Abstract
Comparison of pulsed-field gel electrophoresis patterns (generated with XbaI and BlnI) of Shigella sonnei isolates from Ireland and Italy suggests that two possibly distantly related lineages are present in both countries. Smaller, more closely related groups, including isolates from Ireland and Italy, were also noted. These groups raise the possibility that the dissemination of clonal groups of S. sonnei may have occurred in recent years.
Shigella sonnei is the most prevalent Shigella species in the developed world. Transmission is mainly by direct contact with infected individuals by the fecal-oral route, although outbreaks may occasionally be associated with the consumption of contaminated food or water (1). The spread of S. sonnei is particularly problematic in institutional or crowded settings, such as day care centers, prisons, and military field settings (4, 7).
While there is variation in the number of reported cases of S. sonnei infection from year to year, overall the number of cases in Ireland has declined in recent years, with a decrease from 736 notified shigellosis cases in 1991 to 36 in 2003 (6). A similar pattern has been observed in England and Wales, with a reduction from 16,626 S. sonnei isolates in 1992 to 841 in 2005 (5). Likewise, in southern Italy, a declining, but irregular, trend has been recorded. Between 1991 and 2000, only 56 isolates of S. sonnei were recorded at the southern Italy Centre for Enteric Pathogens (CEPIM) and 28 of these were from an outbreak in Palermo, Sicily (8). This trend towards a very low background rate of isolation of S. sonnei has continued in recent years (only three strains in 2004), with the exception of a large community outbreak lasting several months in Palermo in 2003 (9). In previous decades, 421 isolates were recorded between 1971 and 1980 and 779 for the decade 1991 to 2000. Outbreaks of S. sonnei (one in a refugee camp, one in a small suburban community, and two in day care centers) also occurred in 2001 in Lombardy, northern Italy (see above).
In a previous publication, the National Salmonella Reference Laboratory of Ireland reported that 67 S. sonnei isolates from the west of Ireland in the period from 1998 to 1999 comprise two groups, designated A (n = 53) and B (n = 14) by XbaI pulsed-field gel electrophoresis (PFGE) (3). Group A comprised two variants, A and A1, while group B included five variants, B and B1 to B4. A subsequent report from the southern Italy CEPIM, Palermo, Sicily, studying 74 S. sonnei isolates from various parts of Italy in 2001 to 2003 also described two predominant patterns by PFGE, designated group A (n = 13) and group B (n = 61) (9). Visual examination of the published PFGE patterns suggested some similarity between groups of isolates in both countries, raising the possibility that one or more closely related groups of S. sonnei may have disseminated over a wide area of Europe. However, the PFGE reported in the earlier studies did not use a standardized method, so exact comparison of patterns was difficult. To explore this issue, 30 isolates from Italy were sent to the National Salmonella Reference Laboratory in Ireland to allow direct comparison of patterns generated in a single laboratory. Thirty Italian isolates (from 2001 to 2003) and seven Irish isolates (from 1998 to 1999) were examined using the standardized PulseNet PFGE protocol (10), using XbaI as the primary restriction enzyme as previously described (2). The 7 Irish isolates examined by the PulseNet method in this study represented one example of each unique XbaI pattern previously identified in the study of 67 isolates digested with the same enzyme but with electrophoresis under different run conditions (3). A representative of each of the seven Irish PFGE patterns along with Italian isolates that were indistinguishable or very similar to these patterns was subsequently digested with BlnI. Digestion patterns for Irish and Italian isolates were compared using BioNumerics software.
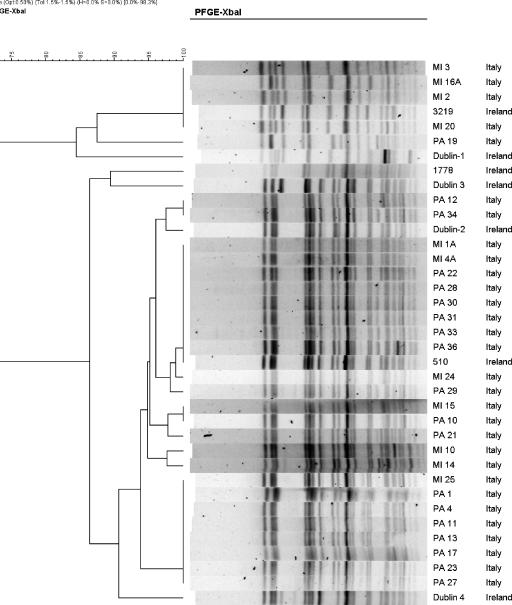
Two clusters were identified in the dendrogram generated (Fig. 1). Five of the Italian isolates clustered with Irish isolates 3219 (representing type A) and Dublin 1 (representing type A1). The seven isolates were 84% similar (Fig. 1). Within this cluster is a group of four Italian isolates (MI 12, MI 13, MI 16A, and MI 20) that are indistinguishable from strain 3219. Isolate 3219 is representative of the type A isolate, which accounted for 49 of the 67 isolates in the previous study of Irish isolates. Twenty-five of the Italian isolates clustered with representatives of the previously designated Irish group B isolates with 86% similarity (Fig. 1). Isolate 510 representing pattern B was indistinguishable from eight Italian isolates (MI 1A, MI 4A, PA 22, PA 28, PA 30, PA 31, PA 33, and PA 36).
