Abstract
The gingival sulcus contains a complex ecosystem that includes many uncultivated bacteria. Understanding the dynamics of this ecosystem in transitions between health and disease is important in advancing our understanding of the bacterial etiology of periodontitis. The objective of this longitudinal study was to examine the stability of bacterial colonization in the gingival crevice and to explore the relationship between shifts in microbial composition and changes in periodontal health status using a comprehensive, quantitative, culture-independent approach. Subgingival plaque samples and periodontal data were collected from 24 subjects over 2 years. Baseline and 2-year plaque samples were analyzed using quantitative ribosomal 16S cloning and sequencing. Ten subjects remained periodontally healthy over 2 years, the periodontal health of seven subjects worsened, and seven subjects showed clinical improvement. Bacterial stability was greatest among healthy, clinically stable subjects and lowest for subjects whose periodontal status worsened (P = 0.01). Higher numbers of species lost or gained were also observed for subjects whose clinical status changed (P = 0.009). This provides evidence that a change in periodontal status is accompanied by shifts within the bacterial community. Based on these data, measures of microbial stability may be useful in clinical diagnosis and prognosis. Regarding individual species, increases in levels of the uncultivated phylotype Veillonella sp. oral clone X042, a gram-negative bacterium and the most common member of the subgingival bacterial community, were associated with periodontal health (P = 0.04), suggesting that this is an important beneficial species. Filifactor alocis, a gram-positive anaerobe, was found at higher levels in subjects with disease (P = 0.01).
The gingival sulcus contains a complex ecosystem formed by resident and transient bacteria, and the critical role of bacteria in the etiology of chronic periodontitis is well established. The etiology is widely thought to be polymicrobial (16), but the role of individual species and their complex interactions with the host is not well understood. The earliest studies were conducted using open-ended cultivation methods. These were followed by a generation of studies employing molecular detection methods such as hybridization assays and PCR to target bacteria identified by cultivation. These less cumbersome methods allowed much larger scale studies to be carried out. The most recent advance has been cloning and sequencing of 16S rRNA genes. This approach has once again allowed an open-ended exploration of bacterial populations and has revealed the presence of many uncultivated species (18). Recent cloning studies have suggested that some of these uncultivated species, as well as unexpected named species, could be important in disease and in health (4, 5, 7). The most recent of these studies (7) used universal bacterial primers, quantitative conditions for cloning and sequencing, and sample sizes large enough to permit statistical analysis. Surprisingly, this study suggested that there are gram-negative bacteria associated with health and that gram-positive bacteria may play a role in disease (7). Further study of these complex bacterial communities is needed.
Little is known about the natural history of fluctuations in subgingival bacterial communities in periodontal health or in transitions between health and disease. Current perspectives have been gained from culturing (9, 12) and targeted molecular approaches examining changes in levels of certain bacterial species, e.g., Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans, and Tannerella forsythia, during changes in health status (12, 13, 17, 22) or in response to therapy (2, 24). However, fluctuations of a small subset of species are not likely to be representative of the changes occurring in a complex microbial community. Understanding the dynamics of this ecosystem in transitions between health and disease is important in advancing our understanding of the bacterial etiology of periodontitis. The elucidation of microbial shifts associated with disease onset and progression has obvious clinical implications, but the discovery of species associated with stable, healthy ecosystems may be equally or more important in the development of therapies for periodontitis. For example, colonization stability has been studied in the gastrointestinal tract, where colonization with normal flora decreases susceptibility to infections by exogenous pathogens such as Salmonella or Clostridium spp. (1, 23). These studies have contributed to the clinical management of gastrointestinal infections using probiotics or microbial replacement therapy (14).
In order to understand microbial fluctuations within a diverse community, it is necessary to study quantitative changes in all species over time. Although cultivation of bacteria can allow detection of previously unknown or unsuspected species (22), the profile is obviously limited to cultivable species. When targeted molecular methods capable of detecting uncultivated species are used, it is not yet technically feasible to study all species in each sample. Cloning and sequencing of 16S rRNA genes offers several advantages for studying the stability of bacterial communities. It has been used for bacterial identification and quantification in many naturally occurring microbial communities (7, 25, 26), and a large database of bacterial sequences is available. This open-ended approach allows detection of every species in a sample, including uncultivated and previously unknown organisms. Comparison of 16S rRNA gene sequences also provides a more accurate identification than phenotypic characterization. Finally, when used under carefully controlled conditions, this method can be quantitative.
The objective of the present study was to examine the stability of bacterial colonization in the gingival crevice and to identify microbial shifts associated with changes in periodontal health by using a comprehensive, open-ended, quantitative molecular approach that allows the examination of the relationship between changes in health status and changes in the predominant subgingival flora, including cultivated and uncultivated species.
MATERIALS AND METHODS
Subject selection.
Subjects for this institutionally approved study were a subgroup of a population that was monitored over 2 years in order to study the colonization stability of oral bacteria. Subjects were recruited from four churches with diverse demographic profiles in Columbus, Ohio. For the study reported here, exclusion criteria were a history of antibiotic therapy in the past 3 months and any history of smoking or diabetes, and inclusion criteria were an age of 40 years or more and the presence of at least 20 teeth. Twenty-five subjects who met these criteria were randomly selected, and 24 of these were included in the statistical analysis.
Periodontal examination and sample collection.
Each subject received a periodontal examination at baseline and after 24 months. Probe depths (PD) were recorded for six sites around each tooth at each visit. Periodontal health was defined as the absence of sites with PD of >4 mm (deep sites) at baseline, and periodontitis was defined as two or more sites with PD of ≥5 mm. Subsequent changes of ≥2 mm in PD at two or more sites were considered indicative of a change in periodontal health. Subgingival plaque samples were collected at each visit on sterile endodontic paper points (Caulk Dentsply). Plaque was collected and pooled from the mesial sulcus of every tooth in the mouth. Samples were placed in a 1.5-ml microcentrifuge tube and frozen.
DNA amplification, cloning, and sequencing.
DNA isolation, cloning, and sequencing of the 16S rRNA gene was carried out using a protocol described previously (7). Briefly, bacterial 16S rRNA genes were amplified from community DNA using 22 cycles of PCR with broad-range eubacterial primers A17 (5′-GTT TGA TCC TGG CTC AG-3′) and 317 (5′-AAG GAG GTG ATC CAG GC-3′). The amplicons were purified and cloned using a TOPO TA cloning kit (Invitrogen, San Diego, CA). The presence of inserts of the correct size (≅1,500 bp) was confirmed by PCR amplification and gel electrophoresis. The products were then purified with a Qiaquick 96 kit (QIAGEN, Valencia, CA) and sequenced with an ABI Prism cycle sequencing kit (BigDye Terminator Cycle Sequencing) using an ABI 3730 instrument.
