Abstract
The genus Leptospira is classified into 13 named species and 4 genomospecies based upon DNA-DNA reassociation studies. Phenotypic tests are unable to distinguish between species of Leptospira, and there is a need for a simplified molecular approach to the identification of leptospires. 16S rRNA gene sequences are potentially useful for species identification of Leptospira, but there are a large number of sequences of various lengths and quality in the public databases. 16S rRNA gene sequences of near full length and bidirectional high redundancy were determined for all type strains of the species of the Leptospiraceae. Three clades were identified within the genus Leptospira, composed of pathogenic species, nonpathogenic species, and another clade of undetermined pathogenicity with intermediate 16S rRNA gene sequence relatedness. All type strains could be identified by 16S rRNA gene sequences, but within both pathogenic and nonpathogenic clades as few as two or three base pairs separated some species. Sequences within the nonpathogenic clade were more similar, and in most cases ≤10 bp distinguished these species. These sequences provide a reference standard for identification of Leptospira species and confirm previously established relationships within the genus. 16S rRNA gene sequencing is a powerful method for identification in the clinical laboratory and offers a simplified approach to the identification of Leptospira species.
Leptospirosis is an acute febrile disease caused by pathogenic spirochetes of the genus Leptospira. The disease is maintained in nature by chronic renal infection of carrier animals and acquired by direct or indirect contact with urine or tissues from infected animals. Traditionally, several hundred serovars of Leptospira were classified into two species, Leptospira interrogans and L. biflexa (13), which contained pathogenic and saprophytic strains, respectively. These species were differentiated by several phenotypic characteristics, including growth in the presence of 8-azaguanine and growth at 13°C (14). Based upon DNA-DNA hybridization data, the genus is now classified into 17 species (1, 20, 22, 25, 32), several of which contain both pathogenic and nonpathogenic serovars. Phenotypic characteristics previously used to differentiate L. interrogans sensu lato and L. biflexa sensu lato (13) are no longer useful in the current classification.
There is a need for a method of identification for Leptospira species that is more widely available than DNA-DNA hybridization but which will yield accurate identification to the species level. Analysis of 16S rRNA gene sequence is now widely used for identification of fastidious bacteria (3), including Leptospira species (10, 23). Leptospire genomes contain two 16S rRNA genes, which are not closely linked but are on chromosome I (21, 26). Numerous 16S rRNA sequences from Leptospira serovars have been deposited in GenBank, but in many cases these are only partial-length sequences. Even though not all species have been sequenced, such sequences are potentially useful for species identification. In the present study, we determined nearly full-length 16S rRNA gene sequences of approximately 1,430 bp from well-characterized type strains and representative serovars of Leptospira species, Turneriella parva, and Leptonema illini to evaluate the use of 16S rRNA gene sequencing for species identification of leptospires.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Leptospira (n = 39 strains), Leptonema (n = 2), and Turneriella (n = 2) strains from the Centers for Disease Control and Prevention collection (Table 1) were maintained in semisolid PLM-5 medium (Serologicals Corp., Norcross, GA) containing 1.5% agar (Difco, Sparks, MD) at room temperature. Subcultures in liquid PLM-5 medium were incubated at 30°C for 7 days. The strains were chosen to represent important species and serovars causing human disease and include the type strain of all described species of Leptospiraceae.
TABLE 1.
