Abstract
Infections caused by Naegleria fowleri, Acanthamoeba spp., and Balamuthia mandrillaris occur throughout the world and pose many diagnostic challenges. To date, at least 440 cases of severe central nervous system infections caused by these amebas have been documented worldwide. Rapid and specific identification of these free-living amebas in clinical samples is of crucial importance for efficient case management. We have developed a triplex real-time TaqMan PCR assay that can simultaneously identify Acanthamoeba spp., B. mandrillaris, and N. fowleri in the same PCR vessel. The assay was validated with 22 well-characterized amebic strains harvested from cultures and nine clinical specimens that were previously characterized by in vitro culture and/or immunofluorescence assay. The triplex assay demonstrated high specificity and a rapid test completion time of less than 5 h from the reception of the specimen in the laboratory. This assay was able to detect one single ameba per sample analyzed, as determined with cerebrospinal fluid spiked with diluted cultured amebas. This assay could become useful for fast laboratory diagnostic assessment of amebic infections (caused by free-living amebas) in laboratories with adequate infrastructure to perform real-time PCR testing.
Naegleria fowleri, Acanthamoeba spp., and Balamuthia mandrillaris are free-living ubiquitous amebas associated with severe disease in humans. N. fowleri is the most virulent of these three amebas and is associated with an acute infection called primary amebic meningoencephalitis (PAM) (21). More than 200 cases of PAM, predominantly in children and young adults, were reported worldwide as of July 2002 (39). PAM is a fulminant infection that typically leads to death within 1 to 2 weeks from the onset of symptoms. Early intervention with amphotericin B has resulted in recovery in a few cases (38), but laboratory diagnosis of this infection had to be performed rapidly to allow efficient treatment.
Acanthamoeba and Balamuthia are phylogenetically related and morphologically similar. Acanthamoeba species are commonly found in the environment (20), and B. mandrillaris has recently been isolated from soil (11, 33). They primarily infect individuals with a compromised immune system, most likely through inhalation or skin wounds. From localized infections at the entry point, these amebas can reach the central nervous system (CNS) by vascular dissemination and cause lesions in the brain. The resulting infection is called granulomatous amebic encephalitis (GAE), which, in contrast to PAM, is a subacute or chronic disease. Symptoms of GAE include confusion, dizziness, drowsiness, headache, seizures, and a decreased level of consciousness. More than 240 GAE infections were reported in the literature as of 2002, most of them with fatal outcomes (39). In addition to GAE, Acanthamoeba infection is associated with keratitis in otherwise healthy persons who wear contact lenses or have suffered trauma to the eye. The number of diagnosed Acanthamoeba keratitis cases has increased dramatically over the last 20 years, and it has been estimated that over 3,000 such cases have occurred in the United States alone (39).
The laboratory diagnosis of PAM currently relies on the demonstration of N. fowleri trophozoites in cerebrospinal fluid (CSF), microscopic or histochemical examination of biopsy or autopsy specimens, or isolation of the organisms in culture. However, since no distinct clinical features differentiate PAM from fulminant bacterial infections, it is often misdiagnosed as such and thus incorrectly managed (5, 12). Laboratory diagnoses of Acanthamoeba infection and B. mandrillaris GAE follow the same rationale (39), but these infections are also frequently misdiagnosed and confused with brain tumors, abscesses, toxoplasmosis, or cysticercosis (20). Previous studies have reported a lack of expertise among pathologists and laboratorians in recognizing morphologically these rare pathogens in clinical material (23, 25). Molecular methods such as PCR offer an attractive alternative to microscopy and culture, since they can be performed by personnel without a high level of expertise in recognizing diagnostic morphological features of amebas. Furthermore, molecular methods are very sensitive and may allow the detection of fewer microorganisms per volume of sample analyzed than morphological methods would. This increases the likelihood of identifying the infective agent in samples in which very small numbers of amebas can be expected, for example, CSF from GAE patients.
Various conventional PCR (which uses gel electrophoresis to determine test results) protocols have been developed for the specific detection of Acanthamoeba (16, 17, 31, 41), N. fowleri (14, 24, 28, 29, 35), and B. mandrillaris (3). These protocols have been applied to the detection of Acanthamoeba in corneal scrapings from keratitis cases (18, 22, 27, 34), for confirmatory laboratory diagnosis of N. fowleri PAM (7, 13), and to detect B. mandrillaris in clinical specimens (42). However, none of the published protocols allows for the simultaneous detection of these parasites in CSF and other clinical specimens. Multiplex detection would be an invaluable laboratory approach for a reliable and efficient method for differential diagnoses of PAM and GAE infections.
This work describes a multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., B. mandrillaris, and N. fowleri in CSF specimens. Real-time PCR is preferred to conventional PCR in clinical laboratories because there is no need for postamplification handling, leading to faster analysis and reduced risk of amplicon contamination (8, 19). Real-time PCR can also provide an estimate of pathogen load (1). The new multiplex real-time PCR assay was evaluated with amebic strains obtained from in vitro cultures and a set of clinical specimens. In addition, the assay was compared to a published TaqMan assay for Acanthamoeba detection (30).
MATERIALS AND METHODS
Microbial strains and cultures.