Sequence analysis.
Partial sequences of 900 to 1,100 bp were obtained from each amplicon. The sequences generated were compared to the GenBank database to identify the closest match using a Time Logic DeCypher TeraBlast server hosted by the Ohio Supercomputer Center. Sequences with low homology to GenBank entries were screened for chimeras using the ChimeraCheck program of Ribosomal Database Project II (http://rdp8.cme.msu.edu/html/). Eleven clones were identified as chimeric sequences and excluded from further analysis. Sequences were aligned and a similarity matrix constructed from the alignments using the method of Jukes and Cantor. Phylogenetic trees were constructed using the neighbor-joining method. MacVector software was used to generate alignments and similarity matrices for phylogenetic analysis.
Stability and modeling variability.
Microbial stability in each subject was computed as the number of bacterial clones that were conserved over 2 years. In order to do this, 100 clones from baseline were compared to 100 clones identified at the 2-year time point. The percentage of clones that were identical at both time points was calculated.
Identifying just 100 clones from each sample introduces some degree of random variability, so a computational model was constructed to estimate this. Two virtual bacterial populations for the model were created, one by pooling the 200 clones identified from the subject with the least diverse bacterial profile, and one consisting of the 200 clones identified from the subject with the most diverse bacterial profile. One hundred clones were randomly selected from each virtual population using a random number generator. Fifty replicate data sets (bootstraps) were generated for each, and the overall average percentage of conservation was calculated. This value was used to adjust for methodological sampling error. Since 75% conservation was the average resulting from sampling of an invariant population, this value was assumed to represent total (100%) conservation of the microbial community. All observed values were normalized using this correction.
Statistical analysis.
Statistical analysis was carried out with JMP (SAS Institute Inc., Cary, NC). Analysis of variance (ANOVA) was used for comparisons of microbial conservation and diversity (number of species present and Shannon-Weiner diversity index) between periodontally stable subjects and subjects whose periodontal health status improved or worsened. ANOVA and chi-square analyses were used for comparisons on demographic variables. Comparisons of the levels of each species as well as of changes in levels (increase or decrease) between the stable healthy group and the groups whose periodontal health improved or worsened were made using the Kruskal-Wallis test.
RESULTS
This study examined the stability of subgingival bacteria over 2 years for a group of 25 subjects. The subjects were divided into three groups based on changes in their clinical periodontal status over this period: stable healthy, improved, or worse. One subject with periodontitis who was stable over the 2-year period was not included in the statistical analysis.
At initial presentation, the mean ages of the stable healthy group, the group whose periodontal health worsened, and the group whose periodontal condition improved were 54 years (standard deviation [SD], 8.05), 48 years (SD, 8.7), and 52.2 years (SD, 9.8), respectively. The difference was not significant by ANOVA. Males formed 85.7% of the improved group, 14.2% of the worsened group, and 30% of the stable group. Although the results were significant (P = 0.02) by chi-square analysis, microbial stability was not significantly different between males and females within any group. The stable group was 72.3% white, while the improved and worse groups were 79% and 84.3% white, respectively. This difference was not statistically significant by chi-square analysis.
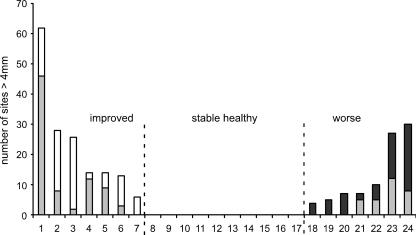
Figure 1 shows the changes in periodontal status of the 24 subjects over 2 years. At initial presentation, the periodontal status of subjects ranged from periodontally healthy to moderate periodontitis. Ten subjects maintained periodontal health for the duration of the study (stable healthy group). Seven subjects had slight to moderate periodontitis at baseline and evidenced improvement in periodontal status at the end of 2 years (improved group). Seven subjects showed an increase in the number of deep sites (worse group). Three of these subjects were healthy at baseline and presented with initial disease at the 2-year time-point, and four showed worsening of slight periodontitis present at baseline. A single additional subject with periodontitis that neither worsened nor improved was not included in the analysis.
FIG. 1.
Clinical stability of 24 subjects over 2 years. For the improved group, the shaded and open bars taken together indicate the number of sites with probe depths of >4 mm (deep sites) at baseline. Open bars represent decreases in the number of deep sites that occurred between the baseline and final sampling times. For the worse group, the shaded bars indicate initial clinical status and the solid bars represent increases in the number of deep sites that occurred between the baseline and final sampling times.
A quantitative approach to cloning and sequencing of 16S rRNA genes that maintained the relative proportions of individual species was used (6). To avoid plateau effects and retain a representative set of amplicons, a low cycle number and a set of broad universal primers were used. One hundred bacterial 16S clones were sequenced from each sample to allow statistical comparisons to be made. Sequence data were obtained from each subject at baseline and at 2 years. Thus, a total of 4,800 clones were identified for this study. A total of 260 species or phylotypes were identified. Table 1 lists these species in the order of their overall frequency and shows the mean levels over 2 years in each sample group for the 136 most common species or phylotypes. Data for the entire set of 260 species are available online (see Table S1A in the supplemental material).
TABLE 1.