Strains studied and their 16S rRNA gene GenBank accession numbers
| Clade | Species | Serovar | Strain | GenBank accession no. | Diffa | Gapsb |
|---|---|---|---|---|---|---|
| Pathogenic | Leptospira interrogans | Icterohaemorrhagiae | RGAT ATCC 43642T | AY631894 | R | |
| Leptospira interrogans | Australis | Ballico | AY996794 | 1 | −1 | |
| Leptospira interrogans | Autumnalis | Akiyami A | AY996791 | 1 | −1 | |
| Leptospira interrogans | Bulgarica | Mallika | AY996792 | 6 | +1, −1 | |
| Leptospira interrogans | Canicola | Hond Utrecht IV | AY996798 | 1 | −1 | |
| Leptospira interrogans | Copenhageni | M 20 | AY996790 | 0 | 0 | |
| Leptospira interrogans | Hardjo | Hardjoprajitno | AY996796 | 1 | −1 | |
| Leptospira interrogans | Hardjo | Lepto-0184 | AY996797 | 1 | −1 | |
| Leptospira interrogans | Pomona | Pomona | AY996800 | 2 | −1 | |
| Leptospira interrogans | Pyrogenes | Salinem | AY996793 | 0 | −1 | |
| Leptospira alexanderi | Manhao 3 | L60T ATCC 700520T | AY631880 | 13 | +1, −1 | |
| Leptospira alexanderi | Manzhuang | A23 | AY996803 | 15 | +3, −1 | |
| Leptospira alexanderi | Nanding | M 6901 | AY996804 | 12 | +2, −1 | |
| Leptospira borgpetersenii | Javanica | Veldrat Batavia 46T ATCC 43292T | AY887899 | 10 | −1 | |
| Leptospira borgpetersenii | Ballum | Mus 127 | AY631884 | 10 | −1 | |
| Leptospira kirschneri | Cynopteri | 3522 CT ATCC 49945T | AY631895 | 1 | −1 | |
| Leptospira kirschneri | Bim | 1051 | AY996802 | 1 | −2 | |
| Leptospira kirschneri | Bim | PUO 1247 | AY996801 | 1 | −1 | |
| Leptospira noguchii | Panama | CZ 214T ATCC 43288T | AY631886 | 9 | −1 | |
| Leptospira santarosai | Shermani | LT 821T ATCC 43286T | AY631883 | 17 | −1 | |
| Leptospira santarosai | Georgia | LT 117 | AY996805 | 15 | −1 | |
| Leptospira weilii | Celledoni | CelledoniT ATCC 43285T | AY631877 | 13 | −1 | |
| Leptospira genomospecies 1 | Sichuan | 79601T ATCC 700521T | AY631881 | 13 | −1 | |
| Intermediate | Leptospira inadai | Lyme | 10T ATCC 43289T | AY631896 | R | |
| Leptospira inadai | Aguaruna | MW 4 | AY631891 | 0 | 0 | |
| Leptospira inadai | Kaup | LT 64-68 | AY631887 | 1 | 0 | |
| Leptospira broomii | Not designated | 5399T ATCC BAA-1107T | AY796065 | 3 | 0 | |
| Leptospira fainei | Hurstbridge | BUT 6T ATCC BAA-1109T | AY631885 | 7 | 0 | |
| Leptospira fainei | Hurstbridge | BKID 6 | AY996789 | 7 | 0 | |
| Nonpathogenic | Leptospira biflexa | Patoc | Patoc IT ATCC 23582T | AY631876 | R | |
| Leptospira biflexa | Andamana | CH 11 | AY631893 | 1 | 0 | |
| Leptospira meyeri | Ranarum | Iowa City FrogT ATCC 43287T | AY631878 | 10 | 0 | |
| Leptospira meyeri | Hardjo | Went 5 | AY631889 | 10 | 0 | |
| Leptospira meyeri | Semaranga | Veldrat Semarang | AY631892 | 8 | 0 | |
| Leptospira wolbachii | Codice | CDCT ATCC 43284T | AY631879 | 6 | 0 | |
| Leptospira wolbachii | Gent | Wa Gent | AY631890 | 6 | 0 | |
| Leptospira genomospecies 3 | Holland | WaZ HollandT ATCC 700522T | AY631897 | 4 | 0 | |
| Leptospira genomospecies 4 | Hualin | LT 11-33T ATCC 700639T | AY631888 | 3 | 0 | |
| Leptospira genomospecies 5 | Saopaulo | Sao PauloT ATCC 700523T | AY631882 | 5 | 0 | |
| Other | Leptonema illini | Illini | 3055T | AY714984 | R | |
| Leptonema illini | Habaki | Habaki | AY996806 | 10 | 0 | |
| Turneriella parva | Parva | HT NCTC 11395T | AY293856 | R | ||
| Turneriella parva | Parva | S-308-81 | AY398688 | 0 | 0 |
Diff, number of base pairs differing from the prototype strain in that clade. R, prototype strain for that clade.
