CAID2 agar plates show specific colors earlier and more defined than on CAID. The new CAID2 chromogenic medium improves the performance of the former CAID and ALB2 for the isolation and identification of C. albicans and other yeast species of clinical relevance, such as C. tropicalis or C. guilliermondii. CAID2 improves the detection of mixed cultures compared with other media such as ALB2, CAID, or CAC. Moreover, CAID2 allows for a better presumptive differentiation between C. albicans and C. dubliniensis among germ tube-producing yeasts, the latter showing a turquoise blue color, while 91.7% of C. albicans isolates develop cobalt blue colonies. Another remarkable feature of CAID2 is that C. tropicalis strains show a pink-violet colony color which facilitates its differentiation from the other Candida species that may develop pink colonies. The new formulation of CAID2 makes this chromogenic agar a good candidate to replace the previous bioMérieux formulations CAID and ALB2 and compares well to CAC performance for the presumptive identification of clinically relevant yeast species.
Acknowledgments
We have been financed in part by the projects GIU05/05 (from the Universidad del País Vasco-Euskal Herriko Unibertsitatea), PI030662/2003, and G03/075 RESITRA (from the Fondo de Investigación Sanitaria del Ministerio de Sanidad de España).
We are grateful to Myriam Müller, Ignacio Urrechaga, and José Manuel Torres, bioMérieux España, for the donation of the ALB2, CAID, and CAID2 agar plates used in this study.
REFERENCES
- 1.Alexander, B. D., E. D. Ashley, L. B. Reller, and S. D. Reed. 2006. Cost savings with implementation of PNA FISH testing for identification of Candida albicans in blood cultures. Diagn. Microbiol. Infect. Dis. 54:277-282. [DOI] [PubMed] [Google Scholar]
- 2.Baumgartner, C., A. M. Freydiere, and Y. Gille. 1996. Direct identification and recognition of yeast species from clinical material by using Albicans ID and CHROMagar Candida plates. J. Clin. Microbiol. 34:454-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernal, S., E. Martin Mazuelos, M. Garcia, A. I. Aller, M. A. Martinez, and M. J. Gutierrez. 1996. Evaluation of CHROMagar Candida medium for the isolation and presumptive identification of species of Candida of clinical importance. Diagn. Microbiol. Infect. Dis. 24:201-204. [DOI] [PubMed] [Google Scholar]
- 4.Cárdenes, C. D., A. J. Carrillo-Muñoz, A. Arias, C. Rodríguez-Álvarez, A. Torres-Lana, A. Sierra, and M. P. Arévalo. 2002. Comparison of Albicans ID2 agar plate with germ tube for presumptive identification of Candida albicans. Diagn. Microbiol. Infect. Dis. 42:181-185. [DOI] [PubMed] [Google Scholar]
- 5.Colombo, A. L., A. S. Melo, R. F. Crespo Rosas, R. Salomao, M. Briones, R. J. Hollis, S. A. Messer, and M. A. Pfaller. 2003. Outbreak of Candida rugosa candidemia: an emerging pathogen that may be refractory to amphotericin B therapy. Diagn. Microbiol. Infect. Dis. 46:253-257. [DOI] [PubMed] [Google Scholar]
- 6.Cooke, V. M., R. J. Miles, R. G. Price, G. Midgley, W. Khamri, and A. C. Richardson. 2002. New chromogenic agar medium for the identification of Candida spp. Appl. Environ. Microbiol. 68:3622-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuétara, M. S., A. Alhambra, and A. del Palacio. 2006. Diagnóstico microbiológico tradicional de la candidiasis invasora en el enfermo crítico no neutropénico. Rev. Iberoam. Micol. 23:4-7. [DOI] [PubMed] [Google Scholar]
- 8.Freydiere, A. M., R. Guinet, and P. Boiron. 2001. Yeast identification in the clinical microbiology laboratory: phenotypical methods. Med. Mycol. 39:9-33. [DOI] [PubMed] [Google Scholar]
- 9.Fricker-Hidalgo, H., S. Orenga, B. Lebeau, H. Pelloux, M. P. Brenier-Pinchart, P. Ambroise-Thomas, and R. Grillot. 2001. Evaluation of Candida ID, a new chromogenic medium for fungal isolation and preliminary identification of some yeast species. J. Clin. Microbiol. 39:1647-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvath, L. L., D. R. Hospenthal, C. K. Murray, and D. P. Dooley. 2003. Direct isolation of Candida spp. from blood cultures on the chromogenic medium CHROMagar Candida. J. Clin. Microbiol. 41:2629-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jabra-Rizk, M. A., T. M. Brenner, M. Romagnoli, A. A. M. A. Baqui, W. G. Merz, W. A. Falkler, and T. F. Meiller. 2001. Evaluation of a reformulated CHROMagar Candida. J. Clin. Microbiol. 39:2015-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanbe, T., T. Arishima, T. Horii, and A. Kikuchi. 2003. Improvements of PCR-based identification targeting the DNA topoisomerase II gene to determine major species of the opportunistic fungi Candida and Aspergillus fumigatus. Microbiol. Immunol. 47:631-638. [DOI] [PubMed] [Google Scholar]