Species and phylotypes for each clinical group, arranged in order of decreasing frequencya
| Overall rank | Species/phylotype(s) | % of total clonesb | Mean % of total clones ± SD for individuals with the following clinical status:
|
||
|---|---|---|---|---|---|
| Stable | Better | Worse | |||
| 1 | Veillonella sp. oral clone X042 | 9.52 | 10.3 ± 4.0 | 8.1 ± 7.5 | 9.8 ± 8.1 |
| 2 | Campylobacter gracilis | 3.65 | 4.5 ± 4.3 | 4.2 ± 4.7 | 1.9 ± 2.6 |
| 3 | Streptococcus mitis | 3.48 | 3.4 ± 3.2 | 3.4 ± 3.2 | 3.5 ± 2.2 |
| 4 | Streptococcus gordonii | 3.21 | 4.5 ± 5.7 | 3.1 ± 1.8 | 1.5 ± 1.7 |
| 5 | Streptococcus oralis | 2.75 | 2.7 ± 2.3 | 3.4 ± 2.9 | 2.1 ± 1.9 |
| 6 | Streptococcus sanguinis | 2.42 | 3.2 ± 3.9 | 1.9 ± 2.4 | 1.9 ± 2.0 |
| 7 | Streptococcus sp. oral strain 12F | 2.42 | 2.7 ± 2.0 | 3.5 ± 3.9 | 1.0 ± 1.7 |
| 8 | Peptostreptococcus sp. oral clone FG014 | 2.17 | 2.1 ± 2.1 | 2.3 ± 2.6 | 2.1 ± 5.1 |
| 9 | Selenomonas infelix | 2.13 | 2.0 ± 1.9 | 1.8 ± 1.4 | 2.7 ± 2.4 |
| 10 | Campylobacter showae | 1.92 | 1.9 ± 2.1 | 2.0 ± 2.9 | 1.9 ± 2.8 |
| 11 | Peptostreptococcus sp. oral clone AJ062 | 1.90 | 0.9 ± 2.3 | 0.5 ± 1.4 | 4.8 ± 17.0 |
| 12 | Streptococcus genomospecies C8 | 1.88 | 2.3 ± 4.0 | 1.7 ± 2.8 | 1.5 ± 4.0 |
| 13 | Streptococcus intermedius | 1.81 | 1.9 ± 1.7 | 1.6 ± 2.3 | 1.9 ± 3.2 |
| 14 | Gemella morbillorum | 1.73 | 1.5 ± 1.7 | 2.4 ± 3.5 | 1.4 ± 1.7 |
| 15 | Gemella haemolysans | 1.63 | 1.9 ± 1.6 | 1.5 ± 1.7 | 1.4 ± 1.8 |
| 16 | Streptococcus pneumoniae | 1.63 | 1.8 ± 2.1 | 1.4 ± 2.2 | 1.6 ± 1.8 |
| 17 | Filifactor alocis | 1.48 | 0.2 ± 0.4 | 1.4 ± 2.9 | 3.4 ± 3.8 |
| 18 | Granulicatella elegans | 1.46 | 1.6 ± 1.6 | 1.6 ± 1.4 | 1.2 ± 1.4 |
| 19 | Peptostreptococcus sp. oral clone BS044 | 1.46 | 1.3 ± 1.8 | 1.7 ± 1.9 | 1.4 ± 1.6 |
| 20 | Veillonella atypica | 1.42 | 1.6 ± 2.1 | 1.2 ± 2.2 | 1.4 ± 1.9 |
| 21 | Streptococcus sp. oral clone 2056B | 1.23 | 1.0 ± 1.4 | 1.6 ± 2.3 | 1.3 ± 2.0 |
| 22 | Peptostreptococcus micros | 1.19 | 1.9 ± 2.8 | 0.7 ± 1.7 | 0.7 ± 1.5 |
| 23 | Neisseria elongata | 1.17 | 1.4 ± 3.7 | 1.3 ± 1.9 | 0.6 ± 2.8 |
| 24 | Streptococcus sp. oral strain 7A | 1.08 | 0.9 ± 1.5 | 1.4 ± 1.7 | 1.4 ± 1.8 |
| 25 | Selenomonas sputigena | 0.98 | 0.8 ± 1.4 | 1.1 ± 2.2 | 1.1 ± 1.9 |
| 26 | Streptococcus sp. oral clone FN051 | 0.96 | 0.7 ± 1.5 | 1.7 ± 2.2 | 0.7 ± 1.5 |
| 27 | Capnocytophaga gingivalis | 0.94 | 0.7 ± 0.9 | 0.9 ± 1.2 | 1.3 ± 1.5 |
| 28 | Campylobacter concisus | 0.90 | 1.3 ± 2.0 | 0.6 ± 1.0 | 0.6 ± 1.2 |
| 29 | Selenomonas sp. oral clone EW051a | 0.88 | 1.0 ± 1.4 | 0.1 ± 0.5 | 1.5 ± 2.2 |
| 30 | Streptococcus anginosus | 0.81 | 0.5 ± 0.9 | 0.6 ± 1.0 | 1.4 ± 1.3 |
| 31 | Megasphaera sp. oral clone BB166 | 0.79 | 0.8 ± 1.2 | 0.9 ± 1.5 | 0.6 ± 1.0 |
| 32 | Eikenella corrodens | 0.73 | 0.8 ± 1.0 | 0.9 ± 1.0 | 0.6 ± 1.2 |
| 33 | Dialister sp. oral clone E2_20 | 0.71 | 0.5 ± 1.0 | 1.1 ± 1.6 | 0.7 ± 1.1 |
| 34 | Streptococcus genomospecies C7 | 0.67 | 0.6 ± 1.2 | 1.3 ± 2.3 | 0.2 ± 0.6 |
| 35 | Neisseria sp. oral clone AP060 | 0.67 | 0.9 ± 1.7 | 0.9 ± 1.5 | 0.2 ± 0.8 |
| 36 | Streptococcus salivarius | 0.63 | 0.8 ± 1.8 | 0.3 ± 0.6 | 0.7 ± 1.4 |
| 37 | Selenomonas sp. oral clone AA024 | 0.63 | 1.1 ± 2.5 | 0.3 ± 0.7 | 0.2 ± 0.8 |
| 38 | Selenomonas sp. oral clone AJ036 | 0.63 | 0.6 ± 0.8 | 0.6 ± 1.0 | 0.6 ± 1.2 |
| 39 | Streptococcus infantis | 0.58 | 0.6 ± 1.0 | 0.5 ± 1.3 | 0.6 ± 1.0 |
| 40 | Dialister sp. oral clone BS095 | 0.58 | 0.7 ± 1.0 | 0.4 ± 0.9 | 0.7 ± 0.8 |
| 41 | Selenomonas noxia | 0.56 | 0.7 ± 1.2 | 0.7 ± 0.9 | 0.3 ± 0.6 |
| 42 | Gemella sanguinis | 0.50 | 0.6 ± 1.9 | 0.6 ± 1.3 | 0.2 ± 0.4 |
| 43 | Dialister pneumosintes | 0.50 | 0.4 ± 0.8 | 0.4 ± 0.8 | 0.8 ± 1.1 |
| 44 | Dialister invisus | 0.50 | 0.7 ± 1.3 | 0.5 ± 0.9 | 0.2 ± 0.4 |
| 45 | Selenomonas sp. oral clone DY027 | 0.48 | 0.5 ± 1.0 | 0.4 ± 0.9 | 0.5 ± 1.1 |
