Abstract
Five different serotypes of Salmonella enterica were implicated in a large outbreak linked to fresh Roma tomatoes served at gas station deli counters in Pennsylvania and nearby states during July 2004: S. enterica serotypes Javiana, Anatum, Thompson, Typhimurium, and Muenchen. One of these serotypes, Anatum, was isolated from both tomatoes and patients. Pulsed-field gel electrophoresis (PFGE) played a key role in identifying the outbreak-associated isolates and distinguishing them from unrelated sporadic isolates. It also demonstrated that the genetic fingerprints of serotype Anatum isolates derived from patients were indistinguishable from those derived from tomatoes. Rapid communication of PFGE fingerprints with other public health laboratories through the Centers for Disease Control and Prevention's PulseNet USA national molecular surveillance network for bacterial food-borne pathogens facilitated the tracking of this outbreak in other states. The work described in this report emphasizes the laboratory's role in core public health functions and services, thereby providing a highly visible example of public health in action.
An estimated 1.4 million cases of nontyphoidal Salmonella infections occur in the United States each year, causing approximately 15,000 hospitalizations and 400 deaths (29). Fresh produce has become an increasingly prominent source of Salmonella outbreaks (1, 3, 5, 11, 12, 14, 23, 24, 27, 31).
Multiple serotypes of Salmonella enterica were implicated in a large Salmonella outbreak that occurred in Pennsylvania and nearby states in the summer of 2004. A case-controlled investigation linked the outbreak to the consumption of Roma tomatoes from the deli counters of a chain of 302 gas station convenience stores in Pennsylvania and four nearby states (chain A) (4). Pulsed-field gel electrophoresis (PFGE) played a key role in identifying the outbreak-associated isolates and distinguishing them from unrelated sporadic isolates.
PFGE is a DNA-based fingerprinting method considered to be the “gold standard” for subtyping numerous bacterial pathogens (7, 8, 10, 16, 22, 25, 28, 30). PFGE involves digesting intact bacterial chromosomes with restriction endonucleases that have specificities for sites in DNA that are relatively uncommon. In preparation for PFGE, bacterial cells are embedded in agarose plugs so that when they are lysed and digested, the DNA remains unsheared. Because the resulting fragments are very large (approximately 20 kbp to 1,135 kbp), a specialized power supply and electrophoresis chamber are used to provide alternating electric fields that facilitate their movement through the gel. PFGE yields restriction fragment band patterns that can be compared by specialized software. Adoption of a highly standardized protocol for PFGE testing developed by PulseNet USA at the Centers for Disease Control and Prevention (CDC) has made lab-to-lab comparisons of PFGE band patterns feasible and informative (8, 26). All 50 state public health laboratories, several city and county public health laboratories, and 8 food safety regulatory laboratories participate in PulseNet USA (9). The assignment of names to particular band patterns by PulseNet USA has made PFGE data useful for epidemiological investigations. PFGE-based surveillance of Salmonella and other food-borne pathogens by the Pennsylvania Department of Health (PADOH) Bureau of Laboratories (BOL) is a critical component of the state's plan for public health preparedness. The purpose of this report is to demonstrate the essential role that PFGE played in the investigation of a large Salmonella outbreak that took place during July 2004.
MATERIALS AND METHODS
Culture growth, identification, and serology.
The Salmonella isolates used for this study were grown, identified, serogrouped, and serotyped by the Bacteriology Section of the BOL using standard procedures (2, 6). They were collected from Pennsylvania patients and Roma tomatoes between 1 and 31 July 2004.
Sample preparation, DNA restriction, PFGE, and gel analysis.
