Abstract
We report the first Shiga toxin 2-producing Acinetobacter haemolyticus strain that was isolated from the feces of a 3-month-old infant with bloody diarrhea. Usual enteropathogenic bacteria were not detected. This finding suggests that any Shiga toxin-producing microorganism capable of colonizing the human gut may have the potential to cause illness.
CASE REPORT
In November 2001, a 3-month-old male infant was admitted at Pereira Rossell Pediatric Hospital with bloody diarrhea of 12 h evolution without fever or other previous pathologies. The patient was treated empirically with intravenous ceftriaxone and hospitalized for 24 h.
Samples of feces were obtained before and after the patient was treated with antibiotics, and they were simultaneously studied at the Microbiology laboratory of the Pereira Rossell Pediatric Hospital and at the Reference laboratory for Shiga toxin-producing Escherichia coli. This last study, done in the context of an institutional program aimed at the regional surveillance of bloody diarrheas and hemolytic-uremic syndrome (HUS) etiology, is the focus of the present article.
The presence of Salmonella, Shigella, enteroinvasive E. coli, and enteropathogenic E. coli, Yersinia enterocolitica, and Campylobacter in fecal samples was studied using standard procedures, as previously described (18). The searching of Shiga toxin-producing E. coli (STEC) was done by selective enrichment protocol (9) on Trypticase soy broth (Bacto, Le Pont de Claix, France) with 2.5 mg/liter sodium tellurite and 0.05 mg/liter cefixime (CT-TSB; BioMérieux, Marcy L′Étoile, France) and further isolation on MacConkey sorbitol (SMAC) plates (Oxoid, Hampshire, England).
The microscopic analysis of the two fecal samples showed a low number of leukocytes (5 to 10 per microscopic field) and did not reveal spiral bacteria that would suggest the presence of Campylobacter spp.
The cultures from the first fecal sample developed well in all inoculated media.
From the second fecal sample, only 12 colonies (all sorbitol negative) were recovered on a directly inoculated SMAC plate after 48 h of culture. From the enrichment broth, we did not recover any microorganism on the SMAC plate.
The microbiological studies did not reveal the presence of usual enteropathogenic bacteria in any of the two samples.
Twenty sorbitol-positive colonies plus a lysate from the confluent zone in the SMAC plate of the first sample (nonfermenting colonies were not recovered) and the 12 sorbitol-negative colonies recovered from the second sample were analyzed by PCR, as previously described (13), to detect the presence of Stx1/Stx2-encoding organisms. This PCR was performed with primers VT1A-F (GAAGAGTCCGTGGGATTACG) and VT1B-R (AGCGATGCAGCTATTAATAA) for stx1 and with primers VT2A-F (TTAACCACACCCACGGCAGT) and VT2B-R (GCTCTGGATGCATCTCTGGT) for stx2.
E. coli K-12 C600 E. coli K-12 C600 (F− thi-1 thr-1 leuB6 lacY1 tonA21 supE44 Δλ Δstx1 Δstx2) and Stx2/Stx1-producing E. coli STEC O157:H7 EDL933 were used as negative and positive controls, respectively, in all PCR assays (10).
Amplified DNA fragments were analyzed through 2% (wt/vol) agarose gel electrophoresis by staining with 0.5 μg/ml ethidium bromide.
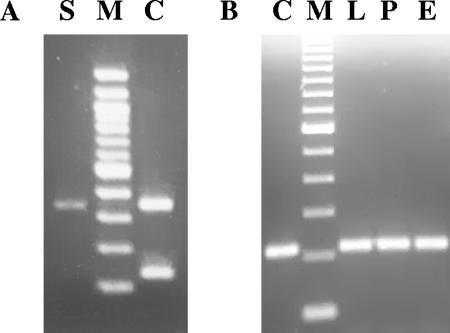
The bacterial lysates derived from the first sample were stx1/stx2 negative, but in the second sample, we detected two strains (DS9B1 and DS9B2) carrying stx2-related sequences (presence of a weak band of 346 bp) (Fig. 1).
FIG. 1.
Agarose gel electrophoresis stained with ethidium bromide of stx2 fragments obtained by PCR from the STAH strain. (A) stx2 PCR, 346 bp. Lanes: S, STAH lysate; M, 100-bp DNA molecular marker (Promega); C, E. coli O157:H7 EDL933 lysate (stx2/stx1 positive control). (B) stx2 PCR, 215 bp. Lanes: C, E. coli K-12 C600 933W lysate (stx2 positive control); M, 100-bp DNA molecular marker (Gibco); L, STAH lysate; P, phage genome DNA; E, DNA obtained by plasmidic extraction.
