Abstract
A multiplex PCR method has been developed to differentiate between the most common clinical serotypes of Salmonella enterica subsp. enterica encountered in Washington State and the United States in general. Six genetic loci from S. enterica serovar Typhimurium and four from S. enterica serovar Typhi were used to create an assay consisting of two five-plex PCRs. The assays gave reproducible results with 30 different serotypes that represent the most common clinical isolates of S. enterica subsp. enterica. Of these, 22 serotypes gave unique amplification patterns compared with each other and the other 8 serotypes were grouped into four pairs. These were further resolved by two additional PCRs. We compared the data from PCR serotyping with conventional serotyping and found that PCR serotyping was nearly as discriminatory as conventional serotyping was. The results from a blind test screening 111 clinical isolates revealed that 97% were correctly identified using the multiplex PCR assay. The assay can be easily performed on multiple samples with final results in less than 5 h and, in conjunction with pulsed-field gel electrophoresis, forms a very robust test method for the molecular subtyping of Salmonella enterica subsp. enterica.
The genus Salmonella belongs to the family Enterobacteriaceae, and many of its strains are important human and animal gastrointestinal pathogens. The genus Salmonella is composed of two species, Salmonella enterica and Salmonella bongori. S. enterica consists of six subgroups, groups I (S. enterica subsp. enterica), II, IIIa, IIIb, IV, and VI. Only S. enterica subsp. enterica is of clinical relevance, and this subspecies includes the pathogen associated with typhoid fever (4). The other subspecies and S. bongori are usually isolated from either the environment or reptiles and therefore are not clinically important (5). Salmonella spp. typically cause an intestinal infection that is accompanied by fever, abdominal cramps, and diarrhea with symptoms lasting over 1 week (20). The main exception is typhoid fever, a systemic infection with serious medical implications (4). Typhoid is common in the developing world, and typical symptoms include severe headaches and high fevers, but not diarrhea. Humans are the only carriers, and infected persons may be asymptomatic. Typhoid fever is spread only via direct human contact or contact with fecal contaminated foodstuffs. The other S. enterica subsp. enterica strains have reservoirs in domestic and wild animals, and the spread to humans is usually by consumption of contaminated foodstuffs. In 2004, the Food-borne Diseases Active Surveillance Network (FoodNet) of the Centers for Disease Control and Prevention (CDC) Emerging Infections Program reported 6,464 isolates of S. enterica subsp. enterica linked to food-borne disease in 10 states (39). The 2003 annual report on Salmonella from the CDC states that 33,589 Salmonella isolates were recorded by public health laboratories (29). On the basis of this and other existing data, it is estimated that there are 1.3 million cases of salmonellosis annually in the United States with 15,000 hospitalizations and 400 deaths (18).
Currently the most common method of typing S. enterica subsp. enterica has been to discriminate isolates on the basis of O (surface polysaccharide) and H (flagellar) antigenic properties. Typing the O antigen denotes the serogroup, and typing the flagellae denotes the serotype. The Vi or capsular antigens are specific to S. enterica serovar Typhi. Currently, this method employs more than 150 O and H antigens for the characterization of over 2,500 Salmonella serovars, of which 1,478 are S. enterica subsp. enterica (5, 29). Typically, the serotype uses either a name or an antigenic formula (5) based on both the serogroup and serotype. At least three antibody-antigen reactions are required to identify a particular Salmonella serovar, and rarer serovars often require many further tests to be correctly typed. The scoring of antigenic formulae uses the Kaufmann-White scheme which is annually updated by the World Health Organization (5, 30). Despite its widespread use, serotyping has deficiencies that limit its utility, including that it often takes 3 or more days to generate a result and approximately 5 to 8% of isolates are partially typed or untyped. This can be caused by several factors which include the blocking of exposure of the surface O antigens due to capsular polysaccharides in mucoid strains and in “rough” strains that produce partially formed O antigens that can cross-react with different O antisera (5). These types of isolates can be partially typed by their H antigens. With H-antigen typing, both flagellar phases must be assayed; this entails testing one flagellar phase at a time. Nonmotile isolates can be partially typed by their O antigens. Further problems with serotyping include that prolonged subculturing can affect the antigenic properties of the strain, highly trained laboratorians are required to type strains accurately, antisera must be available, and the high costs of producing and validating specific antisera to rare antigens are problematic. Delays caused by identification can hinder the response to an outbreak of disease and/or its epidemiologic surveillance.
Over the last 20 years, alternative strategies to replace or complement traditional serotyping methods have been proposed. These include ribotyping (12), ribosomal DNA intergenic spacer amplification (22), random amplification of DNA polymorphism (35), IS200 analysis (13, 38), real-time PCR (21), PCR-single-strand conformation polymorphism analysis (27), amplified fragment length polymorphism (37), sequence analysis (26), multiplex PCR (2), and DNA microarrays (8, 31). Other laboratories have taken protein-based approaches to type Salmonella enterica by methods including protein arrays (7) and mass spectrometry (40). The problems associated with these strategies include reproducibility of results between different laboratories (amplified fragment length polymorphism, IS200 analysis, random amplification of DNA polymorphism, and PCR-single-strand conformation polymorphism analysis), the requirement of specialized equipment, high material costs per sample, and highly trained staff (DNA sequencing, real-time PCR, mass spectrometry, and DNA and protein microarrays).