Gaps, insertion or deletions compared to the prototype strain.
DNA extraction and 16S rRNA gene sequencing.
DNA was extracted from cultures of 43 strains of Leptospiraceae using QIAamp DNA minikits according to the manufacturer's directions (QIAGEN, Valencia, CA). The 16S rRNA genes were amplified from the purified DNA by using the Expand High-Fidelity PCR system (Roche Diagnostics Corp., Indianapolis, IN). Briefly, each 50-μl reaction contained approximately 10 ng of DNA, 2.5 U of polymerase, 1.5 mM MgCl2, 5% (vol/vol) dimethyl sulfoxide, 200 μM deoxynucleoside triphosphates, and 100 nM concentrations of primers fD1 and rP2 corresponding to positions 8 and 1492, respectively, of the Escherichia coli 16S rRNA gene J01695 (Table 2). Amplification was performed on an AB 9700 thermocycler (Applied Biosystems, Foster City, CA) using 94°C for 5 min, followed by 35 cycles of 94°C for 15 s, 50°C for 5 s, and 72°C for 90 s, with a final single extension of 72°C for 5 min, and then held at 4°C. Amplified products were characterized by electrophoresis of 5 μl of each reaction on a 1.2% agarose gel for 30 min at 85 V. Excess nucleotides and primers were inactivated with the ExoSAP method (USB Corp., Cleveland, OH). Cycle sequencing was performed by standard protocols (27), including the use of a 45°C annealing temperature and the 16 sequencing primers listed in Table 2. Primers F785 and R802 were designed in the present study, but the majority are primers from the European rRNA database (31). Sequencing reaction products were purified with magnetic carboxylate beads (Agencourt Bioscience, Beverly MA). Reactions were sequenced on an AB 3100 (Applied Biosystems). Chromatograms were assembled and analyzed in Seqmerge (Wisconsin Package version 10.3; Accelrys, Inc., San Diego, CA) (6).
TABLE 2.
Primers used to determine 16S rRNA gene sequences in Leptospiraceae
| Primer | Sequence | Usea | Directionalityb | Source or reference |
|---|---|---|---|---|
| fD1 | CCGAATTCGTCGACAACAGA GTTTGATCCTGGCTCAG | P | F | 30 |
| rP2 | CCCGGGATCCAAGCTTACGGCTACCTTGTTACGACTT | P | R | 30 |
| fD1-5p | CCGAATTCGTCGACAACAG | S | F | This study |
| BSF8 | AGAGTTTGATCCTGGCTCAG | S | F | 31 |
| F785e | GGATTAGATACCCTGGTA | S | F | This study |
| BSR357 | CTGCTGCCTCCCGTA | S | R | 31 |
| R802e | TACCAGGGTATCTAATCC | S | R | This study |
| BSF343 | TACGGGAGGCAGCAG | S | F | 31 |
| R536e | GTATTACCGCGGCTGCTG | S | R | 17 |
| R536 | GWATTACCGCGGCKGCTG | S | R | 17 |
| F519e | CAGCAGCCGCGGTAATAC | S | F | 17 |
| BSF917 | GAATTGACGGGGRCCC | S | F | 31 |
| BSR926 | CCGTCAATTYYTTTRAGTTT | S | R | 31 |
| BSF1099 | GYAACGAGCGCAACCC | S | F | 31 |
| BSR1114 | GGTTGCGCTCGTTRC | S | R | 31 |
| BSF1391 | TGTACACACCGCCCGTC | S | F | 31 |
| BSR1407 | GACGGGCGGTGTGTRC | S | R | 31 |
| rP2-5p | CCCGGGATCCAAGCTTAC | S | R | This study |
P, PCR; S, sequencing.
F, forward; R, reverse.
Phylogenetic analysis.