The amebic strains obtained by in vitro culture are listed in Table 1. All Naegleria spp. (N. australiensis, N. dunnebackei, N. fowleri, N. gruberi, N. jadini, N. italica, and N. lovaniensis) were grown in a modified Nelson's medium; all Acanthamoeba spp. (A. castellanii, A. culbertsoni, A. healyi, A. polyphaga, and A. rhysodes) were grown in a proteose peptone, yeast extract, glucose, and serum medium. Balamuthia mandrillaris strains were grown either in monkey kidney cells (E6) or in BM3 axenic medium by using conditions described previously (32).
TABLE 1.
Cultured amebas included in the evaluation
| Sample | Genus and/or species | Source(s) (source/gender/state/country)a | Genotype | Result of real-time PCR
|
|
|---|---|---|---|---|---|
| Triplex assay from this study | Assay by Riviere et al. (30) | ||||
| V006 | Acanthamoeba | Brain tissue/F/GA/USA | T1 | Acanthamoeba | Acanthamoeba |
| V062 | Acanthamoeba | Corneal scraping/M/MA/USA | T4 | Acanthamoeba | Acanthamoeba |
| V369 | Acanthamoeba | Brain tissue/M/NE/USA | T10 | Acanthamoeba | Negative |
| V409 | Acanthamoeba | Horse brain/CA/USA | T10 | Acanthamoeba | Negative |
| V499 | Acanthamoeba | Water/NY/USA | T7 | Acanthamoeba | Negative |
| V550 | Acanthamoeba | Skin/CA/USA | T4 | Acanthamoeba | Acanthamoeba |
| V532 | Acanthamoeba | Water/NM/USA | T4 | Acanthamoeba | Negative |
| V039 | B. mandrillaris | Mandrill brain/CA/USA | B. mandrillaris | Negative | |
| V451 | B. mandrillaris | Brain tissue/F/NY/USA | B. mandrillaris | Negative | |
| V416 | B. mandrillaris | Brain tissue/F/Australia | B. mandrillaris | Negative | |
| V020 | N. fowleri | CSF/M/TX/USA | I | N. fowleri | Negative |
| V212 | N. fowleri | CSF/M/Mexico | I | N. fowleri | Negative |
| V511 | N. fowleri | CSF/M/GA/USA | I | N. fowleri | Negative |
| V515 | N. fowleri | CSF/M/AZ/USA | III | N. fowleri | Negative |
| V551 | N. fowleri | CSF/M/Colombia | N. fowleri | Negative | |
| CAMP | N. fowleri | CSF/F/CA/USA | II | N. fowleri | Negative |
| 7615250 | N. lovaniensis | Thermal water/Belgium | Negative | Negative | |
| V419 | N. dunnebackei | Water/CA/USA | Negative | Negative | |
| EG | N. gruberi | Soil/CA/USA | Negative | Negative | |
| 400 | N. jadini | Swimming pool/Belgium | Negative | Negative | |
| PP 397 | N. italica | Water, mud/Italy | Negative | Negative | |
| AB-T-F3 | N. australiensis | Drainage water/Australia | Negative | Negative | |
Some source parameters are omitted as necessary. F, female; M, male; USA, United States of America.
Amebas growing in the log phase of culture were dislodged from the culture tubes by the immersion of the tubes in ice-cold water for 5 min and centrifugation at 500 × g to harvest the cells. The cell pellets were then either directly subjected to DNA extraction or diluted to obtain dilution series. In the latter case, the sediment was dispersed in about 1 ml of ameba saline and counted in a hemacytometer. The suspension with amebas was adjusted to yield 104 cells/ml and was then twofold serially diluted in CSF until the most diluted sample contained less than one ameba per 50 μl of sample. The CSF used in this work represented leftovers from samples submitted to the CDC for diagnosis of neurocysticercosis: CSF samples from individuals that tested negative in the cysticercosis antibody test (37) were pooled together and examined microscopically for the presence of microorganisms before utilization in dilutions and spiking experiments.
Haemophilus influenzae (one strain each of serotypes A through F), Neisseria meningitidis (one strain each of serogroups A, B, C, X, Y, Z, W135, and 29E), Streptococcus pneumoniae M4421, and Streptococcus agalactiae M8224 were used as negative controls. Ten microliters of heat-killed cell suspensions was mixed with 40 μl parasite-free CSF and then subjected to DNA extraction. Other negative controls included CSF samples from four patients with cysticercosis and CSF spiked with DNA extracted from Toxoplasma gondii (environmental isolate).
Clinical specimens.
During the summer and fall of 2005, clinical specimens from nine patients were sent to the CDC for confirmation of the presence of free-living amebas as the causative agent of infections associated with neurological symptoms or for this to be ruled out. Seven of these were CSF samples; these were centrifuged at 1,000 × g for 5 min, all except 50 μl of the supernatant was removed, and the sediment was resuspended in the remaining volume. The other two clinical samples were brain tissue, of which approximately 50 mg per sample was processed with the DNA extraction methods described below.
DNA extraction.