In this work, we describe a simple multiplex PCR method to serotype the 30 most common serovars of clinically relevant S. enterica subsp. enterica. This technique is based upon the PCR detection of genes present in specific serotypes but not others. These genes were selected from analysis of previous work including whole-genome sequencing and comparative genomic hybridization of various S. enterica subsp. enterica serotypes (14, 28, 31, 33). This method is sensitive, reproducible, and cost-effective and can easily be used in conjunction with other routine typing methods, such as pulsed-field gel electrophoresis (PFGE).
MATERIALS AND METHODS
Strains and DNA extraction.
The 30 most common serotypes of S. enterica subsp. enterica from the Washington State Public Health Laboratory (WA PHL) strain collection were used in this study (Table 1). These represent approximately 75% of all serotypes commonly identified by clinical laboratories (29). Each isolate was previously serotyped according to the modified Kauffmann-White scheme (5, 6, 29, 30). All strains were cultured on nutrient agar plates (Difco, Becton Dickson, Franklin Lakes, NJ) with overnight incubation at 36°C. The strains were not routinely subcultured; new samples were grown from frozen stocks when necessary. DNA was prepared from each isolate by either boiling a 100-μl cell suspension containing Instagene Matrix (Bio-Rad, Hercules, CA) for 10 min or by lysing cells in agarose plugs using a previously described method for PFGE analysis of Escherichia coli (16). Gel slices (1 mm × 2 mm) from PFGE plugs containing genomic DNA were liquefied for PCR analysis by melting in 35 μl of 10 mM Tris-HCl (pH 8.0) at 95°C for 10 min.
TABLE 1.
Salmonella enterica subsp. enterica serotypes used in the multiplex PCR and their STM and STY amplification patterns
| Serovar | Amplification pattern
|
|
|---|---|---|
| STM | STY | |
| Agona | 2, 3 | 5 |
| Anatum | 1, 2, 3, 4, 5 | 0 |
| Berta | 2, 3, 5 | 2, 3 |
| Bovismorbificans | 2, 3, 5 | 0 |
| Braenderup | 2, 5 | 0 |
| Brandenburg | 1, 2 | 2 |
| Chester | 1, 2 | 0 |
| Derby | 1, 2, 3, 5 | 5 |
| Dublin | 2, 3, 5 | 3 |
| Enteritidis | 2, 3, 5 | 3 |
| Hadar | 3, 5 | 0 |
| Heidelberg | 1, 2, 4, 5 | 2 |
| Infantis | 2 | 2 |
| Java 1 | 1, 2, 4, 5 | 0 |
| Java 2 | 2, 4 | 0 |
| Javiana | 1, 2 | 2, 5 |
| Mbandaka | 2, 3, 5 | 2, 5 |
| Montevideo | 5 | 2, 4 |
| Munchen | 1, 2, 5 | 0 |
| Newport | 1, 2, 3, 5 | 0 |
| Ohio | 2, 5 | 0 |
| Oranienburg | 2, 5 | 2, 4 |
| Paratyphi B | 1, 2, 4, 5 | 0 |
| Poona 1 | 1, 5 | 1, 2 |
| Poona 2 | 1, 2 | 1, 2 |
| Poona 3 | 1, 5 | 2 |
| Saintpaul | 1, 2, 3, 4, 5 | 0 |
| Stanley | 1, 2, 5 | 2 |
| Thompson | 2, 3, 5 | 5 |
| Typhi | 1 | 1, 2, 3, 5 |
| Typhimurium | 1, 2, 3, 4, 5 | 4 |
| Weltevreden | 1, 2, 4, 5 | 1, 4, 5 |
| Westhampton | 1, 2, 3 | 1, 2, 4, 5 |
Primer design and PCR.
The genomic regions chosen for this serotyping study were derived from previous work that used microarray technology to compare the genomic complement of S. enterica serovar Typhimurium LT2 (STM) and S. enterica serovar Typhi CT18 (STY) to common serotypes of S. enterica subsp. enterica (see Table 2) (31). These loci were chosen for their ability to give unique results and differentiate the clinically most common serotypes of Salmonella. The biological functions and phylogenetic implications of these loci have been discussed in those previous investigations (8, 31, 32, 34). Suitable primers for two multiplex PCRs, each amplifying five different regions, were designed using Primer Express v2.0 (Applied Biosystems, Foster City, CA) and were synthesized by MWG-Biotech Inc. (High Point, NC). The other discriminatory PCRs used primer set STM7, which was derived from serovar Typhimurium LT2, and primer set PT4-1, which was based on data from a comparative analysis including serovar Enteriditis PT4 and serovar Dublin genomes (33). A region unique to the serovar Enteriditis genome was identified, and primers were designed accordingly. For all PCRs, primer concentrations are provided in Table 2. The first multiplex assay (named STM) incorporated loci from serovar Typhimurium, and optimal results were obtained by using Platinum Taq (Invitrogen, Carlsbad, CA) under the following conditions. Reactions were performed in a final volume of 25 μl containing 8 μl of template DNA, 1.4× reaction buffer, 0.2 mM (each) deoxynucleoside triphosphates (dNTPs), 2 mM MgCl2, and 3.5 units of Taq polymerase. The second assay (named STY) was based on four loci in serovar Typhi C18 and one from serovar Typhimurium LT2. With this particular reaction, TaKaRa Taq Hot Start (Fischer Scientific, Pittsburgh, PA) was found to give more consistent results than Platinum Taq. All reactions were performed in a final volume of 25 μl containing 8 μl of template DNA, 1.6× reaction buffer, 0.2 mM dNTPs, 2 mM MgCl2, and 4.0 units of Taq polymerase. The reactions using primer sets STM7 and PT4-1 were performed in a final volume of 25 μl containing 8 μl of template DNA, 1.6× reaction buffer, 0.2 mM dNTPs, 2 mM MgCl2, and 4.0 units of Taq polymerase (Platinum or TaKaRa). All assays used the same cycling parameters, and the reactions were performed in a DNA Engine DYAD Peltier thermal cycler (Bio-Rad, Hercules, CA) under the following conditions: 1 cycle of 94°C for 5 min, followed by 40 cycles of 94°C for 30 seconds, 62°C for 30 seconds, and 72°C for 1 min, and a final extension of 72°C for 5 min.