Sequences were aligned with CLUSTAL X (29) in Mega 3.1 (15) and trimmed to consensus, and a neighbor joining tree was created. Gaps in the aligned sequences were replaced by Ns in BioEdit (Ibis Therapeutics, Carlsbad CA). Evolutionary distances were estimated by using the Jukes and Cantor model in GCG (6).
Nucleotide sequence accession numbers.
DNA sequences have been deposited in the GenBank database with the accession numbers shown in Table 1.
RESULTS
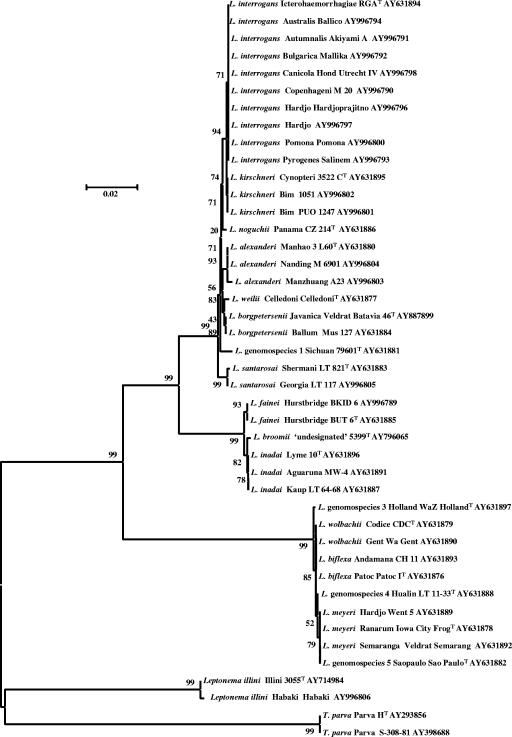
The 16S rRNA gene sequences of 43 strains, representing all 17 species of Leptospira, as well as T. parva and Leptonema illini, were determined. The length of the sequences ranged from 1,422 to 1,432 bp for leptospires and 1,428 to 1,440 bp for T. parva and Leptonema illini. These sequences were compared to each other by alignment and in dendrograms. Different serovars of the same species showed highly similar (only an average of 0.2 bases different of about 1,430) or identical 16S rRNA gene sequences (data not shown). Phylogenetic analysis of 16S rRNA gene sequences confirmed previous reports (10, 22) that the Leptospiraceae form five main clusters of species (Fig. 1). The pathogenic species, exemplified by L. interrogans, form one clade distinct from the nonpathogenic species, exemplified by L. biflexa. A third clade, comprising L. inadai, L. fainei, and L. broomii, was clearly separate from the pathogenic and nonpathogenic clades. These groupings are distinct from the remaining two clades, which include the species T. parva (12, 19) and Leptonema illini (11).
FIG. 1.
Unrooted tree of 43 Leptospiraceae 16S rRNA gene sequences. The scale bar equals the fraction of base pairs that are different.
Within the pathogenic clade, the eight species were readily identified based on consistent differences in 16S rRNA gene sequences (Fig. 2). This clade included L. alexanderi, L. borgpetersenii, L. interrogans, L. kirschneri, L. noguchii, L. santarosai, L. weilii, and Leptospira genomospecies 1. The percentage similarity of sequences within this clade was high, ≥98.6% or 2 to 20/1,431 bp different (i.e., 2 to 20 bp out of 1,1431 bp were different) (Table 3), reaffirming the high degree of species conservation among spirochetes (16, 23).
FIG. 2.
Base differences in the 16S rRNA genes of type strains formatted to emphasize the differences within each of the three clades of Leptospira. Path, pathogenic clade; Int, intermediate clade; NonP, nonpathogenic clade. Vertical numbers show the relative positions in L. interrogans AY631894. Center dots indicate the same base as L. interrogans; a dash indicates a gap. The A at position 57 in L. alexanderi would be inserted between 56 and 57 in the L. interrogans AY631894 sequence. The sequences indicate differences within each clade but do not include all differences between clades. Note that positions 95, 154, 158, 222, and 952 appear twice, and position 1077 appears in all three clades. There are >160 other positions that can be used to differentiate between Leptospira clades.