| 46 | Selenomonas sp. oral clone DS051 | 0.48 | 0.4 ± 0.7 | 1.0 ± 1.7 | 0.1 ± 0.4 |
| 47 | Selenomonas sp. oral clone IK004 | 0.46 | 0.3 ± 0.7 | 0.6 ± 1.7 | 0.5 ± 1.3 |
| 48 | Veillonella sp. oral clone BU083 | 0.46 | 0.4 ± 0.9 | 0.2 ± 0.4 | 0.9 ± 1.9 |
| 49 | Capnocytophaga granulosa | 0.44 | 0.9 ± 1.4 | 0.1 ± 0.5 | 0.1 ± 0.3 |
| 50 | Streptococcus sp. oral clone EK048 | 0.44 | 0.3 ± 0.8 | 0.6 ± 1.4 | 0.5 ± 0.8 |
| 51 | Abiotrophia para-adiacens | 0.42 | 0.6 ± 1.1 | 0.4 ± 0.8 | 0.2 ± 0.6 |
| 52 | Abiotrophia sp. oral clone P4PA_155 | 0.42 | 0.5 ± 0.9 | 0.4 ± 0.8 | 0.5 ± 0.9 |
| 53 | Streptococcus parasanguinis | 0.42 | 0.4 ± 1.2 | 0.7 ± 1.9 | 0.2 ± 0.6 |
| 54 | Selenomonas sp. oral clone DS071 | 0.42 | 0.1 ± 0.2 | 0.6 ± 1.2 | 0.8 ± 1.3 |
| 55 | Selenomonas sp. oral clone EQ054 | 0.42 | 0.5 ± 1.2 | 0.4 ± 0.8 | 0.3 ± 0.6 |
| 56 | Neisseria meningitidis | 0.42 | 0.5 ± 0.8 | 0.2 ± 0.6 | 0.5 ± 0.8 |
| 57 | Campylobacter rectus | 0.42 | 0.1 ± 0.5 | 0.6 ± 1.0 | 0.7 ± 1.1 |
| 58 | Rothia dentocariosa | 0.40 | 0.6 ± 1.1 | 0.4 ± 0.9 | 0.1 ± 0.3 |
| 59 | Streptococcus cristatus | 0.40 | 0.3 ± 0.6 | 0.5 ± 0.9 | 0.4 ± 0.9 |
| 60 | Selenomonas dianae | 0.40 | 0.6 ± 0.9 | 0.1 ± 0.3 | 0.4 ± 0.8 |
| 61 | Capnocytophaga sp. oral clone AH015 | 0.38 | 0.3 ± 0.4 | 0.6 ± 1.3 | 0.4 ± 0.8 |
| 62 | Streptococcus sinensis | 0.38 | 0.3 ± 0.6 | 0.3 ± 0.5 | 0.6 ± 0.8 |
| 63 | Kingella oralis | 0.38 | 0.3 ± 0.7 | 0.9 ± 1.2 | 0.0 ± 0.0 |
| 64 | Neisseria denitrificans | 0.38 | 0.4 ± 0.7 | 0.7 ± 1.3 | 0.1 ± 0.3 |
| 65 | Enterococcus faecalis | 0.35 | 0.6 ± 0.9 | 0.2 ± 0.6 | 0.1 ± 0.4 |
| 66 | Eubacterium brachy | 0.35 | 0.3 ± 0.7 | 0.6 ± 1.0 | 0.2 ± 0.6 |
| 67 | Capnocytophaga sputigena | 0.33 | 0.3 ± 0.7 | 0.5 ± 0.8 | 0.2 ± 0.6 |
| 68 | Eubacterium sp. oral clone EW053 | 0.33 | 0.3 ± 1.0 | 0.4 ± 0.8 | 0.2 ± 0.6 |
| 69 | Selenomonas sp. oral clone EW084 | 0.33 | 0.5 ± 0.1 | 0.2 ± 0.6 | 0.2 ± 0.6 |
| 70 | Eubacterium saphenum | 0.31 | 0.3 ± 0.8 | 0.1 ± 0.3 | 0.6 ± 1.0 |
| 71 | Porphyromonas gingivalis | 0.29 | 0.0 ± 0.0 | 0.2 ± 0.6 | 0.8 ± 2.4 |
| 72 | Streptococcus agalactiae | 0.29 | 0.3 ± 1.1 | 0.3 ± 0.8 | 0.2 ± 0.6 |
| 73 | Selenomonas sp. oral clone DO042 | 0.29 | 0.2 ± 0.4 | 0.5 ± 0.9 | 0.3 ± 0.8 |
| 74 | Streptococcus bovis | 0.27 | 0.2 ± 0.4 | 0.5 ± 1.1 | 0.2 ± 0.4 |
| 75 | Streptococcus suis | 0.27 | 0.3 ± 0.5 | 0.1 ± 0.3 | 0.4 ± 0.8 |
| 76 | Peptostreptococcus sp. oral clone CK035 | 0.27 | 0.4 ± 0.9 | 0.1 ± 0.3 | 0.3 ± 0.8 |
| 77 | Selenomonas-like sp. oral clone CS015 | 0.27 | 0.1 ± 0.2 | 0.3 ± 0.7 | 0.5 ± 0.8 |
| 78 | Selenomonas sp. oral clone CS002 | 0.27 | 0.1 ± 0.2 | 0.6 ± 1.3 | 0.2 ± 0.6 |
| 79 | Corynebacterium matruchotii | 0.25 | 0.2 ± 0.4 | 0.4 ± 0.9 | 0.1 ± 0.5 |
| 80 | Streptococcus sp. oral clone 4093B | 0.25 | 0.2 ± 0.7 | 0.3 ± 0.8 | 0.3 ± 0.5 |
| 81 | Streptococcus sp. oral clone FX003 | 0.25 | 0.3 ± 0.6 | 0.1 ± 0.5 | 0.4 ± 0.9 |
| 82 | Eubacterium sp. oral clone E1-K17 | 0.25 | 0.3 ± 0.7 | 0.2 ± 0.4 | 0.2 ± 0.6 |
| 83 | Treponema sp. strain Smibert-5 | 0.25 | 0.2 ± 0.4 | 0.2 ± 0.8 | 0.4 ± 0.9 |
| 84 | Atopobium rimae | 0.23 | 0.4 ± 0.9 | 0.0 ± 0.0 | 0.2 ± 0.6 |
| 85 | Streptococcus pyogenes | 0.23 | 0.3 ± 0.8 | 0.3 ± 0.6 | 0.1 ± 0.4 |
| 86 | Desulfobulbus sp. oral clone CH031 | 0.23 | 0.3 ± 0.8 | 0.0 ± 0.0 | 0.4 ± 1.2 |
| 87 | Peptostreptococcus sp. oral clone MDA2346-2 | 0.23 | 0.0 ± 0.0 | 0.3 ± 0.6 | 0.5 ± 1.2 |
| 88 | Selenomonas sp. oral clone CS024 | 0.23 | 0.4 ± 0.8 | 0.3 ± 0.0 | 0.3 ± 0.6 |
| 89 | Selenomonas sp. oral clone GT010 | 0.23 | 0.5 ± 1.4 | 0.1 ± 0.0 | 0.1 ± 0.5 |
| 90 | Selenomonas lacticifex | 0.23 | 0.6 ± 1.6 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 91 | Alysiella filiformis | 0.23 | 0.0 ± 0.0 | 0.8 ± 1.9 | 0.0 ± 0.0 |
| 92 | Lautropia sp. oral clone AP009 | 0.23 | 0.1 ± 0.2 | 0.7 ± 1.3 | 0.0 ± 0.0 |