Cultured amebas and spiked and clinical CSF samples were digested with proteinase K in a buffer consisting of 50 mM Tris-HCl (pH 8.5), 1% laureth-12, and 1 mM EDTA as described previously for DNA isolation of protozoa (9). Cell pellets with cultured amebas were suspended in 100 to 400 μl (depending on the pellet size) digestion buffer supplemented with 0.1 μg proteinase K per μl. For clinical CSF samples and serial dilutions of cultured amebas, 50 μl of each sample was mixed with 50 μl of digestion buffer with 0.2 μg proteinase K per μl. The samples were digested for 2 to 16 h at 56°C, followed by 95°C incubation for 10 min to inactivate the proteinase.
The brain tissue specimens were processed in three different ways: (i) digestion with proteinase K for 2 h in the buffer described above, (ii) DNA extraction using QIAamp micro DNA extraction kit (QIAGEN, Valencia, Calif.), and (iii) heating at 95°C for 10 min with no further treatment.
Real-time PCR assay design.
Partial or full-length 18S rRNA gene sequences from 40 Acanthamoeba spp., 5 B. mandrillaris amebas, 5 N. fowleri amebas, and 1 ameba each of N. australiensis, N. italica, N. gruberi, N. jadini, N. clarki, and N. lovaniensis, and the human 18S rRNA gene sequence were retrieved from GenBank and aligned with the GeneStudio alignment suite (GeneStudio Inc., Suwanee, Ga.). The alignment was used to identify regions where all sequences from each targeted species were identical but at the same time differed sufficiently from the other species' sequences to facilitate specific detection. Selected regions were then exported to Primer Express software (Applied Biosystems, Foster City, Calif.) for the design of PCR primers and TaqMan probes, with an emphasis on oligonucleotides that could be combined in a triplex format. The Acanthamoeba primers and probe were designed to detect all Acanthamoeba variants because it is not yet clear which genotypes are pathogenic. The primer and probe sets for B. mandrillaris and N. fowleri were designed specifically to detect these amebas at the species level.
Triplex real-time PCR.
For the detection of Acanthamoeba spp., primers AcantF900 (5′-CCC AGA TCG TTT ACC GTG AA-3′) and AcantR1100 (5′-TAA ATA TTA ATG CCC CCA ACT ATC C-3′) were used to amplify fragments of approximately 180 bp (differing by a few bases depending on the species). The TaqMan probe used was the Cy5-labeled AcantP1000 (5′-Cy5-CT GCC ACC GAA TAC ATT AGC ATG G-BHQ3-3′). For the detection of Balamuthia mandrillaris, primers BalaF1451 (5′-TAA CCT GCT AAA TAG TCA TGC CAA T-3′) and BalaR1621 (5′-CAA ACT TCC CTC GGC TAA TCA-3′) were used to amplify a fragment of 171 bp, which was detected by the 6-carboxyfluorescein-labeled probe BalaP1582 (5′-FAM-AG TAC TTC TAC CAA TCC AAC CGC CA-BHQ1-3′ [where FAM is 6-carboxyfluorescein]). For the detection of Naegleria fowleri, primers NaeglF192 (3′-GTG CTG AAA CCT AGC TAT TGT AAC TCA GT-5′) and NaeglR344 (5′-CAC TAG AAA AAG CAA ACC TGA AAG G-3′) were used to amplify a 153-bp-long fragment, detected by the hexachlorofluorescein (HEX)-labeled probe NfowlP (5′-HEX-AT AGC AAT ATA TTC AGG GGA GCT GGG C-BHQ1-3′). The triplex reaction mix contained 1× Platinum Quantitative PCR Supermix-UDG with ROX (Invitrogen, Carlsbad, Calif.), 0.2 μM of each primer, 0.1 μM of each probe, and 1 μl of DNA in 20-μl total reaction volume. PCRs were performed in a Mx3000P real-time thermocycler (Stratagene, La Jolla, Calif.), with two initial holds at 50°C for 2 min (incubation for uracil-DNA-glycosylase activity) and 95°C for 2 min (activation of Platinum Taq DNA polymerase), respectively, followed by 40 cycles of 95°C for 15 seconds and 63°C for 60 seconds. Fluorescence was measured at the end of each 63°C incubation. The results were analyzed using the Mx3000P version 2.0 software.
Comparison with another Acanthamoeba real-time PCR assay.
The triplex assay was compared to a published assay for Acanthamoeba detection performed with primer set TaqAcF1/TaqAcR1, the probe TaqAcP1 labeled with Cy5, and cycling conditions as described in the original publication with this assay (30). We used the same reagents as in the triplex assay, since this publication does not reveal the PCR reagent compositions.