TABLE 3.
Number of base pair differences in the 16S rRNA genes of type strains of Leptospiraa
| Cladea | Species | No. of bp differences
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. interrogans | L. alexanderi | L. borgpetersenii | L. kirschneri | L. noguchii | L. santarosai | L. weilii | Leptospira genomospecies 1 | L. inadai | L. broomii | L. fainei | L. biflexa | L. meyeri | L. wolbachii | Leptospira genomospecies 3 | Leptospira genomospecies 4 | Leptospira genomospecies 5 | ||
| Path | L. interrogans | 0 | 15 | 11 | 2 | 10 | 18 | 14 | 14 | 72 | 76 | 72 | 205 | 213 | 205 | 207 | 207 | 209 |
| L. alexanderi | 0 | 9 | 13 | 17 | 19 | 13 | 18 | 72 | 76 | 73 | 210 | 218 | 210 | 212 | 212 | 214 | ||
| L. borgpetersenii | 0 | 9 | 11 | 12 | 6 | 15 | 71 | 72 | 71 | 208 | 215 | 208 | 209 | 209 | 212 | |||
| L. kirschneri | 0 | 8 | 16 | 12 | 12 | 70 | 73 | 70 | 205 | 213 | 205 | 207 | 207 | 209 | ||||
| L. noguchii | 0 | 15 | 12 | 14 | 74 | 74 | 71 | 209 | 215 | 209 | 210 | 210 | 214 | |||||
| L. santarosai | 0 | 14 | 20 | 72 | 74 | 74 | 212 | 218 | 212 | 213 | 213 | 215 | ||||||
| L. weilii | 0 | 16 | 76 | 76 | 73 | 209 | 215 | 209 | 210 | 210 | 214 | |||||||
| Leptospira genomospecies 1 | 0 | 71 | 74 | 71 | 210 | 218 | 210 | 212 | 212 | 215 | ||||||||
| Int | L. inadai | 0 | 3 | 7 | 221 | 227 | 220 | 221 | 222 | 224 | ||||||||
| L. broomii | 0 | 4 | 225 | 230 | 224 | 225 | 226 | 227 | ||||||||||
| L. fainei | 0 | 221 | 226 | 220 | 221 | 222 | 224 | |||||||||||
| NonP | L. biflexa | 0 | 10 | 6 | 4 | 3 | 10 | |||||||||||
| L. meyeri | 0 | 10 | 12 | 9 | 6 | |||||||||||||
| L. wolbachii | 0 | 7 | 7 | 10 | ||||||||||||||
| Leptospira genomospecies 3 | 0 | 5 | 12 | |||||||||||||||
| Leptospira genomospecies 4 | 0 | 9 | ||||||||||||||||
| Leptospira genomospecies 5 | 0 | |||||||||||||||||
Path, pathogenic; Int, intermediate; NonP, nonpathogenic. Insertion/deletions or mixed bases are considered mismatches.
The positions of insertions or deletions, as well as differing and mixed bases, are shown in Fig. 2, which includes the positions that differentiate the species within the three Leptospira clades but omits positions that only differentiate between clades. Note that many of the positions that differentiate the species within a specific clade are quite conserved within the other two clades. A total of 35 dissimilar base positions distinguish the type strain 16S rRNA gene sequences of pathogenic Leptospira species, previously identified by DNA-DNA hybridization studies (1) (Fig. 2). The highest 16S rRNA gene sequence similarity between the type strains of species was between those of L. interrogans and L. kirschneri (2/1,432 bp different). However, L. kirschneri serovar Cynopteri strain 3522T and serovar Bim strain PUO247 were identical based on 16S rRNA sequence and were differentiated from L. interrogans serovar Pyrogenes strain Salinem and serovar Bulgarica strain Mallika by a single base difference. Strain RGA of L. interrogans serovar Icterohaemorrhagiae differs from the majority of other strains of this species by one base. In addition to strain RGA, only serovar Copenhageni strain M20 shares an extra G at position 784. All other strains of L. interrogans have only five Gs in this position.