FIG. 1.
PFGE profiles of representative S. sonnei isolates digested with XbaI and analyzed using BioNumerics v.3.5.
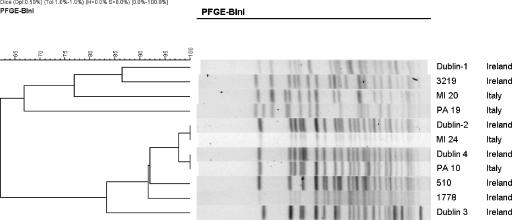
The dendrogram (Fig. 2) derived from the digestion of a subset of 11 isolates with BlnI also reveals two clusters, A (four strains with 66% similarity) and B (seven strains with 83% similarity), that correspond to the clustering of these isolates observed during digestion with XbaI. Of particular note, isolates Dublin 2 (Ireland) and MI24 and PA10 (Italian), which are included in a large group that is more than 95% similar on XbaI, are also more than 95% similar with BlnI.
FIG. 2.
PFGE profiles of representative S. sonnei isolates digested with BlnI and analyzed using BioNumerics v.3.5.
Digestion with a second enzyme is valuable in studies of this nature, as it is easy to overinterpret similarity between isolates digested with a single enzyme. It is a limitation of this study that all isolates were not studied with BlnI; however, this enzyme is prohibitively expensive. This study provides some evidence that S. sonnei isolates from each of two European Union countries form two related groups (A and B) during PFGE digestion with two enzymes. This suggests that two relatively distantly related lineages of S. sonnei (phylogenetically related rather than outbreak/epidemiologically related) may be circulating in Europe. Within these large and relatively distantly related lineages, there are groups, including isolates from both countries, which appear to be more closely related, consistent with relatively recent dissemination of a clonal group of S. sonnei between Ireland and Italy. Further studies of S. sonnei isolates from other regions of Europe should prove to be useful.
Shigella sonnei is now a relatively infrequently detected pathogen in Ireland and Italy. However, as with many other enteric pathogens, it is likely that laboratory-detected cases represent a small minority of affected patients. Networks of the kind that have been developed for some other enteric pathogens have not emerged in Europe for S. sonnei or for other Shigella species. This pilot collaboration suggests that greater collaboration between laboratories across Europe relating to typing of Shigella species may be valuable in defining the epidemiology of shigellosis in Europe. Further typing studies with methods such as multilocus sequence typing in addition to PFGE and with clinical and epidemiological information on the patients are warranted to define the epidemiology of S. sonnei infection in Europe.
Footnotes
Published ahead of print on 23 August 2006.
REFERENCES
- 1.Cimmons, M. 2000. Rapid food-borne pathogen ID system is making a difference. ASM News 66:617-619. [Google Scholar]
- 2.Cormican, M., N. DeLappe, C. O'Hare, G. Doran, D. Morris, G. Corbett-Feeney, et al. 2002. Salmonella enterica serotype Bredeney: antimicrobial susceptibility and molecular diversity of isolates from Ireland and Northern Ireland. Appl. Environ. Microbiol. 68:181-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeLappe, N., F. O'Halloran, S. Fanning, G. Corbett-Feeney, T. Cheasty, and M. Cormican. 2003. Antimicrobial resistance and genetic diversity of Shigella sonnei isolates from western Ireland, an area of low incidence of infection. J. Clin. Microbiol. 41:1919-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DuPont, H. L., M. M. Levine, R. B. Hornick, and S. B. Formal. 1989. Inoculum size in shigellosis and implications for expected mode of transmission. J. Infect. Dis. 159:1126-1128. [DOI] [PubMed] [Google Scholar]
- 5.Health Protection Agency. 2005. Shigella laboratory reports of faecal isolates: England & Wales, 1986-2005 [Online.] http://www.hpa.org.uk/infections/topics_az/shigella/data_ew.htm.
- 6.Health Protection Surveillance Centre. 2003. Notifiable infectious diseases in Ireland, 2003. [Online.] http://www.ndsc.ie/hpsc/NotifiableDiseases/AnnualIDStatistics.
- 7.Hyams, K. C., A. L. Bourgeois, B. R. Merrell, P. Rozmajzl, J. Escamilla, S. A. Thornton, et al. 1991. Diarrheal disease during Operation Desert Shield. N. Engl. J. Med. 325:1423-1428. [DOI] [PubMed] [Google Scholar]
- 8.Mammina, C., A. Aleo, C. Roman, and A. Nastasi. Shigella sonnei biotype g carrying class 2 integrons in southern Italy: a retrospective typing study by pulsed field gel electophoresis. BMC Infect Dis. 6:117. [DOI] [PMC free article] [PubMed]
- 9.Mammina, C., M. Pontello, A. Dal Vecchio, A. Nastasi, and the Shigella sonnei Working Group. 2005. Identification of Shigella sonnei biotype g isolates carrying class 2 integrons in Italy (2001 to 2003). J. Clin. Microbiol. 43:2467-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribot, E. M., M. A. Fair, R. Gautom, D. N. Cameron, S. B. Hunter, B. Swaminathan, et al. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3:59-67. [DOI] [PubMed] [Google Scholar]