RESULTS
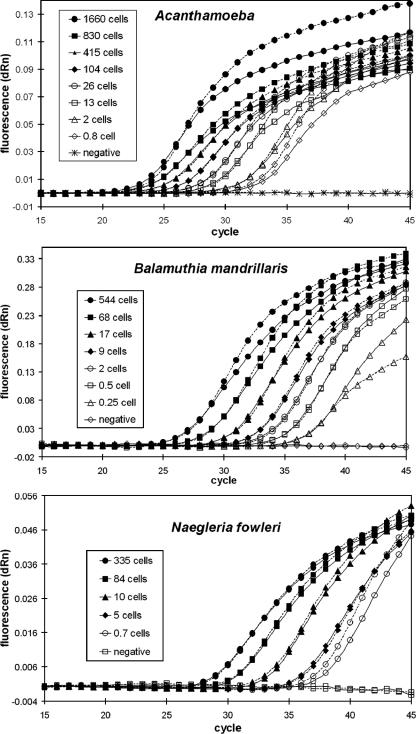
The real-time PCR assay for detection of free-living amebas presented in this study was designed as a multiplex assay for simultaneous detection of Naegleria fowleri, Acanthamoeba spp., and Balamuthia mandrillaris. Since the presence of other oligonucleotides and fluorescent dyes could alter the efficiency of PCR amplification, each set of primers and probe was tested in parallel in an individual format, as well as in a multiplex format, using different concentrations of the three amebas. Figure 1 illustrates the results of these experiments and shows that there was no systematic deviation in the amplification curves when comparing the triplex assay with the single-target assays. No significant difference in amplification efficiency was verified between singleplex and multiplex formats, as measured by the slopes of amplification curves during the exponential phase and the cycle threshold (CT) values obtained with individual samples. In addition, the respective detection limits for the triplex and individual assay formats were identical, since even the most diluted samples were detected in both types of assays.
FIG. 1.
Amplification curves from cultured trophozoites, serially diluted in parasite-free CSF. Negative samples were unspiked CSF. Each sample was amplified in parallel in the single-target assay with just one set of primers and probe (dashed lines) and in the triplex assay containing oligonucleotides for detection of all three variants of amebas (solid lines). Only the fluorescence signal from the relevant probe in the triplex reactions is displayed in each figure (the signals from the other two probes remained below threshold for all samples throughout all runs). All experiments were repeated three times; the average results are displayed. dRn, baseline subtracted fluorescence reading normalized to a reference dye.
Detection limit.
The limit of detection for the triplex assay was determined with cultured trophozoites serially diluted in parasite-free CSF (Fig. 1). The samples with lowest detectable concentrations contained less than 1 ameba per total amount of sample processed. The reason why these samples could be detected is probably associated with the presence of multiple copies of the PCR target gene. Thus, the detection limit for the assay was considered one ameba per sample processed.
Specificity.
The ability of the triplex assay to distinguish between the different amebas was evaluated with cultured trophozoites. Seven strains of Acanthamoeba, including representatives of the clinically relevant genotypes T1, T4, T7, and T10; three strains of B. mandrillaris; and six strains of N. fowleri were used. No cross-reactivity was observed, indicating that the assay accurately identified the three types of pathogenic free-living amebas. The assay did not detect the other Naegleria species, N. gruberi, N. jadini, N. australiensis, N. italica, N. dunnebackei, and N. lovaniensis. The specificity of the assay was further evaluated using CSF spiked with a limited set of other pathogens, as follows, that cause CNS diseases and symptoms similar to what are observed in infections caused by free-living amebas: the bacteria Neisseria meningitidis, Haemophilus influenza, Streptococcus pneumoniae, and Streptococcus agalactiae, and the parasite Toxoplasma gondii. In addition, CSF samples from four patients diagnosed with cysticercosis were also included in the analysis. No fluorescence signal was detected with these negative controls in the triplex PCRs performed.
Comparison with published assay.
The ameba samples used for evaluation of the triplex assay were also tested in the previously published real-time PCR assay for Acanthamoeba (Table 1) (30). This assay failed to detect four of the Acanthamoeba strains, including two clinical isolates, which limits its application as a clinical laboratory diagnosis tool. This result was supported by a comparison of the sequences of the primers and probe with an alignment of 40 Acanthamoeba 18S rRNA gene sequences; the oligonucleotides used in this assay align with a region that is not conserved among Acanthamoeba. However, the specificity of the assay was good, as no reactions with the non-Acanthamoeba samples were detected.
Clinical samples.
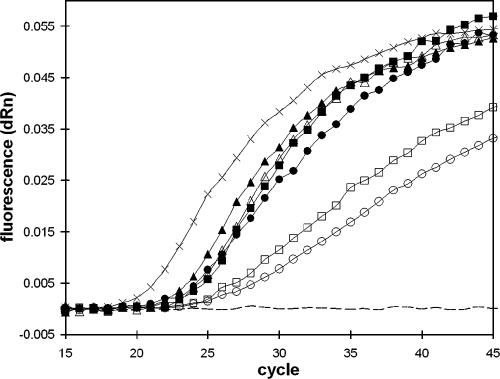
DNA from nine clinical samples (Table 2) was available for analysis; of these nine samples, six were found to be negative and three were positive for N. fowleri. None was positive for Acanthamoeba spp. or B. mandrillaris. For all nine samples, the PCR results were confirmed by immunofluorescence assay (IFA) and in vitro culturing performed on the same specimens. The three N. fowleri-positive samples included one CSF sample and two postmortem brain tissue samples. Figure 2 illustrates the real-time PCR amplification plots from the brain tissue samples from patient 4 and 5. Three different sample-processing protocols were evaluated using these tissue samples: no treatment (except 10-min incubation at 95°C), digestion with proteinase K, and DNA purification using a commercial kit. Remarkably, DNA extraction was not required to get a positive signal in the real-time PCR. However, real-time PCR performed directly on unpurified material from sample 5 led to poor amplification efficiency, as was evident by reduced amplification slopes for the unprocessed and proteinase K-treated sample (Fig. 2). This was probably caused by the presence of blood residues (heme products) that inhibited the amplification process (26). Sample 4 contained less blood than sample 5 did, so sample 4 could be efficiently PCR amplified even without DNA purification.
