Abstract
A national evaluation study was performed in 14 specialized laboratories with the objective of assessing their capacities to provide (i) hepatitis B virus (HBV) viral loads (VL), (ii) HBV genotypes, and(iii) identification of precore/core mutants. The panel consisted of 12 HBV DNA-positive samples with VLs from 2.8 to 9.1 log10 copies/ml, different HBV genotypes (A to F), and 3 mutant and 9 wild-type samples at nucleotide 1896. The coefficients of variation of the mean VLs ranged from 2.4% to 10.4% with the Cobas HBV Monitor assay, from 1.8% to 5.5% with the Cobas TaqMan 48, from 1.5 to 26.2% with RealArt HBV PCR, and from 0 to 7% with branched DNA (bDNA). The Cobas Monitor assay underestimated the VLs of genotype F samples, with differences ranging from 1.4 to 2.4 log10 copies/ml. The accuracies of genotype determinations ranged from 33% to 100%, and those of precore mutant determinations ranged from 25 to 100%. This study showed some drawbacks of two widely used assays: (i) Cobas Monitor has a narrow dynamic range and underestimates genotype F sample VLs and (ii) bDNA shows poor sensitivity and may fail to identify patients with low VLs. With higher performance in terms of analytical sensitivity combined with a larger dynamic range and an ability to quantify the main genotypes equally, real-time PCR methods appear more appropriate for accurate monitoring of HBV DNA quantification. Furthermore, the clinical implications of HBV genotyping and the determination of precore/core mutants need to be clearly stated to justify the standardization of these methods.
Hepatitis B virus (HBV) is responsible for chronic liver disease, with a risk of evolution toward severe complications, such as cirrhosis and hepatocellular carcinoma. The reservoir of chronically infected patients is estimated at 350 million worldwide. An increasing number of studies demonstrate the roles of several biological markers, together with biochemical indices (especially the alanine aminotransferase level), the HBV viral load (VL), the presence of precore/core promoter mutations (20) and drug resistance mutations (38), and genotype (31), in the individual's response to treatment (15). It is expected that HBV VL measurement and HBV sequence polymorphism determination will soon provide clinicians with important information to adapt HBV-specific treatments.
Furthermore, the availability of molecular diagnostic tests for quantification of HBV DNA, especially by real-time PCR methods, is expanding. The main purpose of HBV VL determination is to evaluate the activity of infection and to screen and monitor patients for antiviral treatments. These tools are also useful in diagnosing an HBV infection in some specific situations, such as the screening of blood donations to ensure blood safety with regard to the HBsAg silent phase (window period) (6) and occult HBV infection (37), as well as in cases of atypical serologic patterns involving an envelope mutant (57).
HBV replication kinetics and mechanisms, through a pregenomic RNA intermediate and a reverse transcription step, are important sources of viral diversification. The spontaneous tendency of HBV to accumulate mutations, combined with natural selection pressure, have led to the emergence of eight described genotypes (A to H) defined by a divergence of at least 8% with the complete reference genome sequence (51, 53). It has also been demonstrated that these genotypes show distinct geographic distributions (51, 57). Furthermore, the occurrence of mixed-genotype infections does not seem uncommon (14). To date, HBV genotype determination is mainly useful for epidemiological studies or for tracing a source of contamination. However, differences in the natural history of the disease according to genotype are being explored, and the clinical relevance of genotyping is emerging. Indeed, there is evidence that HBV genotypes influence the HBeAg seroconversion rate: Asian studies have especially demonstrated that genotype B-infected patients seroconvert more rapidly to anti-HBe than those infected with genotype C (33, 59). Furthermore, genotype C seems to be linked to a more severe liver disease than genotype B in Far Eastern studies (13, 19), and the same observation was reported for genotype D versus genotype A in India (55). There are also indications in Europe that the infection outcome is more severe with genotype F than with genotype A or D (50). In addition, the genotype might influence the response to interferon-based treatments (31), with a globally better response for genotype B than C, especially in Asian studies (17, 33, 52). The influence of genotype in the response to nucleoside or nucleotide analogues is probably less important. The clinical significance of precore and core promoter variants associated with HBeAg-negative liver disease is more controversial due to the many host and viral factors. However, HBeAg-negative chronic hepatitis B seems to be associated with a more severe liver disease with a very low rate of spontaneous disease remission and a low sustained response rate to antiviral therapy (40).
Considering the emerging clinical relevance of all of these markers, and before initiating large-scale studies, the Action Coordonnée 11 group of the Agence Nationale de Recherches sur le SIDA et les hépatites B et C initiated an evaluation of HBV VL determination and HBV-genotyping and precore mutation determination methods used in specialized laboratories involved in multicenter clinical trials. The aim of this study was to assess the capacities of these specialized laboratories to provide data on HBV infection through molecular investigations.
MATERIALS AND METHODS
Constitution of the panel and assays used for its selection.
The panel included 12 plasma samples collected from HBV-infected untreated blood donors selected as a subset of subtypes with several ranges of viral loads. The characteristics of these samples are given in Table 1. The VL, the HBV genotype, and the presence of precore/core mutations were determined for each sample.
TABLE 1.