TABLE 2.
Chromosomal regions of S. enterica serovars Typhimurium LT2, Typhi CT18, and Enteriditis (PT4) used to create primers for multiplex PCRa
| Assay | NCBI accession no. | Primer | Reaction concn (pM) | Primer sequence (5′→3′) | Amplicon size (bp) |
|---|---|---|---|---|---|
| STM 1 | AE008729 | STM0716F | 1 | AACCGCTGCTTAATCCTGATGG | 187 |
| STM0716R | 1 | TGGCCCTGAGCCAGCTTTT | |||
| STM 2 | AE008758 | STM1350F | 3 | TCAAAATTACCGGGCGCA | 171 |
| STM1350R | 3 | TTTTAAGACTACATACGCGCATGAA | |||
| STM 3 | AE008735 | STM0839F | 1 | TCCAGTATGAAACAGGCAACGTGT | 137 |
| STM0839R | 1 | GCGACGCATTGTTCGATTGAT | |||
| STM 4 | AE008913 | STM4525F | 1 | TGGCGGCAGAAGCGATG | 114 |
| STM4525R | 1 | CTTCATTCAGCAACTGACGCTGAG | |||
| STM 5 | AE008913 | STM4538F | 2 | TGGTCACCGCGCGTGAT | 93 |
| STM4538R | 2 | CGAACGCCAGGTTCATTTGT | |||
| STY 1 | AL627266 | STY0311F | 0.8 | TGGTATGGTTAAGCGGAGAATGG | 301 |
| STY0312R | 0.8 | GAGAGTCATAGCCCACACCAAAG | |||
| STY 2 | AL627273 | STY0346F | 0.8 | GGCTGGAGCAGCCTTACAAAA | 262 |
| STY0347R | 0.8 | AAGAGTTGCCTGGCTGGTAAAA | |||
| STY 3 | AL627273 | STY2299F | 3 | AATCCCCCCCCCTCAAAAA | 220 |
| STY2300R | 3 | GGTACACGTTTACTGTTTGCTGGA | |||
| STM 6 | AE008879 | STM3845F | 0.8 | ATATCTCATCGTCTCCTTTTCGTGT | 181 |
| STM3845R | 0.8 | GAAGGTCCGGATAGGCATTCT | |||
| STY 4 | AL627273 | STY2349F | 1 | AATTACGGAGCAGCAGATCGAGG | 124 |
| STY2349R | 1 | TGCGGCCAGCTGTTCAAAA | |||
| PT4 | AF370716 | PT4 F | 4 | GGCGATATAAGTACGACCATCATGG | 225 |
| PT4R | 4 | GCACGCGGCACAGTTAAAA | |||
| STM 7 | AE008795 | STM2150F | 4 | CATAACCCGCCTCGACCTCAT | 101 |
| STM2150R | 4 | AGATGTCGTGAGAAGCGGTGG |
The chromosomal regions of S. enterica serovars Typhimurium LT2 (STM), Typhi CT18 (STY), and Enteriditis (PT4) were used to create primers for multiplex PCR.
Sample analysis and scoring.
After PCR amplification, 10 μl of each reaction mixture was separated by electrophoresis in 2.5% agarose (NuSieve 3:1, FMC Bioproducts, Rockland, ME). The STM assay products were run for 60 min at 0.8 mV cm2 in 1× TBE buffer (90 mM Tris, 90 mM borate, 20 mM EDTA [pH 8.0]), and the STY assay products were run for 120 min. DNA was stained with ethidium bromide (1 μg/ml), and the gels were imaged under UV light (AlphaInnotech Corp., San Leandro, CA). PCR amplicon sizes were calculated by comparison to molecular size markers (Hyperladder V; Bioline USA Inc., Randolph, MA).
RESULTS
STM and STY primer optimization.
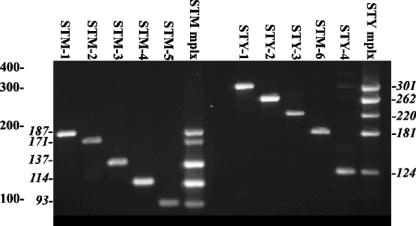
Both the STM and STY assays were designed to be multiplex PCRs, each with five primer pairs. Initially, PCR with each primer pair was optimized to ensure that each amplicon was the correct size and that a sufficient mass of amplicon was generated to permit easy detection after gel electrophoresis (Fig. 1). All of the STM assay primer sets gave a single product which corresponded to the predicted molecular size for each amplicon. All of the STM assay primer sets and STY assay primer sets produced single or multiplexed products of the predicted sizes (Fig. 1).
FIG. 1.