Among the nonpathogenic species, there were 3 to 12/1,422 bp differences between species (Table 3). This clade comprised L. biflexa, L. meyeri, L. wolbachii, and Leptospira genomospecies 3, 4, and 5. The similarity of sequences within this clade was higher than among species of the pathogenic clade (Table 3). The positions of differing and mixed bases are shown in Fig. 2.
The third cluster of Leptospira species comprised L. inadai, L. fainei, and L. broomii (Fig. 1). The sequences in this intermediate cluster were more closely related to the pathogenic cluster than to the nonpathogenic cluster, confirming previous reports of genetic relatedness (10, 22). A total of seven dissimilar bases distinguished these three species (Fig. 2). L. broomii was comparable to a mosaic of L. inadai and L. fainei, with positions 143, 144, 154, and 158 identical to L. inadai and positions 222, 1077, and 1165 identical to L. fainei, suggesting a crossover between positions 158 and 222, but there is no evidence for recombination by horizontal transfer or convergent evolution as a mechanism.
T. parva and Leptonema illini were distant from each other and from Leptospira species (Fig. 1), confirming their lack of relatedness to the genus Leptospira (19). Both species were easily identified and differentiated from Leptospira by their 16S rRNA gene sequences.
DISCUSSION
This study was performed to evaluate the use of 16S rRNA gene sequence analysis for the species identification of leptospires. 16S rRNA gene sequencing is rapidly becoming a common technique for the identification of unknown bacterial isolates, especially those of fastidious organisms such as Leptospira species (3). The identification of Leptospira isolates has been traditionally accomplished by serological methods (7), and the question of species identity was decided by pathogenicity. As a result, all pathogenic serovars were classified as L. interrogans in the past (14).
The adoption of a genotypic classification complicated the identification of leptospires because several serovars are found in more than one species and some species contain both pathogenic and nonpathogenic serovars (18). The determination of serovar is no longer sufficient to assign an isolate to its correct species. Recently, horizontal transfer of outer membrane protein genes has been shown to occur in Leptospira species (9). Horizontal transfer was proposed as the mechanism by which serovar Hardjo antigens are found in strains of both L. interrogans and L. borgpetersenii (5). Similar horizontal gene transfer probably accounts for at least five serovars (Bulgarica, Grippotyphosa, Mwogolo, Paidjan, and Valbuzzi) being shared between L. interrogans and L. kirschneri (18), the two most closely related species based on DNA-based methods and their propensity to cause human disease. There is no reason to suggest a relationship between serovar and species in pathogenic leptospires. In theory, all combinations of pathogenic serovar and species are possible, but the search for all combinations has not been exhaustive as yet. The mosaic-like 16S rRNA gene of L. broomii (compared to those of L. fainei and L. inadai) suggests that there may also have been horizontal transfer of ribosomal genes between leptospires, as has been shown for outer membrane proteins (9) and an intervening sequence (24), but there is no evidence to explain a mechanism.
We sequenced 1,422 to 1,440 bp of the 16S rRNA gene from type strains of Leptospira species, T. parva, and Leptonema illini, in addition to a number of strains representing common serovars. The near-full-length 16S rRNA gene sequences resolved in the present study allow for identification of all Leptospira species. Sequence analysis of 16S rRNA genes is a valuable tool for species identification of isolates (3), but DNA-DNA hybridization is recognized as the definitive methodology for species definition (2, 28). The value of the sequences derived in the present study is greatly enhanced due to the previous characterization of these strains by DNA-DNA hybridization (1, 32). The phylogenetic tree (Fig. 1) confirms the previously described genetic relationships between leptospires, with distinct clades comprised of pathogenic, nonpathogenic, and intermediate species (10, 22).
Only an insertion/deletion and a single base differentiate the type strains of L. interrogans and L. kirschneri (Fig. 2). These species may be confused with each other because they share some serological properties and are frequently encountered as causes of human disease. The close phylogenetic relationship between these two species and between them and L. noguchii was discussed by Haake et al. (9). It is doubtful that these species would have been distinguished by relying on 16S rRNA gene sequence analysis alone without the previous hybridization studies (1, 8, 25). Over-reliance on 16S rRNA gene sequences to choose candidates for DNA hybridization could have also resulted in misidentification of either of these species.