- 13.Koehler, A. P., C. Kai-Cheong, E. T. S. Houang, and A. F. B. Cheng. 1999. Simple, reliable, and cost-effective yeast identification scheme for the clinical laboratory. J. Clin. Microbiol. 37:422-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letscher-Bru, V., M. H. Meyer, A. C. Galoisy, J. Waller, and E. Candolfi. 2002. Prospective evaluation of the new chromogenic medium Candida ID, in comparison with Candiselect, for isolation of molds and isolation and presumptive identification of yeast species. J. Clin. Microbiol. 40:1508-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo, G., and T. G. Mitchell. 2002. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J. Clin. Microbiol. 40:2860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magee, B. B., and P. T. Magee. 2005. Recent advances in the genomic analysis of Candida albicans. Rev. Iberoam. Micol. 22:187-193. [DOI] [PubMed] [Google Scholar]
- 17.Odds, F. C., and R. Bernaerts. 1994. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J. Clin. Microbiol. 32:1923-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Odds, F. C., and A. Davidson. 2000. “Room temperature” use of CHROMagar Candida. Diagn. Microbiol. Infect. Dis. 38:147-150. [DOI] [PubMed] [Google Scholar]
- 19.Pfaller, M. A., A. Houston, and S. Coffmann. 1996. Application of CHROMagar Candida for rapid screening of clinical specimens for Candida albicans, Candida tropicalis, Candida krusei, and Candida (Torulopsis) glabrata. J. Clin. Microbiol. 34:58-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pontón, J., R. Ruchel, K. V. Clemons, D. C. Coleman, R. Grillot, J. Guarro, D. Aldebert, P. Ambroise-Thomas, J. Cano, A. J. Carrillo-Muñoz, J. Gené, C. Pinel, D. A. Stevens, and D. J. Sullivan. 2000. Emerging pathogens. Med. Mycol. 38:225-236. [DOI] [PubMed] [Google Scholar]
- 21.Quindós, G., R. Alonso-Vargas, S. Helou, A. Arechavala, E. Martín-Mazuelos, and R. Negroni. 2001. Evaluación micológica de un nuevo medio de cultivo cromógeno (Candida ID) para el aislamiento e identificación presuntiva de Candida albicans y otras levaduras de interés médico. Rev. Iberoam. Micol. 18:23-28. [PubMed] [Google Scholar]
- 22.Quindós, G., M. T. Ruesga, E. Martín-Mazuelos, R. Salesa, R. Alonso-Vargas, A. J. Carrillo-Muñoz, S. Brena, R. San Millán, and J. Pontón. 2004. In-vitro activity of 5-fluorocytosine against 1,021 Spanish clinical isolates of Candida and other medically important yeasts. Rev. Iberoam. Micol. 21:63-69. [PubMed] [Google Scholar]
- 23.Sahand, I. H., M. D. Moragues, E. Eraso, M. Villar-Vidal, G. Quindós, and J. Pontón. 2005. Supplementation of CHROMagar Candida medium with Pal's medium for rapid identification of Candida dubliniensis. J. Clin. Microbiol. 43:5768-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.San-Millán, R., L. Ribacoba, J. Pontón, and G. Quindós. 1996. Evaluation of a commercial medium for identification of Candida species. Eur. J. Clin. Microbiol. Infect. Dis. 15:153-158. [DOI] [PubMed] [Google Scholar]
- 25.Saunte, D. M., L. Klingspor, S. Jalal, J. Arnau, and M. C. Arendrup. 2005. Four cases of Candida albicans infections with isolates developing pink colonies on CHROMagar Candida plates. Mycoses 48:378-381. [DOI] [PubMed] [Google Scholar]
- 26.Schoofs, A., F. C. Odds, R. Colebunders, M. Ieven, and H. Goossens. 1997. Recognition and identification of Candida dubliniensis isolates from HIV patients: use of specialized isolation media. Eur. J. Clin. Microbiol. Infect. Dis. 16:296-300. [DOI] [PubMed] [Google Scholar]
- 27.Swinne, D., M. Watelle, and N. Nolard. 2005. In vitro activities of voriconazole, fluconazole, itraconazole and amphotericin B against non Candida albicans yeast isolates. Rev. Iberoam. Micol. 22:24-28. [DOI] [PubMed] [Google Scholar]
- 28.Willinger, B., C. Hillowoth, B. Selitsch, and M. Manafi. 2001. Performance of Candida ID, a new chromogenic medium for presumptive identification of Candida species, in comparison to CHROMagar Candida. J. Clin. Microbiol. 39:3793-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willinger, B., and M. Manafi. 1999. Evaluation of CHROMagar Candida for rapid screening of clinical specimens for Candida species. Mycoses 42:61-65. [DOI] [PubMed] [Google Scholar]