Amplification profiles of the primer sets for the STM and STY multiplex assays. S. enterica serovar Typhimurium template DNA was used in the six STM lanes to the left of the gel, with the primer pairs indicated above each lane. (The middle lane was empty.) S. enterica serovar Typhi template DNA was used with the primer pairs indicated above the six lanes to the right. S. enterica serovar Typhimurium and Typhi DNA templates were used to show all of the amplicons produced by the STY multiplex (mplx) assay. The sizes of DNA standards are indicated in base pairs by the leftmost numbers to the left of the gel image, while amplicon sizes (in base pairs) for the STM primer pairs are indicated in italics to the left of the gel and amplicon sizes (in base pairs) for the STY reactions are indicated in italics to the right of the gel.
Optimization experiments demonstrated that the most consistent amplification patterns were obtained for the STM and STY assays with Platinum Taq and TaKaRa Taq, respectively (data not shown).
Application of the STM assay to discriminate different serotypes by PCR.
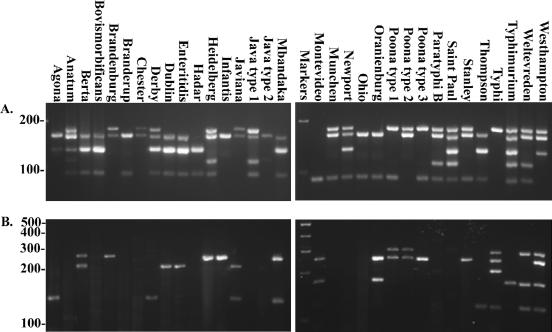
After optimization of the STM assay with S. enterica serovar Typhimurium LT2 genomic DNA, 30 serovars were tested by the multiplex PCR assays (Fig. 2A and Table 1). At least eight isolates from each serotype were tested with the exception of S. enterica serovars Stanley and Montevideo (seven and six isolates, respectively), for a total of 273 isolates. All of the serotypes produced at least one amplicon. From this initial screening, the 30 serovars could be divided into 13 groups on the basis of scoring the presence or absence of appropriately sized amplicons (Table 1). Ten serotypes produced two amplicons. The most common STM multiplex PCR amplicon pattern identified was 2,3,5 amplified from six different serotypes. In addition to the control serovar Typhimurium LT2, all other serovar Typhimurium isolates produced the anticipated 1,2,3,4,5 pattern; S. enterica serovars Anatum and Saintpaul also had this profile. In contrast, S. enterica serotypes Typhi, Montevideo, and Infantis produced only a single product from the maximum of five with the STM assay (Fig. 1 and 2A). Interestingly, different isolates of two serotypes, S. enterica serotypes Poona and Java, consistently produced different STM amplicon patterns. Serotype Java isolates produced either STM pattern 2,4 or 1,2,4,5, and serotype Poona isolates produced either STM pattern 1,2 or 1,5 (Table 1).
FIG. 2.
Agarose gel analysis of STM assay results for the 30 most prevalent S. enterica serotypes indicated above each lane. The sizes of DNA standards are indicated in base pairs to the left of the gel images. STM multiplex primers (A) and STY multiplex primers (B) were used.
Application of the STY assay to discriminate between different serotypes by PCR.
The second assay, STY, also used a set of five primer pairs. Four pairs were designed for loci in the S. enterica serovar Typhi genome, and a fifth primer pair, STM 6, was designed for S. enterica serovar Typhimurium (Table 2). DNAs from the same isolates tested with the STM assay were also screened with the STY assay. Overall, 19 of the serovars tested with the STY assay produced amplicons (Fig. 2B and Table 1). There are 14 different amplification patterns observed from the 30 different serotypes screened with the STY assay. Eleven serotypes were negative for all STY-derived loci (Fig. 2B). Of the remaining serotypes, five produced only two amplicons, one made three amplicons, and only two made four amplicons. Unlike the STM assay, serotype Java was positive for only one amplification product with the STY assay. Serotype Poona, however, was again variable, with the STY assay producing either STY pattern 1,2 or 2 only. No serotype tested produced all five possible STY amplicons. When serotype Typhimurium was screened with the STY assay, the only amplicon produced was for STM 6 (Fig. 2B). Four other serotypes also amplified STM 6, but other amplicons from the STY assay were also generated with these serotypes.
Combining the STM and STY assays to increase serotyping accuracy.
The discriminatory power for serotyping isolates was increased when the amplicon patterns of both the STM and STY assays were combined. With 13 distinct amplification codes for STM and 14 for STY, it was possible to discriminate 22 different serotypes, and four additional pairs of serotypes had the same amplification pattern (Table 1). These pairs were S. enterica serotypes Enteriditis and Dublin, S. enterica serotypes Braenderup and Ohio, S. enterica serotypes Anatum and Saintpaul, and finally S. enterica serotypes Paratyphi B and Java (only those with the STM pattern 1,2,4,5). Serotype Poona was unique in that from the isolates screened there were three different amplification profiles (STM 1,5 STY 1,2; STM 1,2 STY 1,2; and STM 1,5 STY 2). Despite the potential to produce one of three amplification codes, all are still unique for serotype Poona within the group of serotypes tested.