Characteristics of the 12 samples on the panel
| Sample | S-based genotype | HBe statusa | Codon 28 (nt 1896)b | HBV load (log10 copies/ml)c
|
Mean HBV load (log10 copies/ml)d
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cobas Monitor | Cobas TaqMan | Cobas Monitor (n = 9) | Cobas TaqMan (n = 6) | Real Art HBV Artus (n = 2) | In-house real-time PCR (n = 1) | Real-time PCR Dia Tech (n = 1) | bDNAe (n = 2) | Total mean (n = 21) | ||||
| s1 | C | Anti-HBe | V | 4.82 | 4.35 | 4.63 (2.6) | 4.44 (3.5) | 3.94 (6.2) | 4.34 | 4.64 | 4.69 (0) | 4.45 |
| s2 | A | HBeAg | WT | 5.65 | 6.06 | 5.81 (9.9) | 6.05 (2.7) | 5.42 (18.7) | 6.45 | 7.15 | 6.39 (1) | 6.21 |
| s3 | A | HBeAg | WT | 6.66 | 7.02 | 7.35 (6.2) | 7.04 (4.5) | 6.66 (3.7) | 7.53 | 7.87 | 7.33 (0) | 7.30 |
| s4 | A | Anti-HBe | WT | 3.20 | 2.90 | 2.96 (6.6) | 2.92 (3.2) | 2.18 (9.1) | 2.80 | 2.69 | 0 | 2.26 |
| s5 | F | Anti-HBe | V | 2.78 | 4.52 | 2.64 (6.4) | 4.48 (4.8) | 4.08 (3.7) | 4.54 | 4.48 | 5.03 (2) | 4.21 |
| s6 | F | HBeAg | WT | 7.60 | 9.54 | 7.60 (5.3) | 9.30 (5.5) | 8.46 (11.5) | 9.83 | 9.21 | 9.45/>8.00 | 8.98 |
| s8 | D | Anti-HBe | V | 4.72 | 4.68 | 4.67 (2.4) | 4.47 (5.1) | 4.41 (11.9) | 4.53 | 3.45 | 4.98 (0) | 4.42 |
| s9 | A | HBeAg | WT | 8.70 | 8.51 | 9.00 (9.8) | 8.69 (1.8) | 7.28 (21.7) | 9.08 | 8.69 | 8.98/>8 | 8.62 |
| s10 | A | Anti-HBe | WT | 4.00 | 4.38 | 3.98 (3.0) | 4.19 (4.5) | 3.17 (10.9) | 3.76 | 3.91 | 4.43 (7) | 3.91 |
| s11 | B | HBeAg | WT | 9.11 | 9.17 | 9.35 (9.1) | 9.04 (4.1) | 8.07 (4.7) | 9.40 | 9.71 | 9.44/>8.00 | 9.17 |
| s14 | E | HBeAg | WT | 7.40 | 9.25 | 9.32 (9.4) | 9.01 (2.8) | 6.73 (26.2) | 9.41 | 8.28 | 9.11/>8.00 | 8.64 |
| s15 | C | HBeAg | WT | 9.00 | 9.07 | 9.10 (10.4) | 8.96 (2.6) | 8.56 (1.5) | 9.51 | 9.26 | 9.37/>8.00 | 9.13 |
Anti-HBe and HBeAg assays were performed with ETI-EBK antibody and ETI-EBK plus (Dia Sorin, Saluggia, Italy), respectively.
V, precore stop codon mutation (G to A at nucleotide [nt] 1896); WT, wild type as defined by Inno-Lipa.
VL was obtained in laboratory P, which prepared the panel.
Mean VLs were obtained in all participating laboratories (including laboratory P). % CV values are given in parentheses.
For samples for which the mean could not be determined, the VL values are presented separated by a slash.
VL determination was performed twice in two separate runs, with two procedures used according to the manufacturers' instructions: Cobas Amplicor HBV Monitor and the Cobas TaqMan 48 Real-Time PCR System (Roche Molecular Systems, Meylan, France) (limits of detection [LOD], 200 and 35 copies/ml, respectively).
The genotype was determined twice in two separate runs with an in-house method based on the sequence analysis of HBV gene S, using a QIAamp DNA Minikit (QIAGEN, Courtaboeuf, France) for HBV DNA extraction, and a consensus PCR assay amplifying an S gene fragment comprised of nucleotides 256 to 725. Each PCR experiment included negative and positive controls. Double-strand direct sequencing was carried out by the dideoxynucleotide chain terminator method by MWG Biotech (Roissy, France). Several reference sequences of HBV genotypes (A to H) drawn from GenBank were included in the data bank. All sequences were aligned using ClustalW software (56). Phylogenetic analysis was performed using the Phylip package. Distances between sequences were analyzed using the neighbor-joining algorithm based on the Kimura 2 parameter distance estimation method for the nucleotide sequences and on the Dayhoff PAM matrix for the amino acid sequences (48).
Finally, the presence of precore/core mutations was studied twice in two separate runs using a commercially available line probe assay for identification of HBV precore variants (INNO-LiPA HBV Precore; Innogenetics, Gent, Belgium) according to the manufacturer's instructions. For the purpose of the experiments, the recommendation that at least codon 28 (with the presence of the G1896A point mutation) should be analyzed was made in order to cover this most prevalent mutant.
Participating laboratories.
Fourteen laboratories (coded from A to N) participated in the study. All had to perform HBV VL determinations, while HBV genotyping and precore/core mutation determinations were optional.
For HBV VL determination, 9 of the 14 laboratories used one method, three used two methods, and one used three methods. Including the laboratory that had prepared the panel (laboratory “P”) and that retested the panel under the same conditions as the other participants, a total of 21 results of HBV VL determination were given and analyzed. The quantification methods consisted of Cobas HBV Monitor (Roche Molecular Systems) (LOD, 200 copies/ml; linear range, 200 to 2 × 105 copies/ml; laboratories P, A, C, D, F, G, H, I, and M), Cobas TaqMan 48 (Roche Molecular Systems) (LOD, 35 copies/ml; linear range, 170 to 6.4 × 108 copies/ml; laboratories P, B, J, K, L, and N), the RealArt HBV PCR kit (Artus Abbott, Rungis, France) (LOD, 30 copies/ml; linear range, 30 to 3 × 1010 copies/ml; laboratories H and I), HBV RealQuantTM PCR (DiaTech, Jesi, Italy) (laboratory A), and a home-made HBV quantification real-time PCR method (LOD, 50 copies/ml; linear range, 50 to 5 × 108 copies/ml; laboratory L). Finally, two laboratories (E and L) used the branched-DNA (bDNA) signal amplification technology, the HBV bDNA assay (VERSANT HBV 3.0 Assay; Bayer HealthCare-Diagnostics, Tarrytown, New York) (LOD, 2,000 copies/ml; linear range, 2 × 103 to 1 × 108 copies/ml).