Sample preparation, restriction digestion, electrophoresis, and gel staining for PFGE were accomplished following the CDC-standardized procedure used by all PulseNet-certified laboratories as described by Ribot et al. (21) and modified for Salmonella as described by Hunter et al. (13). This procedure is available at the PulseNet website (http://www.cdc.gov/pulsenet/protocols.htm). Restriction endonucleases XbaI, BlnI, and SpeI (Roche Diagnostics Corporation, Indianapolis, IN) were used. The size standard used for all gels was XbaI-digested DNA from Salmonella Braenderup strain H9812 (American Type Culture Collection catalog no. BAA-664), the universal size standard used by all PulseNet laboratories. A Gel Doc 2000 equipped with Quantity One software (Bio-Rad, Hercules, CA) was used for image capture and conversion of gel images to the TIFF file format. Gel images were analyzed using BioNumerics software version 3.5 (Applied Maths, Sint-Martens-Latem, Belgium), the software used by all PulseNet laboratories. The use of the universal size standard permitted normalization and comparison of DNA fingerprints from gel to gel and from lab to lab via BioNumerics.
PulseNet communications.
Analyzed gel images were uploaded to PulseNet's national Salmonella database through a secure server accessible only to PulseNet-certified laboratorians. Notifications regarding clusters of bacterial isolates with similar DNA fingerprints were posted to the PulseNet WebBoard for immediate dissemination to the PulseNet community. Fingerprint pattern names assigned by the CDC's PulseNet team were downloaded from the secure server, added to local databases, and uploaded electronically to the Pennsylvania National Electronic Disease Surveillance System (PA-NEDSS) database used by state epidemiologists. Pattern names include a three-letter code for the serotype (e.g., JGG for serotype Javiana in XbaI pattern name JGGX01.0036), a one-letter two-digit code indicating the enzyme (e.g., X01 for XbaI, A26 for BlnI, or S18 for SpeI), and a four-digit number related to a particular strain pattern (e.g., 0036 in JGGX01.0036). Consecutive four-digit numbers do not indicate relatedness of patterns (9). A cluster/outbreak code, such as 0407PAJGGJAG1, indicates the year (04), the month (07), the state that identified the cluster or outbreak (PA), and the serotype(s) involved (e.g., JGG for serotype Javiana and JAG for serotype Anatum). The code ends in 1c to indicate the first cluster (c stands for cluster) of a particular serotype in the state during the specified month, 2c to indicate the second cluster of that serotype in the state that month, etc. (9). For a well-documented outbreak, such as 0407PAJGGJAG1, the one described in this report, the code ends in a number that is not followed by the letter c. This indicates that isolates designated with this code are part of an outbreak and not just a cluster of similar PFGE fingerprints that may or may not be part of an outbreak (15). In the early stages of this investigation, serotyping and PFGE had linked only serotype Javiana to the unfolding outbreak. Therefore, the original cluster/outbreak code was 0407PAJGG1-c. Subsequently, when serotype Anatum also became implicated and the existence of an outbreak had been established, the code was changed to 0407PAJGGJAG1.
RESULTS
Serotype Javiana, the main cause of the outbreak.
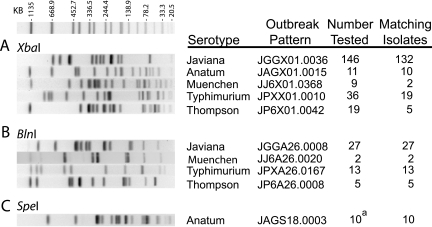
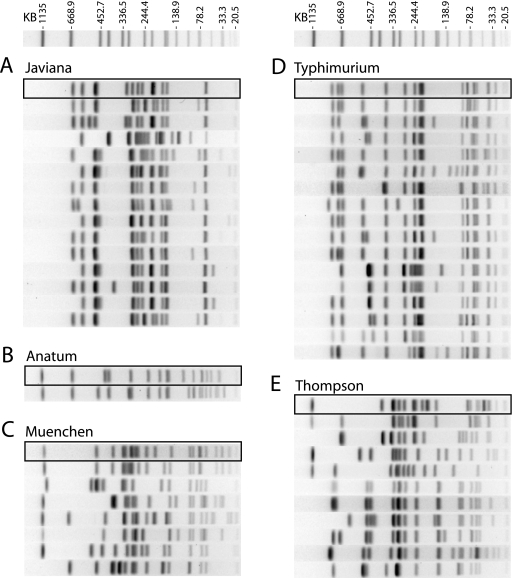
Routine surveillance of Salmonella isolates received in the BOL demonstrated a surge in serotype Javiana isolates collected from patients in early July 2004. PFGE testing of the first 10 of these isolates with XbaI revealed 9 with indistinguishable fingerprint patterns. The CDC assigned pattern name JGGX01.0036 to these isolates (Fig. 1A). During July, a total of 331 serotype Javiana isolates were collected from patients. Testing 146 of them by PFGE identified 132 that matched pattern JGGX01.0036 (Fig. 1A). Digestion of 27 isolates in this group with a second restriction enzyme, BlnI, identified a single pattern shared by all of them. The CDC assigned them the BlnI pattern name JGGA26.0008 (Fig. 1B). Fourteen of the serotype Javiana isolates tested did not match the outbreak pattern and were considered to be unrelated sporadic isolates (Fig. 2A).