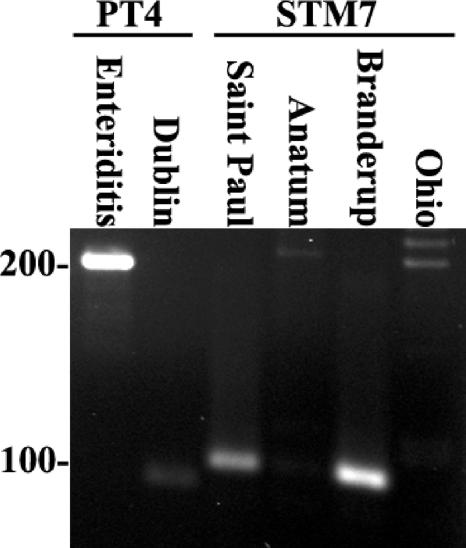
Two other primer sets were developed to further discriminate the serotypes with the same amplicon patterns. A primer set, STM 7, was designed from S. enterica serotype Typhimurium and creates an amplicon of 101 bp (Fig. 3). The STM 7 set was tested on all five pairs and found to distinguish between serotypes Braenderup and Ohio and also between serotypes Anatum and Saintpaul (Fig. 3). The other two pairs, serotypes Enteriditis and Dublin and serotypes Java and Paratyphi B, gave the same amplicon profiles with STM 7.
FIG. 3.
Agarose gel analysis of PT4 and STM7 primer sets used to discriminate paired serotypes. The template DNA used is indicated above each lane, as are primer sets. The sizes of DNA standards are indicated to the left of the gel in base pairs. S. enterica serovar Enteriditis yields the expected 225-bp amplicon, and S. enterica serovar Dublin does not. The 101-bp amplicon is produced only with S. enterica serovars Saintpaul and Braenderup DNA and not with S. enterica serovar Anatum or Ohio DNA.
A unique target for PCR from S. enterica serotype Enteriditis PT4 was designed from previously published subtractive hybridization data (1, 33) and microarray data comparing the genomic complement of S. enterica serotypes Enteriditis and Dublin (33). The PT4 primer set is serotype Enteriditis specific, and therefore, serotypes Enteriditis and Dublin could be differentiated (Fig. 3). Serotypes Java and Paratyphi B were not distinguished by the PT 4 assay. It should be noted that a simple biochemical test for growth in d-tartrate minimal medium can easily distinguish serotype Paratyphi B (no growth) from serotype Java (growth). A PCR assay to discriminate tartrate negative from tartrate positive has been described elsewhere (25).
Accuracy of PCR serotyping compared to serotyping.
The PCR results for the assays were converted to amplification patterns for comparison to traditional serotyping data for the 273 isolates. The effectiveness of differentiating serotype by PCR amplification patterns can be directly compared to serotyping data while assuming that serotyping is 100% accurate (Table 1). The PCR amplicon patterns almost exactly matched the serotyping data. There were two exceptions with the S. enterica serotypes Chester and Infantis where a single isolate in each case produced a different amplicon pattern from the other seven isolates assayed. By repeating DNA extraction and PCR, the amplicon pattern then generated was correct.
Serotyping a panel of Salmonella enterica subsp. enterica isolates in a blind test using the PCR assays.
To determine the accuracy of the PCR assays, 111 strains of S. enterica subsp. enterica serotypes were randomly chosen from the WA PHL stock collection and then typed by both the STM and STY assays (Table 3). The amplicon patterns generated from PCR with the STM and STY assays were compared to the previously established amplicon patterns shown in Table 1. Of the 111 isolates tested, 108 were correctly identified from the results of the STM and STY assays (Table 3), resulting in 97% correct classification. The remaining three isolates tested by PCR gave conflicting results by conventional serotyping identification. Two of the isolates gave amplicon patterns completely different from those already established for the 30 serotypes (Table 1). These were STM 1 STY 0 and STM 1,4,5 STY 2. These isolates were retyped by conventional serotyping methods to establish that there were no errors in the original typing. Neither of the isolates could be accurately retyped, indicating that perhaps there were errors with their original serotyping. The final strain was originally typed as S. enterica serotype Dublin but gave the amplification code STM 1,2,4,5 STY 3; this too is a unique code. However, retyping suggested the isolate was serotype Enteriditis, but the amplification code for this is STM 2,3,5 STY 3. The isolate was tested with the PT4 set to discriminate serotype Enteriditis from serotype Dublin and found to be PT4 negative and therefore was not serotype Enteriditis.
TABLE 3.
Results from blind testing of 111 different S. enterica subsp. enterica serotype isolates using the STM and STY assays to predict their serotype by PCR
| Serotype | No. of isolates
|
|
|---|---|---|
| Testeda | Failedb | |
| Agona | 4 | 0 |
| Anatum | NTc | |
| Berta | NT | |
| Bovismorbificans | NT | |
| Braenderup | NT | |
| Brandenburg | NT | |
| Chester | NT | |
| Derby | NT | |
| Dublin | 1 | 1 |
| Enteritidis | 14 | 0 |
| Hadar | NT | |
| Heidelberg | 8 | 0 |
| Infantis | 6 | 0 |
| Java | 6 | 1 |
| Javiana | 1 | 0 |
| Mbandaka | 1 | 0 |
| Montevideo | 7 | 0 |
| Muenchen | 6 | 0 |
| Newport | 12 | 0 |
| Ohio | NT | |
| Oranienburg | 10 | 1 |
| Paratyphi B | NT | |
| Poona | 1 | 0 |
| Saintpaul | 10 | 0 |
| Stanley | 1 | 0 |
| Thompson | 3 | 0 |
| Typhi | 3 | 0 |
| Typhimurium | 17 | 0 |
| Weltevreden | NT | |
| Westhampton | NT | |
The amplicon pattern generated from each sample was used to predict the serotype by comparing with the patterns in Table 1.
Three isolates produced amplicon patterns that did not correspond with the previously identified amplicon pattern for that serotype (2.7% of all samples tested).
NT, not tested.