Ten laboratories provided HBV-genotyping results. Four used commercially available methods, three used the INNO-Lipa HBV-genotyping test (Innogenetics, Gent, Belgium), and one used the TrueGENE HBV-genotyping test (Bayer Diagnostics, Tarrytown, NY). The other six laboratories used an in-house S gene-sequencing method.
Nine laboratories provided results for the precore/core mutation determination: three used the INNO-LiPA HBV precore assay (Innogenetics, Gent, Belgium), while six provided results obtained from a sequence analysis of the precore/core region of the genome.
Interpretation of the results.
To compare the VL results, the data obtained from all of the laboratories were analyzed as a whole. For clarity of the report, all the results were converted into copies/ml (at the time of study, Roche Diagnostics did not recommend expressing the results with Cobas Monitor as international units) through the appropriate conversion factor provided by the manufacturers, with 1 international unit corresponding to 5.82 copies for the Roche TaqMan assay, to 7.5 copies for the Artus method, to 7 copies for the Dia Tech method, and to 5.6 copies for the bDNA assay. Values converted into log10 units obtained for each panel sample were compared to the mean of all measurement values given by all participants using the same procedure. In addition, methods were compared by comparison of the mean log10 VLs calculated for each sample. A difference of more than 0.5 log10 was considered significant. When a VL was not determined with precision but was above the linear range of the assay, the result was excluded from the calculation of the mean values.
For the analysis of genotype and precore mutation results, the data from the participating laboratories were compared to those obtained in laboratory P.
RESULTS
HBV viral load determination.
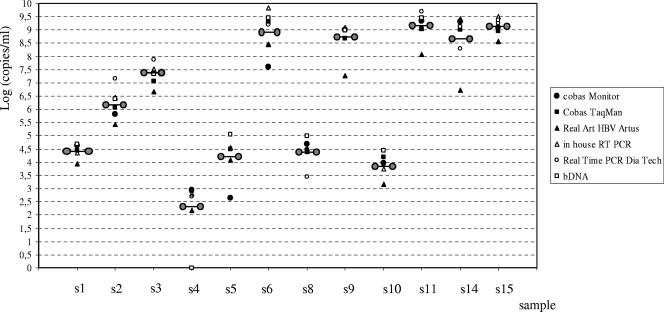
For HBV VL determination, 10 of the 14 laboratories used a single method, 4 used two methods, and 1 used three methods. A total of 21 results of HBV DNA quantification were provided and analyzed. A summary of the results obtained with the different assays is given in Table 1. Figure 1 represents the dispersion of mean HBV viral loads obtained with each assay in tested samples.
FIG. 1.
Dispersion of HBV viral loads, expressed as means, obtained with each assay. The means of viral loads including all results for each sample are symbolized by linked circles.
Nine measures of HBV VL obtained with Cobas Monitor were analyzed for all samples, except for samples s9, s11, s14, and s15, which were not diluted by one laboratory and therefore were not precisely quantified (thus, eight measures were analyzed for these four samples). The coefficients of variation (CV) of the mean VL ranged from 2.4% to 10.4% (Table 1). There was no correlation between the mean VL and the CV. However, the highest CV tended to be observed for the highest VL. Compared with the other methods, the Cobas Monitor assay underestimated the VLs of genotype F samples, with differences ranging from 1.4 to 2.4 and from 0.86 to 2.2 log10 copies/ml for samples s5 and s6, respectively. The interlaboratory comparison showed the tendency of laboratory F to give significantly higher VL values (from 0.63 to 2.14 log10 copies/ml) for samples with VLs above 7 log10 copies/ml (n = 7) (Fig. 1). Significant differences were also observed for two samples quantified in laboratory A (the VL was overestimated, with differences of 0.68 [s6] and 0.59 [s11] log10 copies/ml, respectively) and in four samples (s9, s11, s14, and s15) from laboratory I (underestimation, with differences from 0.54 to 0.70 log10 copies/ml). The four samples involved in these misclassifications had VLs above 7.6 log10 copies/ml.
Three to six measures of VL determined with the Cobas TaqMan were included in the calculation of the mean VL value, depending on the samples. Four and three measures were taken into consideration for samples s6 and s9 and for samples s11, s14, and s15, respectively. The CV of the mean VL ranged from 1.8% to 5.5%. Significant differences were observed for two samples (s6 and s11) that were diluted before being tested in laboratory J (the VL was overestimated, with differences of 0.69 and 0.77 log10 copies/ml, respectively), and two samples (s3 and s6) were underestimated by laboratories K and L, respectively (with differences of 0.62 and 0.54 log10 copies/ml). The four samples involved in these misclassifications had VLs above 7.6 log10 copies/ml.
The results obtained with the Artus procedure in two laboratories showed high CV values (from 1.5 to 26.2%). Six samples were quantified with VL differences of more than 0.5 log10 (from 0.74 to 2.24 log10 copies/ml). Moreover, the genotype E sample was quantified with a significantly lower VL by comparison to the other assays (1.9 log10-copy/ml difference from the mean result).
Determinations of the VL performed with bDNA in two laboratories demonstrated very good reproducibility, with an extremely low CV (calculated for six samples) ranging from 0 to 7%, whatever the VL. However, s4, which had the lowest VL of the panel (2.9 log10 copies/ml), was not detected by the two laboratories using this method (Table 1).
Genotype determination.