| 93 | Granulicatella adiacens | 0.21 | 0.3 ± 0.6 | 0.2 ± 0.6 | 0.1 ± 0.5 |
| 94 | Gemella sp. strain 1754-94 | 0.21 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.6 ± 0.9 |
| 95 | Streptococcus intestinalis | 0.21 | 0.3 ± 0.8 | 0.2 ± 0.6 | 0.1 ± 0.3 |
| 96 | Streptococcus mutans | 0.21 | 0.2 ± 0.7 | 0.1 ± 0.4 | 0.3 ± 0.6 |
| 97 | Streptococcus peroris | 0.21 | 0.2 ± 0.4 | 0.1 ± 0.5 | 0.4 ± 0.7 |
| 98 | Catonella sp. oral clone FL037 | 0.21 | 0.3 ± 0.8 | 0.0 ± 0.0 | 0.4 ± 0.9 |
| 99 | Catonella morbi | 0.21 | 0.1 ± 0.3 | 0.4 ± 0.6 | 0.2 ± 0.6 |
| 100 | Selenomonas noxia-like sp. oral clone CI002 | 0.21 | 0.5 ± 1.1 | 0.0 ± 0.0 | 0.1 ± 0.3 |
| 101 | Selenomonas sp. oral clone GI064 | 0.21 | 0.0 ± 0.0 | 0.7 ± 0.2 | 0.0 ± 0.0 |
| 102 | Neisseria flava | 0.21 | 0.3 ± 0.8 | 0.1 ± 0.4 | 0.2 ± 0.8 |
| 103 | Campylobacter hominis | 0.21 | 0.2 ± 0.5 | 0.4 ± 0.9 | 0.0 ± 0.0 |
| 104 | Corynebacterium sp. oral clone AK153 | 0.19 | 0.2 ± 0.5 | 0.4 ± 0.9 | 0.0 ± 0.0 |
| 105 | Streptococcus hyointestinalis | 0.19 | 0.0 ± 0.0 | 0.2 ± 0.6 | 0.4 ± 0.9 |
| 106 | Streptococcus sp. oral clone CH016 | 0.19 | 0.1 ± 0.2 | 0.5 ± 0.8 | 0.1 ± 0.3 |
| 107 | Streptococcus sp. oral clone BM035 | 0.19 | 0.4 ± 0.8 | 0.0 ± 0.0 | 0.1 ± 0.5 |
| 108 | Eubacterium saburreum | 0.19 | 0.4 ± 0.8 | 0.1 ± 0.3 | 0.0 ± 0.0 |
| 109 | Megasphaera micronuciformis | 0.19 | 0.1 ± 0.5 | 0.2 ± 0.6 | 0.3 ± 0.8 |
| 110 | Selenomonas sp. oral clone EW076 | 0.19 | 0.3 ± 0.6 | 0.1 ± 0.3 | 0.2 ± 0.4 |
| 111 | Selenomonas sp. oral clone DD020 | 0.19 | 0.3 ± 0.6 | 0.1 ± 0.5 | 0.1 ± 0.5 |
| 112 | Veillonella parvula | 0.19 | 0.1 ± 0.3 | 0.2 ± 0.6 | 0.3 ± 0.6 |
| 113 | Neisseria genomospecies P1 clone P4PC_20 | 0.19 | 0.4 ± 1.1 | 0.0 ± 0.0 | 0.1 ± 0.4 |
| 114 | Neisseria pharyngis | 0.19 | 0.3 ± 0.6 | 0.0 ± 0.0 | 0.2 ± 0.4 |
| 115 | Deferribacteres sp. oral clone BH007 | 0.17 | 0.0 ± 0.0 | 0.4 ± 0.9 | 0.1 ± 0.4 |
| 116 | Streptococcus didelphis | 0.17 | 0.4 ± 0.9 | 0.1 ± 0.3 | 0.1 ± 0.3 |
| 117 | Streptococcus oligofermentans | 0.17 | 0.1 ± 0.2 | 0.4 ± 0.6 | 0.1 ± 0.4 |
| 118 | Streptococcus sp. oral clone 3097C | 0.17 | 0.1 ± 0.2 | 0.1 ± 0.5 | 0.4 ± 0.6 |
| 119 | Streptococcus sp. oral clone AY020 | 0.17 | 0.3 ± 0.6 | 0.1 ± 0.4 | 0.1 ± 0.3 |
| 120 | Streptococcus sp. oral clone BW009 | 0.17 | 0.4 ± 0.8 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 121 | Catonella sp. oral clone EZ006 | 0.17 | 0.1 ± 0.2 | 0.2 ± 0.8 | 0.3 ± 0.8 |
| 122 | Johnsonella ignava | 0.17 | 0.2 ± 0.7 | 0.3 ± 0.8 | 0.0 ± 0.0 |
| 123 | Selenomonas sp. oral clone EW079 | 0.17 | 0.2 ± 0.5 | 0.1 ± 0.4 | 0.2 ± 0.6 |
| 124 | Veillonella dispar | 0.17 | 0.2 ± 0.4 | 0.1 ± 0.5 | 0.2 ± 0.4 |
| 125 | Treponema sp. strain VI:G:G47 | 0.17 | 0.0 ± 0.0 | 0.1 ± 0.5 | 0.4 ± 0.9 |
| 126 | Treponema sp. strain V:19:D36 | 0.17 | 0.0 ± 0.0 | 0.3 ± 0.7 | 0.3 ± 0.8 |
| 127 | Deferribacteres sp. oral clone BA121 | 0.15 | 0.2 ± 0.7 | 0.2 ± 1.0 | 0.0 ± 0.0 |
| 128 | Gemella sp. oral strain C24KA | 0.15 | 0.1 ± 0.3 | 0.2 ± 0.6 | 0.1 ± 0.4 |
| 129 | Streptococcus sp. oral clone 2061A | 0.15 | 0.1 ± 0.2 | 0.1 ± 0.3 | 0.4 ± 0.9 |
| 130 | Streptococcus uberis | 0.15 | 0.1 ± 0.2 | 0.3 ± 0.6 | 0.1 ± 0.4 |
| 131 | Dialister sp. oral strain GBA27 | 0.15 | 0.2 ± 0.4 | 0.2 ± 0.4 | 0.1 ± 0.3 |
| 132 | Eubacterium sp. oral clone EI074 | 0.15 | 0.1 ± 0.3 | 0.3 ± 0.6 | 0.1 ± 0.5 |
| 133 | Eubacterium sp. oral clone DO008 | 0.15 | 0.1 ± 0.3 | 0.0 ± 0.0 | 0.4 ± 1.2 |
| 134 | Eubacterium sp. oral clone IR009 | 0.15 | 0.2 ± 0.7 | 0.1 ± 0.3 | 0.1 ± 0.5 |
| 135 | Megasphaera sp. oral clone BU057 | 0.15 | 0.2 ± 0.5 | 0.1 ± 0.5 | 0.1 ± 0.4 |
| 136 | Selenomonas-like sp. oral strain FNA3 | 0.15 | 0.3 ± 0.7 | 0.0 ± 0.0 | 0.1 ± 0.3 |
The 136 most common species are listed here. For a complete listing of all 260 species detected, see Table S1A in the supplemental material.