FIG. 1.
Outbreak PFGE patterns. Fingerprints for the five outbreak serotypes shown were obtained and compared as described in Materials and Methods. The size standard shown above the test fingerprints is Salmonella Braenderup strain H9812 digested with XbaI. Pattern names were assigned by the PulseNet team of the CDC. The number tested refers to isolates collected from patients in July 2004. a, six isolates from tomatoes and four from patients.
FIG. 2.
Comparison of outbreak and sporadic isolates. Fingerprints of outbreak and unrelated sporadic isolates were obtained and compared as described in Materials and Methods. The size standard shown above the test fingerprints is Salmonella Braenderup strain H9812 digested with XbaI. The outbreak fingerprint of each serotype is enclosed in a box, and the fingerprints of the sporadic isolates of each serotype are shown below the outbreak fingerprint. A representative fingerprint is shown for each pattern; duplicate fingerprints were omitted. The number of isolates matching each outbreak pattern is summarized in Fig. 1.
Serotype Anatum and the tomato connection.
A concurrent case-controlled study conducted by the state's Bureau of Epidemiology, with assistance from the CDC, identified Roma tomatoes as the source of the outbreak (4, 19). The Pennsylvania Department of Agriculture, responsible for testing produce, isolated Salmonella from an unopened package of presliced Roma tomatoes obtained from chain A (17). Further testing at the BOL identified the serotype of the tomato-derived Salmonella to be Anatum (20). Serotype Anatum was the only serotype isolated from food collected at chain A. Testing of six tomato-derived serotype Anatum isolates by PFGE, using restriction enzymes XbaI and SpeI, showed all six to be indistinguishable by both enzymes. Testing of serotype Anatum isolates collected from five patients in July revealed fingerprint patterns indistinguishable from the tomato-derived serotype Anatum from four patients and one unrelated sporadic serotype Anatum isolate from the fifth patient (Fig. 2B). The CDC assigned XbaI pattern name JAGX01.0015 (Fig. 1A) and SpeI pattern name JAGS18.0003 (Fig. 1C) to the fingerprints of these serotype Anatum isolates.
Multiple serotypes.
When it became clear that more than one serotype was associated with this outbreak, three additional Salmonella serotypes (Thompson, Typhimurium, and Muenchen) collected from patients in July were targeted for testing by PFGE. Testing with two enzymes identified five serotype Thompson isolates with indistinguishable fingerprints: XbaI pattern JP6X01.0042 (Fig. 1A) and BlnI pattern JP6A26.0008 (Fig. 1B). Among the 19 unrelated serotype Thompson isolates tested, 10 different XbaI patterns were observed (Fig. 2E). PFGE testing of serotype Typhimurium isolates identified 13 with indistinguishable PFGE patterns, XbaI pattern JPXX01.0010 (Fig. 1A) and BlnI pattern JPXA26.0167 (Fig. 1B), and 16 with unrelated XbaI patterns (Fig. 2E). PFGE testing using XbaI and BlnI demonstrated that two of the nine serotype Muenchen isolates were indistinguishable from one another by both enzymes: XbaI pattern JJ6X01.0368 (Fig. 1A) and BlnI pattern JJ6A26.0020 (Fig. 1B). Seven serotype Muenchen isolates were unrelated (Fig. 2C). In all, five different serotypes of Salmonella enterica were implicated in this outbreak, serotypes Javiana, Anatum, Thompson, Typhimurium, and Muenchen, and serotype Javiana was the main cause of the outbreak (4).