TABLE 2.
Results from clinical samples
| Sample no. | Source (gender/animal, state/country)a | Resultb
|
||
|---|---|---|---|---|
| Triplex real-time PCR | IFA | Culture | ||
| 1 | CSF (M, AZ) | Negative | Negative | Negative |
| 2 | CSF (horse, CA) | Negative | Negative | Negative |
| 3 | CSF (F, CO) | Negative | Negative | Negative |
| 4 | Brain tissue (M, OK) | N. fowleri | Positive | N. fowleri |
| 5 | Brain tissue (M, OK) | N. fowleri | Positive | N. fowleri |
| 6 | CSF (M, Japan) | Negative | Negative | Negative |
| 7 | CSF (M, ND) | Negative | Negative | NA |
| 8 | CSF (M, CO) | Negative | Negative | Negative |
| 9 | CSF (M, TX) | N. fowleri | Positive | N. fowleri |
F, female; M, male.
NA, not available.
FIG. 2.
Amplification curves from triplex real-time PCR on brain tissue samples from patient 4 (closed symbols) and 5 (open symbols). Amplification signals from unprocessed tissue are displayed as boxes, those from proteinase-treated tissue as circles, those from DNA extracted from the tissue samples as triangles, those from DNA extracted from cultured N. fowleri (positive control) as multiplication symbols, and those from water (negative control) as a dashed line. Only the fluorescence signal from the N. fowleri-specific probe is displayed (the signals from the other two probes remained below threshold for all samples throughout the run). dRn, baseline subtracted fluorescence reading normalized to a reference dye.
The CT values obtained in the real-time PCR were around 22 for the brain tissue samples (Fig. 2) and 23 for the N. fowleri-positive CSF sample. In contrast, the samples of cultured N. fowleri cells displayed in Fig. 1 had CT values ranging from 28 to 37. Thus, the CT values for the clinical samples were far outside the range of CT values obtained from the samples with known concentrations. Quantitative estimates of the parasitic load therefore could not be performed for any of the three clinical samples.
DISCUSSION
The triplex real-time PCR assay described here used PCR primers and TaqMan probes targeting three regions of the nuclear small subunit ribosomal (18S rRNA) gene. The 18S rRNA gene is a robust diagnostic target for PCR because it is part of a ribosomal repeat unit that is present in multiple copies in each cell. It has been estimated that Acanthamoeba has approximately 600 copies of the ribosomal repeat unit (4), and Naegleria species seem to have several thousand copies (6). Upon disruption, each ameba cell will thus release hundreds or thousands of target molecules, resulting in an assay being deemed to have high sensitivity. The 18S rRNA gene is also considered evolutionarily stable with limited intraspecies sequence variation, which is important for diagnostic identification.
Another reason for targeting the 18S rRNA gene is that it has been extensively studied in free-living amebas for species classification and genotyping. The classification of Acanthamoeba is not simple, since definitions of species by morphological features may not correlate with those based on data from molecular analyses. A substantial sequence variation between different species is observed; based on sequences from the 18S rRNA gene, Acanthamoeba can be divided into 15 sequence types (T1 through T15) (15, 36). There is no consensus as to which species or sequence types are pathogenic to humans, although T4 is most common among keratitis isolates and T1, T7, and T10 among GAE isolates (20). B. mandrillaris is so far the only species of the genus Balamuthia, and all the isolates examined displayed identical nuclear ribosomal sequences (2). The 18S rRNA gene has also been used for classification of species within the genus Naegleria (10). N. fowleri is believed to be the only Naegleria species that is pathogenic for humans, although N. australiensis and N. italica display pathogenicity in mouse models. Different isolates within the species N. fowleri exhibit very similar nuclear 18S rRNA sequences, but genotypic variations at the internal transcribed spacer sequences of isolates have been observed (10, 43). Although isolates can be divided into six genotypes (I, II, III, IV, V, and VI) on the basis of these sequences, no relevant association of these variations with specific biological factors impacting clinical diagnostics has been ascertained. Based on the collective data described above, it is safe to state that the triplex real-time PCR assay designed in this study is species specific for N. fowleri and B. mandrillaris and genus specific for Acanthamoeba.
One main advantage of this triplex assay compared to other available tests is the fact that it is multiplex. By using this approach, it was possible to identify all three pathogenic free-living amebas in the same reaction vessel. The simultaneous detection of Acanthamoeba spp. and B. mandrillaris is especially useful because these amebas are morphologically similar and cause the same types of infections in the CNS and the skin (21). In fact, before B. mandrillaris was recognized as a human pathogen, infections caused by this ameba were sometimes erroneously attributed to Acanthamoeba (40). In addition to use in clinical diagnostics, the multiplex assay may be of value for simultaneous detection of pathogenic free-living amebas in environmental samples. However, the multiplex feature of this assay is optional; if so preferred, the three components can be utilized as single-targeting assays or combined into duplex assays without impacting the quality of the results. This makes this assay adaptable to circumstances that may not require the simultaneous detection of all three free-living amebas for a diagnostic decision.