Values are means of baseline and 24-month samples.
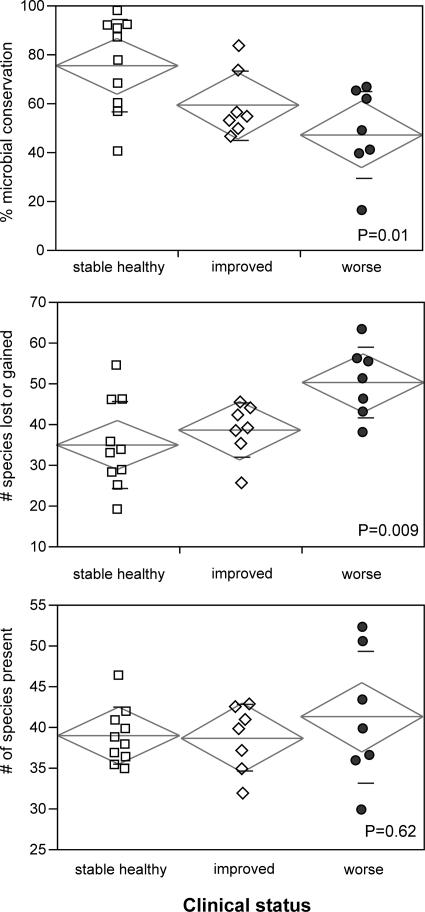
Figure 2 shows the relationship between periodontal and microbial stability. The mean adjusted microbial conservation of the stable healthy group was 75.5% (SD, 18.7%), that of the improved group was 59.2% (SD, 14.1%), and that of the worse group was 47.4% (SD, 17.8%). This difference was statistically significant (P = 0.01) by ANOVA. The number of species lost or gained is shown in the center graph. The improved and worsened groups showed a greater mean number of species lost or gained than the stable group (38.5 [SD, 6.7], 50.3 [SD, 8.6], and 35 [SD, 10.8], respectively; P = 0.009 by ANOVA). However, the complexity of the microbial community, as measured by the total number of species detected (Fig. 2) and by the Shannon-Weiner diversity index, was similar for all groups: the mean number of species detected in each subject in the stable healthy group, the improved group, and the worse group was 39.05 (SD, 3.5), 38.7 (SD, 4.1), and 41.28 (SD, 8.1), respectively. The diversity indices were 3.33 (SD, 0.3), 3.36 (SD, 0.3), and 3.32 (SD, 0.9), respectively. These values were not significantly different by ANOVA.
FIG. 2.
Relationship between periodontal status and microbial stability and diversity for 24 subjects over 2 years. (Top) Relationship between clinical status and microbial conservation. (Center) Relationship between the number of species lost or gained over 2 years and clinical status. (Bottom) Relationship between mean microbial diversity (number of species present in each subject over 2 years) and clinical status. The standard deviation (small lines), group mean (central line in diamond), and 95% confidence intervals (height of diamond) are shown for each group.
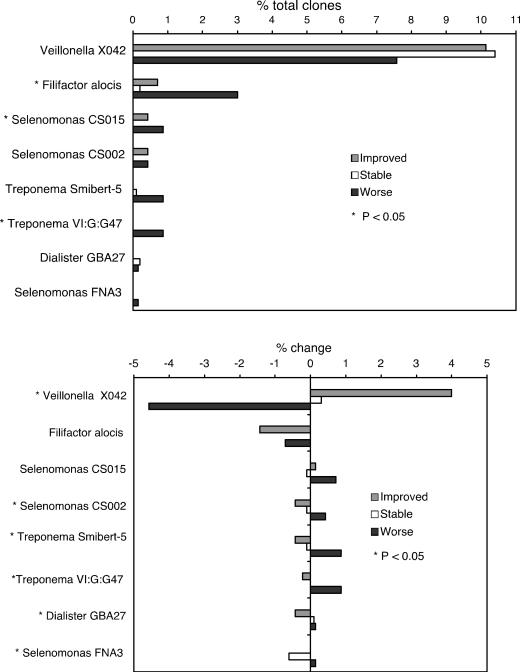
Species or phylotypes that showed a significant relationship to periodontal health status are shown in Fig. 3. Bacterial levels at the 2-year time point for the three groups are shown in the upper graph, and the lower graph of Fig. 3 shows the changes (increases or decreases in levels over 2 years) in the three groups.
FIG. 3.
Mean levels at 2 years (top) and change in levels (mean level at 2 years minus mean level at baseline) (bottom) for species significantly associated with improvement or worsening of periodontitis. (Top) Percentage of total clones accounted for by each species for the stable healthy, improved, and worse groups at the final sampling time. (Bottom) Percentage of change between the initial and final sampling times for the same species and clinical groups. Species showing significant differences (P < 0.05) among the three clinical groups are indicated by an asterisk.
DISCUSSION
Microbial and clinical stability.
A comparison among stable, periodontally healthy subjects and subjects whose periodontal disease status either improved or worsened after 2 years showed that bacterial instability was greatest in the group whose periodontal status worsened (Fig. 2, top). Higher numbers of species changeovers were also observed for subjects whose clinical status was unstable (either improved or worsened) (Fig. 2, center). This provides evidence that changes in periodontal status are associated with shifts in the composition of the bacterial community.
An average of 75.5% conservation of the bacterial community was seen for healthy subjects, with one subject demonstrating 98% conservation. The clinical criteria for change in health status were quite stringent, meaning that some subjects classified as stable may in fact have been undergoing clinical change, so the relationship between clinical and microbial stability may be even stronger than these data indicate. The microbial conservation of a single subject with moderate to severe periodontitis whose disease status remained stable over the 2 years was also analyzed (data not shown), and the adjusted microbial stability for this individual was 73.3%, close to the mean of the stable healthy group (75.5%).
Subjects whose clinical status worsened often showed conservation of less than half the bacterial community, and in one extreme case only 16% of the flora was conserved, but no subject exhibited complete replacement of subgingival bacteria during the 2-year observation period.