To obtain a best performance in the detection of the stx2-related sequences, an additional PCR was done using VT2A-F as the forward primer (13) and stxA2INT (TGCTGTCCGTTGTCATGGA), which was especially designed for this study, as a universal reverse primer, allowing the amplification of an approximately 215-bp fragment of the stxA2 gene. The stxA2INT primer was designed using the consensus sequence derived from a multiple alignment of 54 stx2 homologous sequences (from position 385 to 403 of the stxA2 open reading frame). The reaction showed an intense 215-bp PCR product and confirmed the presence of stx2-related sequences (Fig. 1).
This PCR product was sequenced in the CTAG center (Technical Center for Genetic Analysis, School of Sciences, Montevideo, Uruguay). The sequence was first identified through a search at the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov) and at local databases with the BLASTN search algorithm (1) and then analyzed using the European Molecular Biology Open Software Suite package (14) and CLUSTAL X multiple-sequence alignment software (17). It was submitted to GenBank (see “Nucleotide sequence accession number” below). This DNA sequence corresponded to a small conserved region of the stxA2 locus (E value = 4e−98) and showed 100% identity with the 933W phage stx2-corresponding region (4). Thus, this analysis confirmed the presence of stx2-related sequences in the isolated strains.
On the other hand, specific PCR assays (2, 16) for eae (intimin)- and e-hly (enterohemolysin)-related sequences were negative in both strains.
The microbiological studies of strains DS9B1 and DS9B2 showed that both of them were gram-negative diplococcoid cells that partially resisted destaining. They were cytochrome c oxidase negative, catalase positive, strictly aerobic, nutritionally nonfastidious, nonmotile, non-lactose- and -sorbitol-fermenting, and inactive on glucose, with good growth on MacConkey lactose agar. On the basis of these results, the two strains were recognized as belonging to the genus Acinetobacter. The colonies were convex on sheep blood agar, smooth, mucoid, 1 to 3 mm in diameter, and surrounded by a wide zone of beta-hemolysis. They grew well at 28°C and 37°C but not at 41°C and 44°C, they were able to use citrate as the sole carbon source but not malonate, and they carried the enzyme arginine dehydrolase. Following the scheme described by Bouvet and Grimont (3), these results allowed us to determine that they pertained to the species Acinetobacter haemolyticus.
Antibiotic susceptibilities determined by a standard Kirby-Bauer agar diffusion technique (as published by CLSI [formerly NCCLS]) showed that both strains were susceptible to piperacillin, ceftazidime, cefepime, imipenem, gentamicin, amikacin, ciprofloxacin, trimethoprim-sulfamethoxazole, and tetracycline. They showed “intermediate” results with chloramphenicol, cefotaxime, and ceftriaxone.
To obtain the plasmid profile of DS9B1 and DS9B2, one colony of each strain was cultivated at 37°C overnight with shaking (200 rpm) in 3 ml of Luria-Bertani broth, the plasmids were isolated from the bacterial pellet by lysis with alkali, as previously described (15), and further analyzed through 0.7% agarose gel electrophoresis by staining with ethidium bromide. The plasmid profiles were identical, showing a band of approximately 50 kb and a minor band of 5.5 kb. We observed that the 5.5-kb band was lost after some subcultures and that the 50-kb band was also lost in some clones.
The Shiga toxin production was determined by a cytotoxicity assay on Vero cells (ATTC CRL 1587) in 96-well-bottom microtiter trays inoculated with twofold serial dilutions of filter-sterilized culture supernatants or supernatants obtained by polymyxin extraction (8). Although the cell lysate supernatants were not cytotoxic, the culture supernatants of both strains had the same cytotoxic titer as the Shiga toxin 2-producing E. coli used as positive control (50% cytotoxic dose = 64). This would indicate that, once produced, the toxin was probably released to the medium. The production of Shiga toxin 2 and its activity were confirmed by neutralization assay on Vero cells with BC5BB12 anti-Stx2 monoclonal antibody provided by N. Strockbine and J. J. Wells (CDC). In both assays, E. coli K-12 C600 was used as negative control, and Stx1-producing E. coli STEC O26:H11 H19B and Stx2-producing E. coli K-12 C600 933W were used as positive controls (10, 11).
We observed that many stx2-positive colonies became negative for stx2 PCR after subculture (only 1 of every 15 tested colonies was positive). The in vitro instability of the stx genes together with the spontaneous lysis observed in the colonies of A. haemolyticus at temperatures between 30 to 37°C on solid medium cultures suggested that the stx2 genes were present in the context of an infectious bacteriophage genome.