DISCUSSION
The Molecular Microbiology Laboratory at the BOL is part of the PulseNet food-borne disease surveillance network of public health and food regulatory agency laboratories coordinated by the CDC. This network provides a forum for the rapid communication of PFGE test results among the laboratories of these agencies via WebBoard postings and e-mail messages. A surge in Salmonella isolates collected in Pennsylvania in early July and serotyped as Javiana signaled a possible outbreak. The earliest PFGE testing of these isolates identified a single XbaI pattern and strengthened this tentative conclusion. The BOL made its first posting of PFGE results (nine serotype Javiana isolates with matching XbaI patterns) to the PulseNet WebBoard on 16 July 2004. This posting included the epidemiological information that had been released to the public earlier in the day in a PADOH health advisory (18). Within an hour of the Pennsylvania posting, a neighboring state replied by posting PFGE results that matched the Pennsylvania pattern. Within another hour, the CDC named the pattern JGGX01.0036. Within 4 days, three additional states posted their matching results. Ultimately, nine states posted matching PFGE results to the WebBoard. Subsequent testing with a second enzyme, BlnI, showed a single pattern and increased the probability of a common source of infection.
Three days after the first WebBoard posting of serotype Javiana patient results, the Pennsylvania Department of Agriculture cultured Salmonella from an unopened package of Roma tomatoes removed from a deli counter at chain A. Subsequently, six Salmonella isolates were sent to the BOL for serogrouping, serotyping, and PFGE testing. The serogroup of all six of the tomato-derived isolates was determined to be E1, indicating that they could not be serotype Javiana (serogroup D1). They were subsequently serotyped as Anatum. PFGE testing of the six tomato-derived samples along with five serotype Anatum isolates collected from patients between 15 and 19 July 2004 identified four patient-derived isolates with the same XbaI pattern (JAGX01.0015) as the tomato-derived isolates (Fig. 1A). Further testing with a second enzyme revealed that DNA samples from these isolates could not be cleaved by BlnI. Successful digestion was completed with SpeI, and all were found to share the same SpeI pattern (JAGS18.0003) (Fig. 1C).
A concurrent epidemiological analysis indicated higher-than-expected numbers of three Salmonella serotypes: Thompson, Typhimurium, and Muenchen. Patient interviews indicated that some patients had eaten food from chain A during the outbreak time frame. PFGE testing was the key to distinguishing outbreak-related isolates from unrelated sporadic isolates, thus allowing the Bureau of Epidemiology to focus its investigation and corroborate information obtained from patient interviews.
For epidemiological comparisons, knowing the pattern name of each fingerprint is desirable. However, obtaining a pattern name is a multistep, time-consuming process. After gels are electrophoresed, stained, and photographed, the positions of bands must be assigned via BioNumerics software. Although the software suggests band assignments, these assignments must be evaluated and fine-tuned manually, a time-consuming and somewhat subjective process. After the gels were analyzed, patient demographic information was entered, and the information for each isolate was uploaded to PulseNet's national Salmonella database at the CDC. Once PFGE pattern names were assigned, they were entered into PA-NEDSS. This was also a multistep process. To bridge the time lag inherent in this process, e-mail was used to communicate results directly to the Bureau of Epidemiology when particular isolates appeared to match outbreak patterns.