The accuracies of genotype determinations ranged from 33% to 100% (Table 2). Of the 10 laboratories that provided results, 5 correctly identified all samples; 3 gave an erroneous result for a unique sample; 2 gave the correct genotype for all samples but in association with one or two other genotypes in eight and one samples, respectively; 3 did not provide results for one or two samples due to a failure at the amplification or sequencing steps—twice for sample s4 (genotype A; 2.9 log10 copies/ml) and once for samples s5 and s15 (genotype F, 4.5 log10 copies/ml, and genotype C, 9.0 log10 copies/ml).
TABLE 2.
Performances of the laboratories in HBV genotyping
| Parameter | S-based genotype | Viral load determined by TaqMan (log10 copies/ml) | Results for laboratorya:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B (456-798)b | C (377-794) | D (456-798) | E (456-798) | G (250-1280) | H (301-1022) | J (301-1022) | K (459-988) | L (458-830) | M (253-1060) | |||||
| Sample assignment | ||||||||||||||
| s1 | C | 4.35 | C/D | C | C | C | C | C | C | C | C | C | ||
| s2 | A | 6.06 | A/E | A | A | A | A | A | A | A | A | A | ||
| s3 | A | 7.02 | A/C | A | A | A | A | A | A | A | A | G | ||
| s4 | A | 2.90 | A | A | A | Neg | A | A | A | Neg | A | A | ||
| s5 | F | 4.52 | D/F | Neg | F | F | F | F | F | F | F | F | ||
| s6 | F | 9.54 | F | F | F | F | F | F | F | F | F | F | ||
| s8 | D | 4.68 | D | D | D | D/F | D | D | D | E | D | D | ||
| s9 | A | 8.51 | A/C | A | A | A | A | A | A | A | A | A | ||
| s10 | A | 4.38 | A/C/E | F/G | A | A | A | A | A | A | A | A | ||
| s11 | B | 9.17 | A/B | B | B | B | B | B | B | B | B | B | ||
| s14 | E | 9.25 | E | E | E | E | E | E | E | E | E | E | ||
| s15 | C | 9.07 | C/D | Ind | C | C | C | C | C | C | C | C | ||
| No. (%) of correct genotypes | 4 (33.3) | 9 (75.0) | 12 (100) | 10 (83.3) | 12 (100) | 12 (100) | 12 (100) | 10 (83.3) | 12 (100) | 11 (91.7) | ||||
| No. of incorrect genotypes | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | ||||
| No. of coinfections | 8 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| No. of negative or indeterminate results | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | ||||
Genotypes were determined with Inno-Lipa HBV genotyping test (Innogenetics, Gent, Belgium; laboratories B, D, and E), an in-house direct sequencing method (laboratories C, G, H, J, K, and M), and the Trugene HBV-genotyping test (Bayer Diagnostics, Tarrytown, NY; laboratory L). Neg, negative.
Position in the nucleotide sequence of the amplified segment in the S gene of HBV.
Of the 120 expected correct results, 104 (86.7%) were accurate. Among the 16 incorrect results, 9 involved mixed-genotype results, 3 were erroneous genotypes, and 4 were PCR amplification or sequencing failures. Only two samples (genotypes F and E, with VLs of 9.5 log10 copies/ml in both cases) were correctly identified by all of the laboratories.
Precore mutants.
The accuracies of precore mutant determinations ranged from 25 to 100% (Table 3). Of the nine laboratories that provided results, two correctly identified all of the tested samples (12/12 for laboratory L and 8/8 for laboratory N), one gave erroneous results for 8 samples, one provided a dual wild-type (WT) and 1896 variant (V) infection, and four missed 1 to 3 samples due to a failure in PCR amplification, while two declared a failure in the identification of the 28 codon in 2 or 3 samples.
TABLE 3.
Performances of the laboratories in the identification of precore mutants (position 1896)
| Parameter | S-based genotyping | HBe status | Viral load determined by TaqMan (log10 copies/ml) | Results for laboratorya:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pb | Bb | Eb | Lb | Gc | Hc | Jc | Kc | Mc | Nc | ||||||||
| Sample assignment | |||||||||||||||||
| s1 | C | Anti-HBe | 4.35 | V | V | V | V | V | V | V | V | V | V | ||||
| s2 | A | HBeAg | 6.06 | WT | V | WT | WT | WT | WT | WT | WT | WT | NT | ||||
| s3 | A | HBeAg | 7.02 | WT | V | WT | WT | WT | WT | WT | WT | WT | WT | ||||
| s4 | A | Anti-HBe | 2.90 | WT | WT | Neg | WT | WT | WT | WT | Neg | Neg | WT | ||||
| s5 | F | Anti-HBe | 4.52 | V | V | V | V | Ind | V | Neg | V | V | V | ||||
| s6 | F | HBeAg | 9.54 | WT | V | WT | WT | Ind | WT | WT | WT | WT | NT | ||||
| s8 | D | Anti-HBe | 4.68 | V | V | NT | V | V | V | V | V | V | V | ||||
| s9 | A | HBeAg | 8.51 | WT | V | WT | WT | WT | Ind | WT | WT | WT | WT | ||||
| s10 | A | Anti-HBe | 4.38 | WT | V | WT | WT | Ind | WT | WT | WT | WT | WT | ||||
| s11 | B | HBeAg | 9.17 | WT | V | WT/V | WT | WT | Ind | WT | WT | WT | WT | ||||
| s14 | E | HBeAg | 9.25 | WT | V | WT | WT | WT | WT | WT | WT | Neg | NT | ||||
| s15 | C | HBeAg | 9.07 | WT | V | WT | WT | WT | WT | WT | NT | Neg | NT | ||||
| No. (%) of correct results | 3/12 (25) | 9/12 (75) | 12/12 (100) | 9/12 (75) | 10/12 (83) | 11/12 (92) | 10/12 (83) | 9/12 (75) | 8/12 (67) | ||||||||
| No. of false results | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||||
| No. of negative or untested samples | 0 | 2 | 0 | 0 | 0 | 1 | 2 | 3 | 4 | ||||||||
| No. of indeterminate results | 0 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | ||||||||
V, precore stop codon mutation (G to A at nucleotide 1896); Neg, negative; Ind, indeterminate; NT, not tested.