The phage propagation was made from exponential cultures of stx2 PCR-positive colonies (optical density at 600 nm [OD600] = 0.5) in Luria-Bertani broth supplemented with 0.01 M MgSO4, 0.01 M CaCl2, and 0.2% maltose, induced with mitomycin C, and incubated at 37°C with shaking (200 rpm) long enough to show the lightening of the culture due to phage bacterial lysis. To precipitate the phage particles, the supernatants were cleaned by centrifugation, filtered (0.22-μm pore size), and supplemented with 2 M NaCl and then with 20% (wt/vol) polyethylene glycol 8000. The mixes were incubated for 1 h at 4°C. The particles were harvested by centrifugation (25,000 × g, 30 min, 4°C) and suspended in magnesium sulfate buffer. These crude stocks enriched with bacteriophages were then dropped onto double-layer plate agar cultures, as previously described (15), resulting in nontranslucent, irregular lysis zones with shiny edges after 18 h at 37°C. We used E. coli K-12 K600 as the recipient strain (10).
These phage stocks were stx2 PCR positive. Double-layer agar plating assays were made to isolate individual phage plaques (15) and then obtain high-titer stocks. Approximately 50% of the enriched plaques analyzed via stx2 PCR revealed the presence of the target genes.
After RNase and proteinase K treatment (Sigma-Aldrich, Germany), the DNA phage genome was purified from stx2 PCR-positive high-titer pure stocks with phenol-chloroform and isopropanol precipitation, as previously described (15). It showed a DNA band of approximately 50 kb in 0.7% agarose gel electrophoresis stained with ethidium bromide. Remarkably, this DNA molecule coincided with the lambdoid bacteriophages genome DNA size.
In view of the similar molecular size of the phage genomic DNA obtained from high-titer phage stocks and the DNA recovered by plasmid extraction from the A. haemolyticus strains, we suspected that both genetic materials could be the same. We studied by PCR the presence of the stx2-related sequences in these DNA samples, and we found that both types of sample were stx2 PCR positive (Fig. 1). We also observed that the colonies that lost the 50-kb DNA band after subculture became negative for stx2 PCR, for Vero cell cytotoxic assay, and for the presence of stx2-encoding bacteriophages.
A. haemolyticus has been described mainly as an environmental bacteria and an opportunistic, multiresistant intrahospital human pathogenic bacteria (7).
A few bacterial species and/or pathogenic clones can produce Shiga toxin (Stx) and have been associated with cases of bloody diarrhea, thrombocytopenic thrombotic purpura, and HUS in humans (12).
Stx belongs to a defined protein subfamily, the RNA N-glycosidases, that directly targets the cell ribosome machinery, inhibiting cell protein synthesis (5). These toxins can be classified into 2 antigenic groups, Stx-1 and Stx-2, that include (especially Stx-2) an important number of genotypic variants (12). In most cases, the Stx coding genes (stxAB) are included in the lysis cassette in the context of the lambdoid lysogenic bacteriophage genomic DNA (11).
This is the first report of a Shiga toxin-producing Acinetobacter haemolyticus (STAH) strain, and it is also the first report of its association with bloody diarrhea. As the isolated A. haemolyticus strain produces Shiga toxin, a relevant virulence factor, and as usual enteric bacterial pathogens associated with bloody diarrhea were not detected in this case, it is possible that this strain was the etiological agent. The production of Shiga toxin is directly implicated in the generation of bloody diarrheas, and the immunological condition of the patient (due to his age) could have made him especially susceptible to the infection. This finding suggests that any Shiga toxin-producing microorganism capable of colonizing the human gut may have the potential to cause human illness and life-threatening human diseases such as HUS. Although we do not detect the presence of intimin and enterohemolysin, we cannot discard the possibility that other virulence factors could be present in the STAH strain.
On the other hand, we think that there is a chance that another Shiga toxin-producing microorganism which we failed to isolate was implicated in the described case and it might be possible that Acinetobacter haemolyticus had acquired in vivo the Stx2-producing phenotype via horizontal transfer in the gut lumen of the patient. Extrachromosomal DNA analysis, isolation of stx2-encoding phage and purification of its genome, specially designed PCR tests, and the observed in vitro instability of the stx-related sequences suggested that the Shiga toxin genes of STAH are carried in the context of an infective bacteriophage. Nevertheless, to confirm the presence of a phage carrying the stx2 gene, we should make lisogenization assays with the purified bacteriophage to isolate an Stx2-producing recipient strain.