Another important aspect of this real-time PCR approach is the short time needed for turnaround of results. In the case of the two brain tissue samples, the confirmatory result of a suspected N. fowleri infection was obtained within 5 h of receiving the sample in the laboratory. This included time for DNA extraction to remove potential PCR inhibitors likely to be found in blood-containing samples like brain tissue. For samples which do not need elaborate DNA extraction procedures, such as CSF, this time frame can be further shortened to 2 to 3 h. In situations in which CSF is collected from patients still alive and under evaluation, fast turnaround times in the laboratory can make all the difference. The high number of fatal PAM cases is partly due to the high frequency of misdiagnosis of the infection as bacterial meningitis and subsequent treatment with antibacterial drugs that do not affect the amebas (12). The current algorithm for laboratory diagnosis of PAM includes approaches that are labor-intensive and may lack the sensitivity and speed required to reveal N. fowleri as the cause of infection before it is too late to institute appropriate therapy. The real-time PCR assay presented here can therefore be extremely useful as a fast and sensitive complement to existing diagnostic methods.
The triplex real-time PCR assay did not detect any cases of GAE among the nine clinical samples included in this study. These results matched the outcomes of IFA and cultures performed on the same samples. It remains to be determined to what extent Acanthamoeba and Balamuthia cells are present in the CSF of GAE patients. However, with the detection limit of the real-time PCR assay described here being one ameba per sample processed, it is possible that this tool will be useful in clarifying this point. To maximize the sensitivity of the PCR, we recommend that a clinical CSF sample should first be centrifuged and the cell-containing pellet used for PCR analysis as well as other tests. This means that as long as a CSF sample (any volume) collected from a patient contains at least one free-living ameba (B. mandrillaris, Acanthamoeba sp., or N. fowleri), this triplex assay will be able to correctly identify it. In addition, this real-time PCR does not require the unique expertise involved in morphology-based tests but can be performed in any laboratory with adequate infrastructure for real-time PCR testing. With this in mind, the real-time PCR assay described here should be a valuable addition to existing laboratory methods for diagnosing GAE infections.
The CT values obtained from the three clinical samples positive for N. fowleri did not allow quantitative estimates of the number of parasites present in the samples. Further studies are ongoing in our laboratory in order to clarify the circumstances in which this technique can be used to determine the load of parasites in cases of infections.
To our knowledge, the only previously published real-time PCR assay for detection of free-living amebas is a TaqMan assay specific for Acanthamoeba species causing keratitis (30). A comparison between this assay and the multiplex assay revealed that the former seems to lack the ability to identify genotypes T10 and T7. Likewise, it did not detect an environmental T4 isolate. The low number of isolates tested in this work makes it difficult to draw conclusions about the specificity of this assay, but it is clearly not as broadly reactive as the multiplex assay. The reason for this is that the Acanthamoeba assay targets a variable region of the 18S rRNA gene. The original publication describing this assay (30) states that the assay design was based on an alignment of sequences from six Acanthamoeba strains; this was obviously not enough to cover all clinically relevant strains. However, the assay may still be useful for detecting keratitis isolates.
In conclusion, we have described a fast and sensitive real-time PCR assay for the simultaneous detection of the free-living amebas Acanthamoeba spp., B. mandrillaris, and N. fowleri. We anticipate that this assay, due to its speed and sensitivity, could be useful for confirmatory diagnosis of CNS infections caused by free-living amebas in a timely manner and perhaps increase the likelihood of patient survival, especially in cases of PAM infections.
Acknowledgments
We are indebted to Marianna Wilson (Division of Parasitic Diseases, NCID, CDC) for providing us with parasite-free CSF samples and CSF from cysticercosis patients and to Patricia Wilkins and Susanna Schmink (Meningitis and Special Pathogens Branch, Bacterial and Mycotic Diseases, NCID, CDC) for the bacterial strains used as negative controls. We also thank Brent Barnes (a Ferguson EID fellow) for laboratory assistance and Dennis Juranek for his critical review of the manuscript.
The use of trade names is for identification only and does not imply endorsement by the Public Health Service or by the U.S. Department of Health and Human Services.