In summary, pulsed-field gel electrophoresis was essential for discriminating between outbreak-related isolates and unrelated sporadic isolates for each of the five serotypes that were epidemiologically linked to this large multiserotype and multistate outbreak. It also established the connecting thread needed to track the outbreak in other states. Because of its high degree of standardization, the PulseNet USA PFGE protocol and communications network provided the means to compare the current data among state public health laboratories and archived data from the national Salmonella database. The work described in this report emphasizes the laboratory's role in core public health functions and services, thereby providing a highly visible example of public health in action.
Acknowledgments
We gratefully acknowledge the excellent work of James M. Tait, Bacteriology Section of the BOL, in identifying, serogrouping, and serotyping the Salmonella isolates described in this report. We thank the CDC for supplying Salmonella Braenderup strain H9812.
REFERENCES
- 1.Altekruse, S. F., M. L. Cohen, and D. L. Swerdlow. Emerging foodborne diseases. 1997. Emerg. Infect. Dis. 3:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner, F., and A. C. McWhorten-Murlin. 1998. Identification and serotyping of Salmonella. National Salmonella Reference Laboratory, Centers for Disease Control and Prevention, Atlanta, Ga.
- 3.Center for Science in the Public Interest. 2005. Outbreak alert! Closing the gaps in our federal food-safety net. [Online.] http://www.cspinet.org/new/pdf/outbreakalert2005.pdf.
- 4.Centers for Disease Control and Prevention. 2005. Outbreaks of Salmonella infections associated with eating Roma tomatoes-United States and Canada, 2004. Morb. Mortal. Wkly. Rep. 54:325-328. [PubMed] [Google Scholar]
- 5.Cummings, K., E. Barrett, J. C. Mohle-Boetani, J. T. Brooks, J. Farrar, T. Hunt, A. Fiore, K. Komatsu, S. B. Werner, and L. Slutsker. 2001. A multistate outbreak of Salmonella enterica serotype Baildon associated with domestic raw tomatoes. Emerg. Infect. Dis. 7:1046-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farmer, J. 1995. Enterobacteriaceae: introduction and identification, p. 438-449. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. American Society for Microbiology, Washington, D.C.
- 7.Fitzgerald, C., L. O. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerdlow, R. Flahart, J. Sexton, and P. I. Fields. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerner-Smidt, P., J. Kincaid, K. Kubota, K. Hise, S. B. Hunter, M. A. Fair, D. Norton, A. Woo-Ming, T. Kurzynski, M. J. Sotir, M. Head, K. Holt, and B. Swaminathan. 2005. Molecular surveillance of Shiga toxigenic Escherichia coli O157 by PulseNet USA. J. Food Prot. 68:1926-1931. [DOI] [PubMed] [Google Scholar]
- 9.Gerner-Smidt, P., K. Hise, J. Kincaid, S. Hunter, S. Rolando, E. Hyytiä-Trees, E. M. Ribot, B. Swaminathan, and the PulseNet Taskforce. 2006. PulseNet USA: a five-year update. Foodborne Pathog. Dis. 3:9-19. [DOI] [PubMed] [Google Scholar]
- 10.Healy, M., J. Huong, T. Bittner, M. Lising, S. Frye, S. Raza, R. Schrock, J. Manry, A. Renwick, R. Nieto, C. Woods, J. Versalovic, and J. R. Lupski. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedberg, C. W., F. J. Angulo, K. E. White, C. W. Langkop, W. L. Schell, M. G. Stobierski, A. Schuchat, J. M. Besser, S. Dietrich, L. Helsel, P. M. Griffin, J. W. McFarland, and M. T. Osterholm. 1999. Outbreaks of salmonellosis associated with eating uncooked tomatoes: implications for public health. Epidemiol. Infect. 122:385-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horby, P. W., S. J. O'Brien, G. K. Adak, C. Graham, J. I. Hawker, P. Hunter, C. Lane, A. J. Lawson, R. T. Mitchell, M. H. Reacher, E. J. Threlfall, L. R. Ward, and the PHLS Outbreak Investigation Team. 2003. A national outbreak of multiresistant Salmonella enterica serovar Typhimurium definitive phage type (DT) 104 associated with consumption of lettuce. Epidemiol. Infect. 130:169-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter, S. B., P. Vauterin, M. A. Lambert-Fair, M. S. Van Duyne, K. Kubota, L. Graves, D. Wrigley, T. Barrett, and E. Ribot. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston, L. M., L. A. Jaykus, D. Moll, M. C. Martinez, J. Anciso, B. Mora, and C. L. Moe. 2005. A field study of the microbiological quality of fresh produce. J. Food Prot. 68:1840-1847. [DOI] [PubMed] [Google Scholar]
- 15.Joyner, M. 2005. Use of WebBoard. [Online.] http://www.aphl.org/conferences/epidemiologist_conference_2005/files/4_Joyner.pdf.