InnoLiPA HBV precore (Innogenetics, Gent, Belgium).
In-house direct sequencing method.
Of the 108 expected correct results, 82 (76%) were accurate. Among the 26 incorrect results, 9 were erroneous results (V instead of WT in eight samples and one dual V-WT infection), 12 were false-negative results or untested samples, and 5 were due to an indeterminate result.
DISCUSSION
Among the techniques described in recent years for quantifying HBV DNA by PCR (16, 24, 29), some have been based on semiautomatic assays and were described as providing good sensitivity for the detection of HBV DNA (39, 41). More recently, introduction of real-time PCR technology for HBV viral load measurement has offered even better characteristics, with an overall larger range of quantification and improved sensitivity (1, 5, 23, 26, 36, 58). Although these two technologies have been assessed and compared within different single-center laboratories, an evaluation of such techniques through a multicenter quality control study was missing. The strength of such an evaluation is to provide information on what may happen to a patient who may be followed over time in different laboratories using in-house or commercially available techniques. In terms of the technical handling characteristics of the six studied HBV DNA quantification methods, one of the major inconveniences involved the Cobas Monitor PCR system, which has the narrowest dynamic range and requires dilution of samples in cases of high VL to artificially extend the test linearity. This drawback has been pointed out previously (43, 58). Due to the error rate linked to the dilution process, a weakness is introduced in the accuracy of the technique, especially for active replicative HBV carriers with VLs that may reach over 9 log10 copies/ml. It is likely that the highest CV in interlaboratory comparison observed in our study with the Cobas Monitor for samples with a high VL reflects this phenomenon.
In the performances of the HBV DNA quantification methods, we observed an underestimation of the VLs of genotype F samples with Cobas Monitor. Although genotype F is mainly present in Central and South America (4, 7), this failure of Cobas Monitor (58) may have an impact on the monitoring of HBV infection, particularly if the genotype is not determined before VL quantification. Thus, we recommend not using this assay for the monitoring of patients infected by a genotype F strain.
The Artus System tended to provide VL results lower than those of the other assays, especially for the genotype E samples. However, the high CV observed between the results of the two laboratories that used this assay could suggest interlaboratory variability. Further extended studies of the Artus Real Time PCR system (as well as the other real-time PCR methods used by only one participating laboratory) are needed to assess the performances of these new methods of HBV DNA quantification.
Although the observed differences were not significant compared with the other assays, the bDNA assay tended to give higher VL results, as previously reported (25). Moreover, the reproducibility rate was very high (46). However, due to its low sensitivity, linked to the principle of this assay (49), the bDNA assay had a high failure rate for the two samples of the panel with the lowest VLs.
The results obtained here and in other studies have demonstrated that the real-time PCR methods offer several advantages in terms of analytical sensitivity, with lower limits of detection and larger dynamic ranges of quantification. The good performances of real-time PCR methods and the availability of automatic platforms will justify the use of these tests for the diagnosis of HBV infection in the near future, especially for atypical serological patterns, as in occult infection (2, 18), in HBsAg carriers with a low level of replication (22, 42), or for treatment monitoring in order to identify resistant mutants that could be difficult to demonstrate with methods exhibiting a low limit of detection.
Although correct results were obtained for 50% of the participants for the entire panel of HBV genotypes, the data analysis revealed a few discrepancies, essentially due to poor amplification sensitivities of the samples with the lowest VLs and sometimes due to misclassification. Indeed, the false-negative results obtained with samples s4 and s5 were probably due to their low VLs rather than to their genotypes. The misclassification of samples s3 and s10, both containing a genotype A strain and classified as G and F/G by two laboratories using an in-house method, may be explained by the close phylogenic proximity of A and G genotypes in the amplified region (3, 54). However, the possibility that these two samples were really coinfected with the two genotypes cannot be excluded, since that seems to be the case for most subjects infected with genotype G. More surprisingly (and contrary to data provided by three other laboratories using the same technology), laboratory B reported 67% mixed infections with the Inno-Lipa method. This method has been shown to be very sensitive for identification of mixed-genotype infections, but the reported rate of mixed infections is usually between 0.1 and 10% (30, 45). Clearly, Inno-Lipa allows rapid detection of HBV genotypes in diagnostic laboratories, but extensive comparison studies of this assay with cloning experiments are needed to establish the reality of the mixed infections and to clearly exclude any issue of specificity.
Contrary to the results that we have obtained for HCV genotyping (35), the fact that no reference genome database was provided to all the participants did not seem to have influenced the genotyping results obtained by direct sequencing. The reason for this finding is probably the absence of subtype definition for HBV, permitting a less precise phylogenic analysis without consequence for the final genotyping classification.
In the precore/core region, the most common naturally occurring HBV mutations are G1896A, which creates a stop codon in the precore gene (9, 11), and an A1762T-G1764A dual mutation in the core promoter region, which is responsible for a down-regulation of HBcAg production (9). Some of the participating laboratories did not routinely perform the determination of these mutations (only nine provided results). Besides laboratory B, which gave 75% incorrect results for an unknown reason (a nonspecific hybridization with the line probe assay could be involved), incorrect answers were mainly due to an absence of amplification or an indeterminate result independent of the method used. This is in accordance with the fact that the determination of precore/core mutations is not performed on a routine basis and that only a few laboratories are accustomed to the procedure. Indeed, the usefulness of identification of these HBV mutations is still being debated (10, 12, 32, 34). Moreover, the relationship between these mutants and the response to antiviral treatment remains controversial (9, 20, 28, 34) and could be due to other factors, such as genotypes or recruitment bias (21, 27, 44). Thus, it does not seem crucial to determine the existence of these mutations in order to evaluate the severity of the disease and to manage antiviral treatments.