Lysogeny has been proven to occur in vivo (6), and it has important implications for the evolution of new pathogenic strains. Treatment with certain antibiotics can induce the lytic cycle of the lambdoid phage carrying stx genes and can subsequently cause the phage infection of formerly non-toxin-producing E. coli and other bacterial species. This may increase the production and release of Shiga toxin in the gut lumen, aggravating the disease. Therefore, we speculate that some conditions inherent to STAH strain bacterial fitness (such as tolerance to frequently used antibiotics, particularly ceftriaxone, and stx2-encoding phage infection permissiveness) may have promoted its survival and the subsequent acquisition in vivo of the Stx2-producing phenotype in the gut lumen of the treated patient.
This might be another argument against indiscriminate treatment of bloody diarrhea, especially in the cases that involve Shiga toxin-producing organisms.
Nucleotide sequence accession number.
The sequence determined in this study was submitted to the GenBank database under accession number DQ344636.
Acknowledgments
We thank Jorge Blanco et al. and Marta Rivas et al. for providing reference strains, Nancy Strockbine and J. J. Wells for sending monoclonal antibodies, and Monica Tous et al. for providing Vero cells.
This work was partially supported by the CDC through the Manuel Pérez Foundation (Medicine Faculty, Uruguay), by CEGETEC (Technological Management Center, Uruguay), and by CSIC (University Research Committee, Uruguay).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. L. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco, M., J. E. Blanco, A. Mora, J. Rey, J. M. Alonso, J. Hermoso, M. P. Alonso, G. Dahbi, E. A. Gonzalez, M. I. Bernardez, and J. Blanco. 2003. Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)-producing Escherichia coli isolates from healthy sheep in Spain. J. Clin. Microbiol. 41:1351-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvet, P. J., and P. A. D. Grimont. 1986. Taxonomy of the Acinetobacter genus with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov., and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bact. 36:228-240. [Google Scholar]
- 4.Datz, M., C. Janetzki-Mittmann, S. Franke, F. Gunzer, H. Schmidt, and H. Karch. 1996. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl. Environ. Microbiol. 62:791-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endo, Y., K. Tsurugi, T. Yutsudo, Y. Takeda, T. Ogasawara, and K. Igarashi. 1988. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes: RNA N-glycosidase activity of the toxins. Eur. J. Biochem. 171:45-50. [DOI] [PubMed] [Google Scholar]
- 6.Gamage, S., J. E. Strasser, C. L. Chalk, and A. A. Weiss. 2003. Non-pathogenic Escherichia coli O157:H7 can contribute to the production of Shiga toxin. Infect. Immun. 71:3107-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gospodarek, E., and I. Kania. 1992. Occurrence of species from Acinetobacter genus in clinical material and other sources. Med. Dosw. Mikrobiol. 44:41-48. [PubMed] [Google Scholar]
- 8.Karmali, M. A., M. Petric, C. Lim, R. Cheung, and G. S. Arbus. 1985. Sensitive method for detecting low numbers of verotoxin-producing Escherichia coli in mixed cultures by use of colony sweeps and polymyxin extraction of verotoxin. J. Clin. Microbiol. 22:614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudva, I. T., P. G. Hatfield, and C. J. Hodve. 1995. Effect of diet on the shedding of Escherichia coli O157:H7 in a sheep model. Appl. Environ. Microbiol. 61:1363-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Brien, A., and R. K. Holmes. 1987. Shiga and Shiga-like toxins. Microbiol. Rev. 51:206-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Brien, A. D., J. W. Newland, F. Miller, R. K. Holmes, W. Smith, and S. B. Formal. 1984. Shiga-like toxin converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 12.Paton, C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollard, D. R., W. M. Johnson, H. Lior, S. D. Tyler, and K. R. Rozee. 1990. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain reaction. J. Clin. Microbiol. 28:540-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS 2.9.0.0: The European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Schmidt, H., L. Beutin, and H. Karch. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres, M. E., M. C. Pírez, F. Schelotto, G. Varela, V. Parodi, F. Allende, E. Falconi, L. Dell′Acqua, P. Gaione, M. V. Méndez, A. M. Ferrari, A. Montano, E. Zaneta, A. M. Acuña, H. Chiparelli, and E. Ingold. 2001. Etiology of children's diarrhea in Montevideo, Uruguay: associated pathogens and unusual isolates. J. Clin. Microbiol. 39:2134-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]