DISCUSSION
Serotyping of S. enterica subsp. enterica by conventional methods has identified over 1,478 different serotypes (5). These are based on the detection of the cellular (lipopolysaccharide) O-specific and H1 and H2 flagellar antigens. The recent use of microarray analysis for the genomic comparison of common serotypes of S. enterica subsp. enterica has led to the identification of core genes and a series of variable genes (3, 8, 31, 33). By targeting the variability and stability of specific genes across serotypes, we were able to create a highly specific method using multiplex and conventional PCR to serotype the common clinical isolates of S. enterica subsp. enterica. Previous studies that have employed PCR to type S. enterica subsp. enterica have focused on the allelic variance of flagellar genes from the first and second phases, and although accurate, they are currently limited to the number of strains that can be typed (9, 11, 19). In addition to these, there is a PCR protocol specific for S. enterica subsp. enterica, but this method does not differentiate large numbers of serotypes (14). In this assay we can easily discriminate over 30 of the most clinically relevant serotypes of S. enterica subsp. enterica isolated in the United States. From data presented in the 2003 CDC annual summary for Salmonella in the United States, the method described herein could be used to serotype 19 of the top 20 serotypes, representing 76.6% of all isolates in the United States (29). The only serotype of this group which cannot be determined is Salmonella I 4,[5],12:i:-. This is thought to be a monophasic S. enterica serotype Typhimurium, and with this assay, it gives the same pattern as serotype Typhimurium, namely, STM 1,2,3,4,5 STY 4 (10). Microarray analysis of European monophasic strains showed six major deletions, including one encompassing the fljAB region (15). However, strains of Salmonella I 4,[5],12:i:- isolated in the United States appear to be different and are monophasic for some other reason, since fljB (and hin and iroB) are present in strains of Salmonella I 4,[5],12:i:- tested in this laboratory (data not shown).
All of the serotypes tested could be discriminated with the STM and STY PCR assays. The differentiation between S. enterica serotypes Enteriditis and Dublin is an excellent example of this where genomic comparisons between significant numbers of different isolates revealed a small region of serotype Enteriditis that is completely absent in serotype Dublin and thus suitable for distinguishing these two serotypes (1, 33). Other serotypes already known to be very similar included S. enterica serotypes Java and Paratyphi B; serotype Java is a subgroup of serotype Paratyphi B, and our assay reflected this by showing that some serotype Java isolates have the same amplicon code as that of serotype Paratyphi B. These two can however be easily distinguished using biochemical methods or PCR (25). Other secondary discriminatory regions do not have to be as highly specific; the pairs of serotypes Braenderup and Ohio and serotypes Anatum and Saintpaul were discriminated using the target gene, STM 7, designed from serotype Typhimurium. These methods could be used to rapidly identify highly similar serotypes which were previously thought to be very distinct on the basis of their antigenic profiles, e.g., serotypes Anatum and Saintpaul. In addition to different serotypes having the same amplicon pattern; it is interesting to note that some serotypes have more than one amplicon code with the STM assay. Serotypes Java and Poona both had two or more different codes which may reflect intraserovar variation.
The main problem associated with PCR-based serotyping is the inability to discriminate false positives, i.e., the misidentification of rare serotypes that create the same amplicon pattern as the more common ones. To further assist discrimination in these circumstances, we propose that these multiplex PCR assays be performed in tandem with PFGE analysis of the sample. The diagnostic aim from a clinical perspective is that the isolate can be both serotyped and subtyped by PFGE simultaneously in a real-time scenario, i.e., within 1 or 2 days of receiving the sample. Each sample can be entered for conventional serotyping and also be putatively serotyped by PCR concurrently with the PFGE analysis. A common problem encountered with PFGE is that the isolate is subtyped much sooner than it is serotyped. This delay affects the uploading of the pattern to a national database and can impede epidemiologic analysis and outbreak detection. With PCR serotyping, there is no delay waiting for the conventional serotyping data on the isolate; a presumptive serotype can be established with later confirmation (if necessary) by conventional serotyping. Additionally, the preparations of purified chromosomal DNA in agarose plugs used for restriction enzyme digestion and PFGE are easily liquefied for use in PCR applications and give very satisfactory results. Therefore, a single sample preparation can yield high-quality DNA for both subtyping and serotyping by multiplex PCR.
Once given a putative serotype, the PFGE profile can be compared to previous patterns of the same serotype in the laboratory database or PulseNet, a national PFGE program coordinated by the CDC in which all PFGE profiles of S. enterica subsp. enterica (and other bacterial enteric pathogens) from all public health laboratories are stored. There are now over 148,473 XbaI and BlnI patterns in the S. enterica subsp. enterica database. However, because the data set is so large, it is essential that the serotype is known in order to download serotype-specific patterns only. Therefore, by applying a presumptive serotype by multiplex PCR, the rapid identification of a subtype can now be done using the national database. If no pattern is found to match in either the local or PulseNet database, then the isolate is either a new subtype, or the conventional serotyping result may reveal an error in the preliminary PCR serotyping. Recently, the PulseNet network has been expanded to include data from Europe (PulseNet Europe), Asia (PulseNet Asia), and the USDA through VetNet (http://www.ars.usda.gov/main/site_main.htm?modecode=66120508) (17, 24, 36). Therefore, on a global scale, many laboratories could effectively apply this method to their test regimens. The effectiveness of the rapid access and sharing PFGE information have already shown great promise in identifying and curtailing global outbreaks of food-borne diseases (23). Combining these methods with the immunologic screening of isolates using the O antigen would further increase the accuracy of serotyping but without any significant increase in the time taken for identification.