One may conclude from this multicenter study that the large majority of expert laboratories routinely use commercial assays for HBV VL determinations (8, 47). The study also showed some drawbacks with two widely used assays: (i) Cobas Monitor has a narrow dynamic range and underestimates genotype F sample VLs and (ii) bDNA shows poor sensitivity and may therefore fail to identify patients with low VLs. With higher performance in terms of analytical sensitivity combined with a larger dynamic range and an ability to quantify the main genotypes equally, the real-time PCR methods appear more appropriate for accurate monitoring of HBV DNA quantification. Furthermore, the clinical implications of HBV genotyping, as well as the determination of precore/core mutants, need to be clearly stated to justify the standardization of these methods. A problem that could justify the intensive use of appropriate diagnostic tools would be the emergence of a high number of drug-resistant mutants.
Acknowledgments
This work was supported by a grant from the Agence Nationale de Recherches sur le SIDA et les Hepatites B et C.
We thank Véronique Descamps, Annie Girault, Joelle Lerable, Christine Portal, and Annabelle Servant-Delmas for their technical assistance.
REFERENCES
- 1.Aliyu, S. H., M. H. Aliyu, H. M. Salihu, S. Parmar, H. Jalal, and M. D. Curran. 2004. Rapid detection and quantitation of hepatitis B virus DNA by real-time PCR using a new fluorescent (FRET) detection system. J. Clin. Virol. 30:191-195. [DOI] [PubMed] [Google Scholar]
- 2.Allain, J. P. 2004. Occult hepatitis B virus infection: implications in transfusion. Vox. Sang. 86:83-91. [DOI] [PubMed] [Google Scholar]
- 3.Arauz-Ruiz, P., H. Norder, B. H. Robertson, and L. O. Magnius. 2002. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 83:2059-2073. [DOI] [PubMed] [Google Scholar]
- 4.Arauz-Ruiz, P., H. Norder, K. A. Visona, and L. O. Magnius. 1997. Genotype F prevails in HBV infected patients of Hispanic origin in Central America and may carry the precore stop mutant. J. Med. Virol. 51:305-312. [PubMed] [Google Scholar]
- 5.Beuselinck, K., M. van Ranst, and J. van Eldere. 2005. Automated extraction of viral-pathogen RNA and DNA for high-throughput quantitative real-time PCR. J. Clin. Microbiol. 43:5541-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas, R., E. Tabor, C. C. Hsia, D. J. Wright, M. E. Laycock, E. W. Fiebig, L. Peddada, R. Smith, G. B. Schreiber, J. S. Epstein, G. J. Nemo, and M. P. Busch. 2003. Comparative sensitivity of HBV NATs and HBsAg assays for detection of acute HBV infection. Transfusion 43:788-798. [DOI] [PubMed] [Google Scholar]
- 7.Blitz, L., F. H. Pujol, P. D. Swenson, L. Porto, R. Atencio, M. Araujo, L. Costa, D. C. Monsalve, J. R. Torres, H. A. Fields, S. Lambert, C. Van Geyt, H. Norder, L. O. Magnius, J. M. Echevarria, and L. Stuyver. 1998. Antigenic diversity of hepatitis B virus strains of genotype F in Amerindians and other population groups from Venezuela. J. Clin. Microbiol. 36:648-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunetto, M. R., and F. Bonino. 2004. Treatment of chronic hepatitis B: from research to clinical practice via the consensus conferences. Curr. Pharm. Des. 10:2063-2075. [DOI] [PubMed] [Google Scholar]
- 9.Brunetto, M. R., M. Giarin, G. Saracco, F. Oliveri, P. Calvo, G. Capra, A. Randone, M. L. Abate, P. Manzini, M. Capalbo, et al. 1993. Hepatitis B virus unable to secrete e antigen and response to interferon in chronic hepatitis B. Gastroenterology 105:845-850. [DOI] [PubMed] [Google Scholar]
- 10.Brunetto, M. R., M. M. Giarin, F. Oliveri, E. Chiaberge, M. Baldi, A. Alfarano, A. Serra, G. Saracco, G. Verme, H. Will, et al. 1991. Wild-type and e antigen-minus hepatitis B viruses and course of chronic hepatitis. Proc. Natl. Acad. Sci. USA 88:4186-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carman, W. F., M. R. Jacyna, S. Hadziyannis, P. Karayiannis, M. J. McGarvey, A. Makris, and H. C. Thomas. 1989. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet ii:588-591. [DOI] [PubMed] [Google Scholar]
- 12.Chan, H. L., N. W. Leung, M. Hussain, M. L. Wong, and A. S. Lok. 2000. Hepatitis B e antigen-negative chronic hepatitis B in Hong Kong. Hepatology 31:763-768. [DOI] [PubMed] [Google Scholar]
- 13.Chan, H. L., M. L. Wong, A. Y. Hui, L. C. Hung, F. K. Chan, and J. J. Sung. 2003. Hepatitis B virus genotype C takes a more aggressive disease course than hepatitis B virus genotype B in hepatitis B e antigen-positive patients. J. Clin. Microbiol. 41:1277-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, B. F., C. J. Liu, G. M. Jow, P. J. Chen, J. H. Kao, and D. S. Chen. 2006. Evolution of hepatitis B virus in an acute hepatitis B patient co-infected with genotypes B and C. J. Gen. Virol. 87:39-49. [DOI] [PubMed] [Google Scholar]
- 15.Chen, C. J., H. I. Yang, J. Su, C. L. Jen, S. L. You, S. N. Lu, G. T. Huang, and U. H. Iloeje. 2006. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 295:65-73. [DOI] [PubMed] [Google Scholar]