REFERENCES
- 1.Bell, A. S., and L. C. Ranford-Cartwright. 2002. Real-time quantitative PCR in parasitology. Trends Parasitol. 18:337-342. [PubMed] [Google Scholar]
- 2.Booton, G. C., J. R. Carmichael, G. S. Visvesvara, T. J. Byers, and P. A. Fuerst. 2003. Genotyping of Balamuthia mandrillaris based on nuclear 18S and mitochondrial 16S rRNA genes. Am. J. Trop. Med. Hyg. 68:65-69. [PubMed] [Google Scholar]
- 3.Booton, G. C., J. R. Carmichael, G. S. Visvesvara, T. J. Byers, and P. A. Fuerst. 2003. Identification of Balamuthia mandrillaris by PCR assay using the mitochondrial 16S rRNA gene as a target. J. Clin. Microbiol. 41:453-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byers, T. J., E. R. Hugo, and V. J. Stewart. 1990. Genes of Acanthamoeba: DNA, RNA and protein sequences. J. Protozool. 37:17S-25S. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2003. Primary amebic meningoencephalitis—Georgia, 2002. Morb. Mortal. Wkly. Rep. 52:962-964. [PubMed] [Google Scholar]
- 6.Clark, C. G., and G. A. Cross. 1987. rRNA genes of Naegleria gruberi are carried exclusively on a 14-kilobase-pair plasmid. Mol. Cell. Biol. 7:3027-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cursons, R., J. Sleigh, D. Hood, and D. Pullon. 2003. A case of primary amoebic meningoencephalitis: North Island, New Zealand. N. Z. Med. J. 116:U712. [PubMed] [Google Scholar]
- 8.da Silva, A. J., and N. J. Pieniazek. 2003. Latest advances and trends in PCR-based diagnostic methods, p. 397-412. In D. Dionisio (ed.), Textbook-atlas of intestinal infections in AIDS. Springer-Verlag Italia, Milan.
- 9.da Silva, A. J., D. A. Schwartz, G. S. Visvesvara, H. de Moura, S. B. Slemenda, and N. J. Pieniazek. 1996. Sensitive PCR diagnosis of infections by Enterocytozoon bieneusi (microsporidia) using primers based on the region coding for small-subunit rRNA. J. Clin. Microbiol. 34:986-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Jonckheere, J. F. 2002. A century of research on the amoeboflagellate genus Naegleria. Acta Protozool. 41:309-342. [Google Scholar]
- 11.Dunnebacke, T. H., F. L. Schuster, S. Yagi, and G. C. Booton. 2004. Balamuthia mandrillaris from soil samples. Microbiology 150:2837-2842. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, L. 2001. Protozoa from other body sites, p. 106-131. In L. Garcia (ed.), Diagnostic medical parasitology, 4th ed. ASM Press, Washington, D.C.
- 13.Hara, T., and T. Fukuma. 2005. Diagnosis of the primary amoebic meningoencephalitis due to Naegleria fowleri. Parasitol. Int. 54:219-221. [DOI] [PubMed] [Google Scholar]
- 14.Kilvington, S., and J. Beeching. 1995. Development of a PCR for identification of Naegleria fowleri from the environment. Appl. Environ. Microbiol. 61:3764-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohsler, M., B. Leitner, M. Blaschitz, R. Michel, H. Aspock, and J. Walochnik. 2006. ITS1 sequence variabilities correlate with 18S rDNA sequence types in the genus Acanthamoeba (Protozoa: Amoebozoa). Parasitol. Res. 98:86-93. [DOI] [PubMed] [Google Scholar]
- 16.Kong, H. H., and D. I. Chung. 1996. PCR and RFLP variation of conserved region of small subunit ribosomal DNA among Acanthamoeba isolates assigned to either A. castellanii or A. polyphaga. Korean J. Parasitol. 34:127-134. [DOI] [PubMed] [Google Scholar]
- 17.Lai, S., M. Asgari, and H. R. Henney, Jr. 1994. Non-radioactive DNA probe and polymerase chain reaction procedures for the specific detection of Acanthamoeba. Mol. Cell Probes 8:81-89. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann, O. J., S. M. Green, N. Morlet, S. Kilvington, M. F. Keys, M. M. Matheson, J. K. Dart, J. I. McGill, and P. J. Watt. 1998. Polymerase chain reaction analysis of corneal epithelial and tear samples in the diagnosis of Acanthamoeba keratitis. Investig. Ophthalmol. Vis. Sci. 39:1261-1265. [PubMed] [Google Scholar]
- 19.Mackay, I. M. 2004. Real-time PCR in the microbiology laboratory. Clin. Microbiol. Infect. 10:190-212. [DOI] [PubMed] [Google Scholar]
- 20.Marciano-Cabral, F., and G. Cabral. 2003. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 16:273-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez, A. J., and G. S. Visvesvara. 1997. Free-living, amphizoic and opportunistic amebas. Brain Pathol. 7:583-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathers, W. D., S. E. Nelson, J. L. Lane, M. E. Wilson, R. C. Allen, and R. Folberg. 2000. Confirmation of confocal microscopy diagnosis of Acanthamoeba keratitis using polymerase chain reaction analysis. Arch. Ophthalmol. 118:178-183. [DOI] [PubMed] [Google Scholar]
- 23.McCool, J. A., E. V. Spudis, W. McLean, J. White, and G. S. Visvesvara. 1983. Primary amebic meningoencephalitis diagnosed in the emergency department. Ann. Emerg. Med. 12:35-37. [DOI] [PubMed] [Google Scholar]