In this work we describe a fast, accurate, and cost-effective method that can accurately discriminate between serotypes of the most common clinical isolates of S. enterica subsp. enterica. The method can be applied in any laboratory with access to PCR and agarose gel electrophoresis. The method takes only 5 h to perform from start to finish. The very simple nature of the assays means that other panels of test genes can be added in addition to the STM and STY assays, as more information is released on the genetic complement of the rarer serotypes of S. enterica subsp. enterica. For example, by combining the STM and STY assays, there are a total of 210 (1,024) amplicon codes that can be generated. This does not cover the known 1,478 serotypes even if each serotype gave a unique amplicon code (5). However, if a third five-target multiplex reaction was designed to run in conjunction with STM and STY assays, then the number of amplicon codes increases to 215 (32,768), which is 25 times more than the current number of reported serotypes.
Acknowledgments
We thank Donna Green and Jennifer Breezee at the WA PHL for technical assistance and Jennifer Bauer-Gardner, Russ Turpin, and Jonathan Cudnik at the USDA for technical assistance. We thank Charlene Jackson at the USDA for helpful discussions and critical review.
The Association of Public Health Laboratories (APHL) provided administration and support for the International Emerging Infectious Diseases (IEID) fellowship for Seonghan Kim. The CDC provided funding for the IEID fellowship and the Epidemiology and Laboratory Capacity (ELC) grant.
The mention of trade names or commercial products in the manuscript is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 30 August 2006.
REFERENCES
- 1.Agron, P. G., R. L. Walker, H. Kinde, S. J. Sawyer, D. C. Hayes, J. Wollard, and G. L. Andersen. 2001. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar Enteritidis. Appl. Environ. Microbiol. 67:4984-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, J., M. Sota, A. B. Vivanco, I. Perales, R. Cisterna, A. Rementeria, and J. Garaizar. 2004. Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J. Clin. Microbiol. 42:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anjum, M. F., C. Marooney, M. Fookes, S. Baker, G. Dougan, A. Ivens, and M. J. Woodward. 2005. Identification of core and variable components of the Salmonella enterica subspecies I genome by microarray. Infect. Immun. 73:7894-7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhan, M. K., R. Bahl, and S. Bhatnagar. 2005. Typhoid and paratyphoid fever. Lancet 366:749-762. [DOI] [PubMed] [Google Scholar]
- 5.Bopp, C. A., F. W. Brenner, P. I. Fields, J. G. Wells, and N. A. Stockbine. 2003. Escherichia, Shigella, and Salmonella, p. 654-671. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 6.Brenner, F. W. 1998. Modified Kaufmann-White scheme, vol. 1. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, Ga.
- 7.Cai, H. Y., L. Lu, C. A. Muckle, J. F. Prescott, and S. Chen. 2005. Development of a novel protein microarray method for serotyping Salmonella enterica strains. J. Clin. Microbiol. 43:3427-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan, K., S. Baker, C. C. Kim, C. S. Detweiler, G. Dougan, and S. Falkow. 2003. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA microarray. J. Bacteriol. 185:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echeita, M. A., S. Herrera, J. Garaizar, and M. A. Usera. 2002. Multiplex PCR-based detection and identification of the most common Salmonella second-phase flagellar antigens. Res. Microbiol. 153:107-113. [DOI] [PubMed] [Google Scholar]
- 10.Echeita, M. A., S. Herrera, and M. A. Usera. 2001. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:- appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 39:2981-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Echeita, M. A., and M. A. Usera. 1998. Rapid identification of Salmonella spp. phase 2 antigens of the H1 antigenic complex using “multiplex PCR.” Res. Microbiol. 149:757-761. [DOI] [PubMed] [Google Scholar]
- 12.Esteban, E., K. Snipes, D. Hird, R. Kasten, and H. Kinde. 1993. Use of ribotyping for characterization of Salmonella serotypes. J. Clin. Microbiol. 31:233-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ezquerra, E., A. P. Burnens, K. Frith, M. Costas, and J. Stanley. 1993. Molecular genotype analysis of Salmonella bovismorbificans. Mol. Cell Probes 7:45-54. [DOI] [PubMed] [Google Scholar]
- 14.Farrell, J. J., L. J. Doyle, R. M. Addison, L. B. Reller, G. S. Hall, and G. W. Procop. 2005. Broad-range (pan) Salmonella and Salmonella serotype typhi-specific real-time PCR assays: potential tools for the clinical microbiologist. Am. J. Clin. Pathol. 123:339-345. [DOI] [PubMed] [Google Scholar]
- 15.Garaizar, J., S. Porwollik, A. Echeita, A. Rementeria, S. Herrera, R. M. Wong, J. Frye, M. A. Usera, and M. McClelland. 2002. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 40:2074-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerner-Smidt, P., J. Kincaid, K. Kubota, K. Hise, S. B. Hunter, M. A. Fair, D. Norton, A. Woo-Ming, T. Kurzynski, M. J. Sotir, M. Head, K. Holt, and B. Swaminathan. 2005. Molecular surveillance of shiga toxigenic Escherichia coli O157 by PulseNet USA. J. Food Prot. 68:1926-1931. [DOI] [PubMed] [Google Scholar]