No significant relationship was observed between the total number of species present and microbial or clinical stability or disease status, although a much larger range was observed among subjects whose periodontal health worsened. This does not lend support to the model, based on cultivation, that periodontal health is associated with flora of low complexity while a more diverse flora is seen in disease (8, 10, 11). It should be noted that identification of just 100 clones from each sample does not provide detection of all species present in the samples; the actual number of species would be higher.
Altogether, these findings suggest that changes in periodontal health are accompanied by a shift within oral bacterial communities, as well as the corollary that a stable clinical state is reflected in a stable oral microbiota. An understanding of this relationship could potentially be useful for the development of diagnostic and prognostic tests for periodontitis. Understanding of the triggers for these microbial shifts will be important for the development of therapeutic approaches. There are many possible factors that could drive these bacterial shifts, such as changes in oral hygiene practices or host-associated biological modulators, or acquisition of new strains or species of bacteria. How these factors influence oral bacterial communities and act as triggers for change is not well understood and deserves further study.
Bacteria associated with health or disease.
The data were analyzed at the level of species to identify species and phylotypes associated with health or disease. Nonparametric statistics were employed because of the skewed distributions typically observed with bacterial counts. Due to the sample size and the exploratory nature of the study, α levels were set at 0.05 without corrections for multiple tests to allow identification of candidates for further study.
The distribution of the genera in the three sample groups was consistent with that found in a previous study using a similar methodology (7), although the previous study design involved site-specific disease samples rather than the whole-mouth strategy used in the current study. This could have resulted in some dilution of the disease-associated bacteria in the present study, since only a subset of the sampled sites were affected by periodontitis, but the previous study showed similar profiles for deep and shallow sites in the same mouth (7). The most numerous genera overall were Streptococcus, Veillonella, Selenomonas, Campylobacter, and Peptostreptococcus. This is similar to earlier observations using cultivation (22) but more closely approaches observations made using molecular methods (7). The relationship to disease was examined for both absolute levels and amounts of change over 2 years for all species detected. Species with significant relationships are shown in Fig. 3.
Candidate pathogenic species.
Bacterial profiles differed considerably among subjects, as observed here and in a previous study using a similar approach (7). This suggests that the etiology of periodontitis is heterogeneous. Because of this complexity, well-accepted statistical approaches relying on mean values do not completely represent the data, and the large changes observed in a small number of subjects may be diluted in the mean values shown in Fig. 3 and Table 1. Much larger sample sizes may be needed for a complete understanding of the bacterial etiology of periodontitis.
F. alocis accounted for 1.5% of all clones, and higher levels of F. alocis were seen in the group whose periodontal health worsened (Fig. 3, top). This gram-positive anaerobe is closely related to the peptostreptococci and has recently been associated both with periodontitis (3, 5-7) and with endodontic infections (19) by molecular detection methods.
A few other candidate pathogens were identified (Fig. 3), all less numerous than F. alocis. Due to the large number of species examined and the low clone numbers of numerically minor species, it is expected that some associations could be due to random chance, and these candidates require further investigation to establish their disease association. Three uncultivated phylotypes belonging to the genus Selenomonas, oral clones CS015, CS002, and FNA3, were all associated with periodontitis. These findings corroborate previous studies in which selenomonads have been shown to be associated with disease by both cultivation (21, 22) and molecular methods (7, 20). The genus Dialister accounted for 2.3% of all clones, and decreases in levels of oral strain GBA27 were associated with improving health. This is consistent with the previous association of other members of this genus with periodontitis (7, 20-22). Two members of the genus Treponema, Smibert-5 and VI:G:G47, were associated with disease. Treponemes, in particular Treponema denticola, have been associated with disease in many previous studies (7, 20-22), and the genus includes many uncultivated and uncharacterized species that are disease associated (15).
Peptostreptococci did not show a statistically significant association with disease in this study. This was surprising, because a previous study using a similar methodology (7) showed a strong relationship between levels of peptostreptococci and periodontitis. A close look at the data from the previous study reveals that high levels of peptostreptococci in 3 of the 15 subjects with chronic periodontitis largely accounted for the association. In the current study, 65% of the baseline flora in one of the seven subjects in the “worse” group was Peptostreptococcus sp. oral clone FG014. At the 2-year time point, this proportion dropped to 14%, while levels of Peptostreptococcus sp. oral clone AJ062 increased to 22% of total bacteria. The implications of the direction of change (increase or decrease) are not clear-cut, since the temporal relationship between disease activity and measurable clinical destruction is unclear. The least complicated expectation would be that pathogens would appear and increase as disease progresses. However, a complex chain of bacterial events may occur during shifts from health to disease (or the reverse). For this reason, disappearance of pathogens important in disease initiation could be observed in subjects with progressing disease. These findings lend support to a model of periodontitis as a heterogeneous disease, with peptostreptococci playing an important etiological role in some, but not all, individuals. These findings also indicate that large sample sizes may be needed in future studies to elucidate the complex, heterogeneous microbial etiology of periodontitis.
The species traditionally regarded as periodontal pathogens, such as Porphyromonas gingivalis and Tannerella forsythia, were detected but did not show a statistically significant association with disease. This is not surprising, because they constitute a small fraction of total bacteria, and this study was not powered to examine the strength of association of numerically minor species.
Candidate beneficial species.
Veillonella sp. oral clone X042 was the most common bacterium detected overall, accounting for 9.5% of all clones. Levels of this phylotype increased when periodontal health improved and decreased when the clinical status worsened. This phylotype was the most prevalent bacterium in a previous study using quantitative clonal analysis as well, and higher levels were significantly associated with health in that study (7). This strong association with health suggests that monitoring of the levels and changes in levels of this phylotype may prove clinically useful. Exploration of the role of this apparently beneficial phylotype in the plaque biofilm could also have important implications for microbial replacement therapy or probiotics. It has been observed for other infectious diseases that a shift from health to disease is associated not only with an increase in levels of pathogenic species but also with a decrease in levels of protective species. Studies on microbial stability in the gastrointestinal tract have shown that adequate colonization of the intestine with health-compatible bacteria has been shown to decrease susceptibility to infection with pathogenic organisms such as Salmonella (14). When large numbers of resident bacteria saturate an ecological niche, they create resistance to colonization by exogenous pathogens. This “colonization resistance” conferred by the presence of beneficial species is considered an important barrier function of gut commensals and may be important in the gingival crevice as well.
In summary, over a 2-year period, bacterial stability was greatest among clinically stable subjects and lowest in the group whose periodontal status worsened. Higher numbers of species lost or gained were also observed for subjects whose clinical status changed. This provides evidence that changes in periodontal status are accompanied by shifts within the bacterial community. Although the triggers for these bacterial and health shifts are not well understood, these data suggest that measures of microbial stability may be useful in clinical diagnosis and prognosis. Regarding individual species, increases in levels of the uncultivated phylotype Veillonella sp. oral clone X042, a gram-negative bacterium and the most common member of the subgingival bacterial community, were associated with periodontal health, suggesting that this is an important beneficial species. F. alocis, a gram-positive anaerobe, was found at high levels in subjects with disease. These findings suggest that the etiology of periodontitis is more complex than a previous model associating gram-positive bacteria with health and implicating gram-negative bacteria as the causative agents in disease.