Redundant bidirectional sequencing utilizing 16 primer extension reactions per isolate allowed a greater consistency in resolution of mixed bases or insertion/deletions, as found in L. alexanderi serovars Manhao 3, Manzhuang, and Nanding; L. interrogans serovar Bulgarica; and L. kirschneri serovar Bim. Sequencing the first 500 bases or ignoring mixed bases, while effective for some of the genus Leptospira, would not allow the differentiation between the nonpathogens Leptospira genomospecies 5 and L. meyeri serovar Hardjo or Ranarum (data not shown). Because just one dissimilar base in 1,432 (0.07%) at position 94 and an insertion/deletion at position 784 differentiate the type strains of L. kirschneri and L. interrogans, sequences encompassing at least these positions are required in order to assure correct identification and differentiation of these two species.
Intraspecies distances sometimes exceed the interspecies distances for the 16S rRNA genes of Leptospira. A single base difference differentiated many strains of L. interrogans and L. kirschneri, so phylogenetic representation may be less meaningful than sequence identity over the variable positions. Searching for matching sequences may be performed with public databases by using BLAST or with the specialized 16S database RDPII using Seqmatch (4). BLAST gives higher scores for sequences that are equal in length or shorter than the reference but identical across the region. Seqmatch gives higher scores using mixed bases from sequences of PCR amplification products of multiple operons with a genome and is probably the preferred approach for the identification of unknown isolates.
We have shown that all recognized species of the Leptospiraceae can be identified by using a standardized 16S rRNA gene sequencing approach (27) with universal primers. Early Leptospira sequences in the public databases were frequently short (often stopping at positions where indels occur), were sometimes of poor quality due to early sequencing methods, or were derived from incompletely characterized isolates. With the addition of high-quality sequences, 16S rRNA gene sequencing has become an accurate tool for identification of all described leptospires.
Identification and characterization of leptospiral isolates is based upon polyphasic analysis, with both serological and molecular characterization being essential. For public health purposes it has become essential to identify not only the serovar but also the species of isolates in order to accurately track the transmission of leptospires during outbreaks. An effective vaccine against leptospirosis will probably be composed of serovar- and species-specific antigens. An accurate determination of the burden of disease will depend on both species identification and serovar determination to aid in vaccine development. In combination with the use of a standardized pulsed-field gel electrophoresis approach for serovar identification (R. Galloway and P. N. Levett, Abstr. Int. Conf. Emerg. Infect. Dis. 2004, abstr. 214, 2004), a transition from a serological to molecular identification and characterization of leptospires is now possible.
REFERENCES
- 1.Brenner, D. J., A. F. Kaufmann, K. R. Sulzer, A. G. Steigerwalt, F. C. Rogers, and R. S. Weyant. 1999. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int. J. Syst. Bacteriol. 49:839-858. [DOI] [PubMed] [Google Scholar]
- 2.Busse, H. J., E. B. Denner, and W. Lubitz. 1996. Classification and identification of bacteria: current approaches to an old problem. J. Biotechnol. 47:3-38. [DOI] [PubMed] [Google Scholar]
- 3.Clarridge, J. E. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Peña-Moctezuma, A., D. M. Bulach, T. Kalambaheti, and B. Adler. 1999. Comparative analysis of the LPS biosynthetic loci of the genetic subtypes of serovar Hardjo: Leptospira interrogans subtype Hardjoprajitno and Leptospira borgpetersenii subtype Hardjobovis. FEMS Microbiol. Lett. 177:319-326. [DOI] [PubMed] [Google Scholar]
- 6.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dikken, H., and E. Kmety. 1978. Serological typing methods of leptospires, p. 259-307. In T. Bergan and J. R. Norris (ed.), Methods in microbiology, vol. 11. Academic Press, London, England. [Google Scholar]