- 16.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pennsylvania Department of Agriculture. 2004. Ag Department announces positive test in Salmonella investigation. [Online.] http://www.agriculture.state.pa.us/agriculture/cwp/view.asp?A=390&Q=130777.
- 18.Pennsylvania Department of Health. 2004. Salmonella javiana gastroenteritis outbreak in Pennsylvania. [Online.] http://www.dsf.health.state.pa.us/health/CWP/view.asp?A=171&QUESTION_ID=238983.
- 19.Pennsylvania Department of Health. 2004. PA health secretary says study suggests Salmonella outbreak linked to Roma tomatoes. [Online.] http://www.dsf.health.state.pa.us/health/CWP/view.asp?A=190&QUESTION_ID=238157.
- 20.Pennsylvania Department of Health. 2004. Second Salmonella serotype with human cases is now associated with tomatoes eaten at Sheetz. [Online.] http://www.dsf.health.state.pa.us/health/CWP/view.asp?A=171&QUESTION_ID=238969.
- 21.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross, T. L., W. G. Merz, M. Farkosh, and K. C. Carroll. 2005. Comparison of an automated repetitive sequence-based PCR microbial typing system to pulsed-field gel electrophoresis for analysis of outbreaks of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5642-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sivapalasingam, S., C. R. Friedman, L. Cohen, and R. V. Tauxe. 2004. Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 67:2342-2353. [DOI] [PubMed] [Google Scholar]
- 24.Srikantiah, P., D. Bodager, B. Toth, T. Kass-Hout, R. Hammond, S. Stenzel, R. M. Hoekstra, J. Adams, S. Van Duyne, and P. S. Mead. 2005. Web-based investigation of multistate salmonellosis outbreak. Emerg. Infect. Dis. 11:610-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stranden, A., R. Frei, and A. F. Widmer. 2003. Molecular typing of methicillin-resistant Staphylococcus aureus: can PCR replace pulsed-field gel electrophoresis? J. Clin. Microbiol. 41:3181-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swaminathan, B., T. J. Barrett, S. B. Hunter, R. V. Tauxe, and the CDC PulseNet Task Force. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tauxe, R. V. 1997. Emerging foodborne diseases: an evolving public health challenge. Emerg. Infect. Dis. 3:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turabelidze, D., M. Kotetishvili, A. Kreger, J. G. Morris, Jr., and A. Sulakvelidze. 2000. Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. J. Clin. Microbiol. 38:4242-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voetsch, A. C., T. J. Van Gilder, F. J. Angulo, M. M. Farley, S. Shallow, R. Marcus, P. R. Cieslak, V. C. Deneen, R. V. Tauxe, and the Emerging Infections Program FoodNet Working Group. 2004. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin. Infect. Dis. 38(Suppl. 3):S127-S134. [DOI] [PubMed] [Google Scholar]
- 30.Werner, G., R. J. Willems, B. Hildebrandt, I. Klare, and W. Witte. 2003. Influence of transferable genetic determinants on the outcome of typing methods commonly used for Enterococcus faecium. J. Clin. Microbiol. 41:1499-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhuang, R.-Y., L. R. Beuchat, and F. J. Angulo. 1995. Fate of Salmonella montevideo on and in raw tomatoes as affected by temperature and treatment with chlorine. Appl. Environ. Microbiol. 61:2127-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]