- 16.Chen, T., J. M. Luk, S. T. Cheung, W. C. Yu, and S. T. Fan. 2000. Evaluation of quantitative PCR and branched-chain DNA assay for detection of hepatitis B virus DNA in sera from hepatocellular carcinoma and liver transplant patients. J. Clin. Microbiol. 38:1977-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chien, R. N., C. T. Yeh, S. L. Tsai, C. M. Chu, and Y. F. Liaw. 2003. Determinants for sustained HBeAg response to lamivudine therapy. Hepatology 38:1267-1273. [DOI] [PubMed] [Google Scholar]
- 18.Conjeevaram, H. S., and A. S. Lok. 2001. Occult hepatitis B virus infection: a hidden menace? Hepatology 34:204-206. [DOI] [PubMed] [Google Scholar]
- 19.Duong, T. N., N. Horiike, K. Michitaka, C. Yan, M. Mizokami, Y. Tanaka, K. Jyoko, K. Yamamoto, H. Miyaoka, Y. Yamashita, N. Ohno, and M. Onji. 2004. Comparison of genotypes C and D of the hepatitis B virus in Japan: a clinical and molecular biological study. J. Med. Virol. 72:551-557. [DOI] [PubMed] [Google Scholar]
- 20.Erhardt, A., U. Reineke, D. Blondin, W. H. Gerlich, O. Adams, T. Heintges, C. Niederau, and D. Haussinger. 2000. Mutations of the core promoter and response to interferon treatment in chronic replicative hepatitis B. Hepatology 31:716-725. [DOI] [PubMed] [Google Scholar]
- 21.Funk, M. L., D. M. Rosenberg, and A. S. Lok. 2002. World-wide epidemiology of HBeAg-negative chronic hepatitis B and associated precore and core promoter variants. J. Viral Hepat. 9:52-61. [DOI] [PubMed] [Google Scholar]
- 22.Ganem, D., and A. M. Prince. 2004. Hepatitis B virus infection—natural history and clinical consequences. N. Engl. J. Med. 350:1118-1129. [DOI] [PubMed] [Google Scholar]
- 23.Garson, J. A., P. R. Grant, U. Ayliffe, R. B. Ferns, and R. S. Tedder. 2005. Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J. Virol. Methods 126:207-213. [DOI] [PubMed] [Google Scholar]
- 24.Gerken, G., J. Gomes, P. Lampertico, M. Colombo, T. Rothaar, M. Trippler, and G. Colucci. 1998. Clinical evaluation and applications of the Amplicor HBV Monitor test, a quantitative HBV DNA PCR assay. J. Virol. Methods 74:155-165. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert, T. 2003. Exploring the dynamics of power: a Foucauldian analysis of care planning in learning disabilities services. Nurs. Inq. 10:37-46. [DOI] [PubMed] [Google Scholar]
- 26.Gordillo, R. M., J. Gutierrez, and M. Casal. 2005. Evaluation of the COBAS TaqMan 48 real-time PCR system for quantitation of hepatitis B virus DNA. J. Clin. Microbiol. 43:3504-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grandjacques, C., P. Pradat, L. Stuyver, M. Chevallier, P. Chevallier, C. Pichoud, M. Maisonnas, C. Trepo, and F. Zoulim. 2000. Rapid detection of genotypes and mutations in the pre-core promoter and the pre-core region of hepatitis B virus genome: correlation with viral persistence and disease severity. J. Hepatol. 33:430-439. [DOI] [PubMed] [Google Scholar]
- 28.Hadziyannis, S. J., G. V. Papatheodoridis, E. Dimou, A. Laras, and C. Papaioannou. 2000. Efficacy of long-term lamivudine monotherapy in patients with hepatitis B e antigen-negative chronic hepatitis B. Hepatology 32:847-851. [DOI] [PubMed] [Google Scholar]
- 29.Hendricks, D. A., B. J. Stowe, B. S. Hoo, J. Kolberg, B. D. Irvine, P. D. Neuwald, M. S. Urdea, and R. P. Perrillo. 1995. Quantitation of HBV DNA in human serum using a branched DNA (bDNA) signal amplification assay. Am. J. Clin. Pathol. 104:537-546. [DOI] [PubMed] [Google Scholar]
- 30.Hussain, M., C. J. Chu, E. Sablon, and A. S. Lok. 2003. Rapid and sensitive assays for determination of hepatitis B virus (HBV) genotypes and detection of HBV precore and core promoter variants. J. Clin. Microbiol. 41:3699-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janssen, H. L., M. van Zonneveld, H. Senturk, S. Zeuzem, U. S. Akarca, Y. Cakaloglu, C. Simon, T. M. So, G. Gerken, R. A. de Man, H. G. Niesters, P. Zondervan, B. Hansen, and S. W. Schalm. 2005. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet 365:123-129. [DOI] [PubMed] [Google Scholar]
- 32.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2003. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 124:327-334. [DOI] [PubMed] [Google Scholar]
- 33.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2004. Hepatitis B virus genotypes and spontaneous hepatitis B e antigen seroconversion in Taiwanese hepatitis B carriers. J. Med. Virol. 72:363-369. [DOI] [PubMed] [Google Scholar]
- 34.Kidd-Ljunggren, K., M. Oberg, and A. H. Kidd. 1997. Hepatitis B virus X gene 1751 to 1764 mutations: implications for HBeAg status and disease. J. Gen. Virol. 78:1469-1478. [DOI] [PubMed] [Google Scholar]