- 8.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 9.Haake, D. A., M. A. Suchard, M. M. Kelley, M. Dundoo, D. P. Alt, and R. L. Zuerner. 2004. Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J. Bacteriol. 186:2818-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hookey, J. V., J. Bryden, and L. Gatehouse. 1993. The use of 16S rDNA sequence analysis to investigate the phylogeny of Leptospiraceae and related spirochaetes. J. Gen. Microbiol. 139:2585-2590. [DOI] [PubMed] [Google Scholar]
- 11.Hovind-Hougen, K. 1979. Leptospiraceae, a new family to include Leptospira Noguchi 1917 and Leptonema gen. nov. Int. J. Syst. Bacteriol. 29:245-251. [Google Scholar]
- 12.Hovind-Hougen, K., W. A. Ellis, and A. Birch-Andersen. 1981. Leptospira parva sp. nov.: some morphological and biological characters. Zentbl. Bakteriol. Mikrobiol. Hyg. A 250:343-354. [PubMed] [Google Scholar]
- 13.Johnson, R. C., and S. Faine. 1984. Leptospira, p. 62-67. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore, Md. [Google Scholar]
- 14.Kmety, E., and H. Dikken. 1993. Classification of the species Leptospira interrogans and history of its serovars. University Press Groningen, Groningen, The Netherlands.
- 15.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 16.Kunin, V., D. Ahren, L. Goldovsky, P. Janssen, and C. A. Ouzounis. 2005. Measuring genome conservation across taxa: divided strains and united kingdoms. Nucleic Acids Res. 33:616-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S rRNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levett, P. N. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levett, P. N., R. E. Morey, R. Galloway, A. G. Steigerwalt, and W. A. Ellis. 2005. Reclassification of Leptospira parva Hovind-Hougen et al., 1982 as Turneriella parva gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 55:1497-1499. [DOI] [PubMed] [Google Scholar]
- 20.Levett, P. N., R. E. Morey, R. L. Galloway, and A. G. Steigerwalt. 2006. Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int. J. Syst. Evol. Microbiol. 56:671-673. [DOI] [PubMed] [Google Scholar]
- 21.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El-Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-De-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. De Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérolat, P., R. J. Chappel, B. Adler, G. Baranton, D. M. Bulach, M. L. Billinghurst, M. Letocart, F. Merien, and M. S. Serrano. 1998. Leptospira fainei sp. nov., isolated from pigs in Australia. Int. J. Syst. Bacteriol. 48:851-858. [DOI] [PubMed] [Google Scholar]
- 23.Postic, D., N. Riquelme-Sertour, F. Merien, P. Pérolat, and G. Baranton. 2000. Interest of partial 16S rDNA gene sequences to resolve heterogeneities between Leptospira collections: application to L. meyeri. Res. Microbiol. 151:333-341. [DOI] [PubMed] [Google Scholar]
- 24.Ralph, D., and M. McClelland. 1994. Phylogenetic evidence for horizontal transfer of an intervening sequence between species in a spirochete genus. J. Bacteriol. 176:5982-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramadass, P., B. D. W. Jarvis, R. J. Corner, D. Penny, and R. B. Marshall. 1992. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int. J. Syst. Bacteriol. 42:215-219. [DOI] [PubMed] [Google Scholar]
- 26.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 27.Sacchi, C. T., A. M. Whitney, M. W. Reeves, L. W. Mayer, and T. Popovic. 2002. Sequence diversity of Neisseria meningitidis 16S rRNA genes and use of 16S rRNA gene sequencing as a molecular subtyping tool. J. Clin. Microbiol. 40:4520-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. Grimont, P. Kampfer, M. C. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wuyts, J., Y. Van de Peer, T. Winkelmans, and R. De Wachter. 2002. The European database on small subunit rRNA. Nucleic Acids Res. 30:183-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasuda, P. H., A. G. Steigerwalt, K. R. Sulzer, A. F. Kaufmann, F. Rogers, and D. J. Brenner. 1987. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int. J. Syst. Bacteriol. 37:407-415. [DOI] [PubMed] [Google Scholar]