- 24.McLaughlin, G. L., M. H. Vodkin, and H. W. Huizinga. 1991. Amplification of repetitive DNA for the specific detection of Naegleria fowleri. J. Clin. Microbiol. 29:227-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakawa, G. J., T. McCalmont, J. Altman, G. H. Telang, M. D. Hoffman, G. R. Kantor, and T. G. Berger. 1995. Disseminated acanthamebiasis in patients with AIDS. A report of five cases and a review of the literature. Arch. Dermatol. 131:1291-1296. [PubMed] [Google Scholar]
- 26.Panaccio, M., and A. Lew. 1991. PCR based diagnosis in the presence of 8% (v/v) blood. Nucleic Acids Res. 19:1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasricha, G., S. Sharma, P. Garg, and R. K. Aggarwal. 2003. Use of 18S rRNA gene-based PCR assay for diagnosis of Acanthamoeba keratitis in non-contact lens wearers in India. J. Clin. Microbiol. 41:3206-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelandakis, M., S. Serre, and P. Pernin. 2000. Analysis of the 5.8S rRNA gene and the internal transcribed rs in Naegleria spp. and in N. fowleri. J. Eukaryot. Microbiol. 47:116-121. [DOI] [PubMed] [Google Scholar]
- 29.Reveiller, F. L., P. A. Cabanes, and F. Marciano-Cabral. 2002. Development of a nested PCR assay to detect the pathogenic free-living amoeba Naegleria fowleri. Parasitol. Res. 88:443-450. [DOI] [PubMed] [Google Scholar]
- 30.Riviere, D., F. M. Szczebara, J. M. Berjeaud, J. Frere, and Y. Hechard. 2006. Development of a real-time PCR assay for quantification of Acanthamoeba trophozoites and cysts. J. Microbiol. Methods 64:78-83. [DOI] [PubMed] [Google Scholar]
- 31.Schroeder, J. M., G. C. Booton, J. Hay, I. A. Niszl, D. V. Seal, M. B. Markus, P. A. Fuerst, and T. J. Byers. 2001. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J. Clin. Microbiol. 39:1903-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuster, F. L. 2002. Cultivation of pathogenic and opportunistic free-living amebas. Clin. Microbiol. Rev. 15:342-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuster, F. L., T. H. Dunnebacke, G. C. Booton, S. Yagi, C. K. Kohlmeier, C. Glaser, D. Vugia, A. Bakardjiev, P. Azimi, M. Maddux-Gonzalez, A. J. Martinez, and G. S. Visvesvara. 2003. Environmental isolation of Balamuthia mandrillaris associated with a case of amebic encephalitis. J. Clin. Microbiol. 41:3175-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma, S., G. Pasricha, D. Das, and R. K. Aggarwal. 2004. Acanthamoeba keratitis in non-contact lens wearers in India: DNA typing-based validation and a simple detection assay. Arch. Ophthalmol. 122:1430-1434. [DOI] [PubMed] [Google Scholar]
- 35.Sparagano, O. 1993. Differentiation of Naegleria fowleri and other Naegleriae by polymerase chain reaction and hybridization methods. FEMS Microbiol. Lett. 110:325-330. [DOI] [PubMed] [Google Scholar]
- 36.Stothard, D. R., J. M. Schroeder-Diedrich, M. H. Awwad, R. J. Gast, D. R. Ledee, S. Rodriguez-Zaragoza, C. L. Dean, P. A. Fuerst, and T. J. Byers. 1998. The evolutionary history of the genus Acanthamoeba and the identification of eight new 18S rRNA gene sequence types. J. Eukaryot. Microbiol. 45:45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsang, V. C., J. A. Brand, and A. E. Boyer. 1989. An enzyme-linked immunoelectrotransfer blot assay and glycoprotein antigens for diagnosing human cysticercosis (Taenia solium). J. Infect. Dis. 159:50-59. [DOI] [PubMed] [Google Scholar]
- 38.Vargas-Zepeda, J., A. V. Gomez-Alcala, J. A. Vasquez-Morales, L. Licea-Amaya, J. F. De Jonckheere, and F. Lares-Villa. 2005. Successful treatment of Naegleria fowleri meningoencephalitis by using intravenous amphotericin B, fluconazole and rifampicin. Arch. Med. Res. 36:83-86. [DOI] [PubMed] [Google Scholar]
- 39.Visvesvara, G. S. 2003. Pathogenic and opportunistic free-living amebae, p. 1981-1989. In P. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 40.Visvesvara, G. S., A. J. Martinez, F. L. Schuster, G. J. Leitch, S. V. Wallace, T. K. Sawyer, and M. Anderson. 1990. Leptomyxid ameba, a new agent of amebic meningoencephalitis in humans and animals. J. Clin. Microbiol. 28:2750-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vodkin, M. H., D. K. Howe, G. S. Visvesvara, and G. L. McLaughlin. 1992. Identification of Acanthamoeba at the generic and specific levels using the polymerase chain reaction. J. Protozool. 39:378-385. [DOI] [PubMed] [Google Scholar]
- 42.Yagi, S., G. C. Booton, G. S. Visvesvara, and F. L. Schuster. 2005. Detection of Balamuthia mitochondrial 16S rRNA gene DNA in clinical specimens by PCR. J. Clin. Microbiol. 43:3192-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, L., R. Sriram, G. S. Visvesvara, and L. Xiao. 2003. Genetic variations in the internal transcribed r and mitochondrial small subunit rRNA gene of Naegleria spp. J. Eukaryot. Microbiol. 50(Suppl.):522-526. [DOI] [PubMed] [Google Scholar]