Supplementary Material
Acknowledgments
This study was supported by Public Health Service grant DE10467 from the National Institute of Dental and Craniofacial Research.
Footnotes
Supplemental material for this article is available at http://jcm.asm.org/.
REFERENCES
- 1.Cebra, J. J. 1999. Influences of microbiota on intestinal immune system development. Am. J. Clin. Nutr. 69:1046S-1051S. [DOI] [PubMed] [Google Scholar]
- 2.Cullinan, M. P., S. M. Hamlet, B. Westerman, J. E. Palmer, M. J. Faddy, and G. J. Seymour. 2003. Acquisition and loss of Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans and Prevotella intermedia over a 5-year period: effect of a triclosan/copolymer dentifrice. J. Clin. Periodontol. 30:532-541. [DOI] [PubMed] [Google Scholar]
- 3.Dahlen, G., and A. Leonhardt. 2006. A new checkerboard panel for testing bacterial markers in periodontal disease. Oral Microbiol. Immunol. 21:6-11. [DOI] [PubMed] [Google Scholar]
- 4.de Lillo, A., V. Booth, L. Kyriacou, A. J. Weightman, and W. G. Wade. 2004. Culture-independent identification of periodontitis-associated Porphyromonas and Tannerella populations by targeted molecular analysis. J. Clin. Microbiol. 42:5523-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutter, G., U. Schlagenhauf, G. Valenza, M. Horn, S. Burgemeister, H. Claus, and U. Vogel. 2003. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 149:67-75. [DOI] [PubMed] [Google Scholar]
- 6.Kumar, P. S., A. L. Griffen, J. A. Barton, B. J. Paster, M. L. Moeschberger, and E. J. Leys. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338-344. [DOI] [PubMed] [Google Scholar]
- 7.Kumar, P. S., A. L. Griffen, M. L. Moeschberger, and E. J. Leys. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43:3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Listgarten, M. A. 1976. Structure of the microbial flora associated with periodontal health and disease in man. A light and electron microscopic study. J. Periodontol. 47:1-18. [DOI] [PubMed] [Google Scholar]
- 9.Listgarten, M. A., J. Slots, A. H. Nowotny, J. Oler, J. Rosenberg, B. Gregor, and P. Sullivan. 1991. Incidence of periodontitis recurrence in treated patients with and without cultivable Actinobacillus actinomycetemcomitans, Prevotella intermedia, and Porphyromonas gingivalis: a prospective study. J. Periodontol. 62:377-386. [DOI] [PubMed] [Google Scholar]
- 10.Loe, H., E. Theilade, and S. B. Jensen. 1965. Experimental gingivitis in man. J. Periodontol. 36:177-187. [DOI] [PubMed] [Google Scholar]
- 11.Loesche, W. J., and S. A. Syed. 1978. Bacteriology of human experimental gingivitis: effect of plaque and gingivitis score. Infect. Immun. 21:830-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacFarlane, T. W., W. M. Jenkins, W. H. Gilmour, J. McCourtie, and D. McKenzie. 1988. Longitudinal study of untreated periodontitis (II). Microbiological findings. J. Clin. Periodontol. 15:331-337. [DOI] [PubMed] [Google Scholar]
- 13.Machtei, E. E., R. Dunford, E. Hausmann, S. G. Grossi, J. Powell, D. Cummins, J. J. Zambon, and R. J. Genco. 1997. Longitudinal study of prognostic factors in established periodontitis patients. J. Clin. Periodontol. 24:102-109. [DOI] [PubMed] [Google Scholar]
- 14.Madden, J. A., S. F. Plummer, J. Tang, I. Garaiova, N. T. Plummer, M. Herbison, J. O. Hunter, T. Shimada, L. Cheng, and T. Shirakawa. 2005. Effect of probiotics on preventing disruption of the intestinal microflora following antibiotic therapy: a double-blind, placebo-controlled pilot study. Int. Immunopharmacol. 5:1091-1097. [DOI] [PubMed] [Google Scholar]
- 15.Moter, A., C. Hoenig, B. K. Choi, B. Riep, and U. B. Gobel. 1998. Molecular epidemiology of oral treponemes associated with periodontal disease. J. Clin. Microbiol. 36:1399-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishihara, T., and T. Koseki. 2004. Microbial etiology of periodontitis. Periodontol. 2000 36:14-26. [DOI] [PubMed] [Google Scholar]
- 17.Papapanou, P. N., V. Baelum, W. M. Luan, P. N. Madianos, X. Chen, O. Fejerskov, and G. Dahlen. 1997. Subgingival microbiota in adult Chinese: prevalence and relation to periodontal disease progression. J. Periodontol. 68:651-666. [DOI] [PubMed] [Google Scholar]
- 18.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siqueira, J. F., Jr., and I. N. Rocas. 2003. Detection of Filifactor alocis in endodontic infections associated with different forms of periradicular diseases. Oral Microbiol. Immunol. 18:263-265. [DOI] [PubMed] [Google Scholar]
- 20.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 21.Tanner, A., H. D. Bouldin, and M. F. Maiden. 1989. Newly delineated periodontal pathogens with special reference to Selenomonas species. Infection 17:182-187. [DOI] [PubMed] [Google Scholar]
- 22.Tanner, A., M. F. Maiden, P. J. Macuch, L. L. Murray, and R. L. Kent, Jr. 1998. Microbiota of health, gingivitis, and initial periodontitis. J. Clin. Periodontol. 25:85-98. [DOI] [PubMed] [Google Scholar]
- 23.van der Waaij, D. 1989. The ecology of the human intestine and its consequences for overgrowth by pathogens such as Clostridium difficile. Annu. Rev. Microbiol. 43:69-87. [DOI] [PubMed] [Google Scholar]
- 24.van Steenberghe, D., B. Rosling, P. O. Soder, R. G. Landry, U. van der Velden, M. F. Timmerman, E. F. McCarthy, G. Vandenhoven, C. Wouters, M. Wilson, J. Matthews, and H. N. Newman. 1999. A 15-month evaluation of the effects of repeated subgingival minocycline in chronic adult periodontitis. J. Periodontol. 70:657-667. [DOI] [PubMed] [Google Scholar]
- 25.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou, X., S. J. Bent, M. G. Schneider, C. C. Davis, M. R. Islam, and L. J. Forney. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150:2565-2573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.