- 18.Hardnett, F. P., R. M. Hoekstra, M. Kennedy, L. Charles, and F. J. Angulo. 2004. Epidemiologic issues in study design and data analysis related to FoodNet activities. Clin. Infect. Dis. 38(Suppl. 3):S121-S126. [DOI] [PubMed] [Google Scholar]
- 19.Herrera-Leon, S., J. R. McQuiston, M. A. Usera, P. I. Fields, J. Garaizar, and M. A. Echeita. 2004. Multiplex PCR for distinguishing the most common phase 1 flagellar antigens of Salmonella spp. J. Clin. Microbiol. 42:2581-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohmann, E. L. 2001. Nontyphoidal salmonellosis. Clin. Infect. Dis. 32:263-269. [DOI] [PubMed] [Google Scholar]
- 21.Hoorfar, J., P. Ahrens, and P. Radstrom. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 38:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen, M. A., and R. J. Hubner. 1996. Use of homoduplex ribosomal DNA spacer amplification products and heteroduplex cross-hybridization products in the identification of Salmonella serovars. Appl. Environ. Microbiol. 62:2741-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirk, M. D., C. L. Little, M. Lem, M. Fyfe, D. Genobile, A. Tan, J. Threlfall, A. Paccagnella, D. Lightfoot, H. Lyi, L. McIntyre, L. Ward, D. J. Brown, S. Surnam, and I. S. Fisher. 2004. An outbreak due to peanuts in their shell caused by Salmonella enterica serotypes Stanley and Newport—sharing molecular information to solve international outbreaks. Epidemiol. Infect. 132:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto, M., Y. Suzuki, H. Nagano, J. Yatsuyanagi, H. Kurosawa, K. Kobayashi, K. Yamaoka, K. Horikawa, J. Kudaka, J. Terajima, H. Watanabe, and Y. Miyazaki. 2005. Evaluation of pulsed-field gel electrophoresis analysis performed at selected prefectural institutes of public health for use in PulseNet Japan. Jpn. J. Infect. Dis. 58:180-183. [PubMed] [Google Scholar]
- 25.Miko, A., B. Guerra, A. Schroeter, C. Dorn, and R. Helmuth. 2002. Molecular characterization of multiresistant d-tartrate-positive Salmonella enterica serovar Paratyphi B isolates. J. Clin. Microbiol. 40:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mortimer, C. K., T. M. Peters, S. E. Gharbia, J. M. Logan, and C. Arnold. 2004. Towards the development of a DNA-sequence based approach to serotyping of Salmonella enterica. BMC Microbiol. 4:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nair, S., T. K. Lin, T. Pang, and M. Altwegg. 2002. Characterization of Salmonella serovars by PCR-single-strand conformation polymorphism analysis. J. Clin. Microbiol. 40:2346-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 29.Perch, M., C. R. Braden, R. Bishop, P. Fields, R. Plikaytis, and R. V. Tauxe. 2003. Salmonella surveillance summary, 2003. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, Ga.
- 30.Popoff, M. Y., J. Bockemuhl, and A. McWhorter-Murlin. 1992. Supplement 1991 (no. 35) to the Kauffmann-White scheme. Res. Microbiol. 143:807-811. [DOI] [PubMed] [Google Scholar]
- 31.Porwollik, S., E. F. Boyd, C. Choy, P. Cheng, L. Florea, E. Proctor, and M. McClelland. 2004. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J. Bacteriol. 186:5883-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porwollik, S., and M. McClelland. 2003. Lateral gene transfer in Salmonella. Microbes Infect. 5:977-989. [DOI] [PubMed] [Google Scholar]
- 33.Porwollik, S., C. A. Santiviago, P. Cheng, L. Florea, and M. McClelland. 2005. Differences in gene content between Salmonella enterica serovar Enteritidis isolates and comparison to closely related serovars Gallinarum and Dublin. J. Bacteriol. 187:6545-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porwollik, S., R. M. Wong, and M. McClelland. 2002. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:8956-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shangkuan, Y. H., and H. C. Lin. 1998. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella typhi and other Salmonella species. J. Appl. Microbiol. 85:693-702. [DOI] [PubMed] [Google Scholar]
- 36.Swaminathan, B., T. J. Barrett, S. B. Hunter, and R. V. Tauxe. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torpdahl, M., and P. Ahrens. 2004. Population structure of Salmonella investigated by amplified fragment length polymorphism. J. Appl. Microbiol. 97:566-573. [DOI] [PubMed] [Google Scholar]
- 38.Uzzau, S., M. Hovi, and B. A. Stocker. 1999. Application of ribotyping and IS200 fingerprinting to distinguish the five Salmonella serotype O6,7:c:1,5 groups: Choleraesuis sensu stricto, Choleraesuis var. Kunzendorf, Choleraesuis var. Decatur, Paratyphi C, and Typhisuis. Epidemiol. Infect. 123:37-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vugia, D., A. Cronquist, J. Hadler, M. Tobin-D'Angelo, D. Blythe, K. Smith, K. Thornton, D. Morse, P. Cieslak, T. Jones, R. Varghese, J. Guzewich, F. Angulo, P. Griffin, R. Tauxe, and J. Dunn. 2005. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 sites, United States, 2004. Morb. Mortal. Wkly. Rep. 54:352-356. [PubMed] [Google Scholar]
- 40.Wilkes, J. G., L. Rushing, R. Nayak, D. A. Buzatu, and J. B. Sutherland. 2005. Rapid phenotypic characterization of Salmonella enterica strains by pyrolysis metastable atom bombardment mass spectrometry with multivariate statistical and artificial neural network pattern recognition. J. Microbiol. Methods 61:321-334. [DOI] [PubMed] [Google Scholar]