- 35.Laperche, S., F. Lunel, J. Izopet, S. Alain, P. Deny, G. Duverlie, C. Gaudy, J. M. Pawlotsky, J. C. Plantier, B. Pozzetto, V. Thibault, F. Tosetti, and J. J. Lefrere. 2005. Comparison of hepatitis C virus NS5b and 5′ noncoding gene sequencing methods in a multicenter study. J. Clin. Microbiol. 43:733-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leb, V., M. Stocher, E. Valentine-Thon, G. Holzl, H. Kessler, H. Stekel, and J. Berg. 2004. Fully automated, internally controlled quantification of hepatitis B virus DNA by real-time PCR by use of the MagNA Pure LC and LightCycler instruments. J. Clin. Microbiol. 42:585-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, C. J., S. C. Lo, J. H. Kao, P. T. Tseng, M. Y. Lai, Y. H. Ni, S. H. Yeh, P. J. Chen, and D. S. Chen. 2006. Transmission of occult hepatitis B virus by transfusion to adult and pediatric recipients in Taiwan. J. Hepatol. 44:39-46. [DOI] [PubMed] [Google Scholar]
- 38.Locarnini, S., A. Hatzakis, J. Heathcote, E. B. Keeffe, T. J. Liang, D. Mutimer, J. M. Pawlotsky, and F. Zoulim. 2004. Management of antiviral resistance in patients with chronic hepatitis B. Antivir. Ther. 9:679-693. [PubMed] [Google Scholar]
- 39.Lopez, V. A., E. J. Bourne, M. W. Lutz, and L. D. Condreay. 2002. Assessment of the COBAS Amplicor HBV Monitor Test for quantitation of serum hepatitis B virus DNA levels. J. Clin. Microbiol. 40:1972-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcellin, P., C. Castelnau, M. Martinot-Peignoux, and N. Boyer. 2005. Natural history of hepatitis B. Minerva Gastroenterol. Dietol. 51:63-75. [PubMed] [Google Scholar]
- 41.Marin, I. J., M. Poljak, K. Seme, J. Meglic-Volkar, M. Maticic, G. Lesnicar, and V. Brinovec. 2001. Comparative evaluation of semiautomated COBAS AMPLICOR hepatitis B virus (HBV) monitor test and manual microwell plate-based AMPLICOR HBV MONITOR test. J. Clin. Microbiol. 39:758-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinot-Peignoux, M., N. Boyer, M. Colombat, R. Akremi, B. N. Pham, S. Ollivier, C. Castelnau, D. Valla, C. Degott, and P. Marcellin. 2002. Serum hepatitis B virus DNA levels and liver histology in inactive HBsAg carriers. J. Hepatol. 36:543-546. [DOI] [PubMed] [Google Scholar]
- 43.Noborg, U., A. Gusdal, E. K. Pisa, A. Hedrum, and M. Lindh. 1999. Automated quantitative analysis of hepatitis B virus DNA by using the Cobas Amplicor HBV monitor test. J. Clin. Microbiol. 37:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orito, E., M. Mizokami, H. Sakugawa, K. Michitaka, K. Ishikawa, T. Ichida, T. Okanoue, H. Yotsuyanagi, S. Iino, et al. 2001. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Hepatology 33:218-223. [DOI] [PubMed] [Google Scholar]
- 45.Osiowy, C., and E. Giles. 2003. Evaluation of the INNO-LiPA HBV genotyping assay for determination of hepatitis B virus genotype. J. Clin. Microbiol. 41:5473-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pawlotsky, J. M., A. Bastie, C. Hezode, I. Lonjon, F. Darthuy, J. Remire, and D. Dhumeaux. 2000. Routine detection and quantification of hepatitis B virus DNA in clinical laboratories: performance of three commercial assays. J. Virol. Methods 85:11-21. [DOI] [PubMed] [Google Scholar]
- 47.Perrillo, R. P. 2005. Current treatment of chronic hepatitis B: benefits and limitations. Semin. Liver Dis. 25(Suppl. 1):20-28. [DOI] [PubMed] [Google Scholar]
- 48.Phylip, F. J. 1989. Phylogeny Interference Package. Cladistics 5:164-166. [Google Scholar]
- 49.Pichoud, C., F. Berby, L. Stuyver, M. A. Petit, C. Trepo, and F. Zoulim. 2000. Persistence of viral replication after anti-HBe seroconversion during antiviral therapy for chronic hepatitis B. J. Hepatol. 32:307-316. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez-Tapias, J. M., J. Costa, A. Mas, M. Bruguera, and J. Rodes. 2002. Influence of hepatitis B virus genotype on the long-term outcome of chronic hepatitis B in western patients. Gastroenterology 123:1848-1856. [DOI] [PubMed] [Google Scholar]
- 51.Schaefer, S. 2005. Hepatitis B virus: significance of genotypes. J. Viral Hepat. 12:111-124. [DOI] [PubMed] [Google Scholar]
- 52.Seo, Y., S. Yoon, K. Hamano, M. Nakaji, Y. Yano, M. Katayama, T. Ninomiya, Y. Hayashi, and M. Kasuga. 2004. Early response to interferon alpha treatment and long-term clinical outcome in Japanese patients with chronic HBV genotype C infection. Int. J. Mol. Med. 13:75-79. [PubMed] [Google Scholar]
- 53.Simmonds, P., and S. Midgley. 2005. Recombination in the genesis and evolution of hepatitis B virus genotypes. J. Virol. 79:15467-15476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Rossau. 2000. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 81:67-74. [DOI] [PubMed] [Google Scholar]
- 55.Thakur, V., R. C. Guptan, S. N. Kazim, V. Malhotra, and S. K. Sarin. 2002. Profile, spectrum and significance of HBV genotypes in chronic liver disease patients in the Indian subcontinent. J. Gastroenterol. Hepatol. 17:165-170. [DOI] [PubMed] [Google Scholar]
- 56.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weber, B. 2005. Genetic variability of the S gene of hepatitis B virus: clinical and diagnostic impact. J. Clin. Virol. 32:102-112. [DOI] [PubMed] [Google Scholar]
- 58.Weiss, J., H. Wu, B. Farrenkopf, T. Schultz, G. Song, S. Shah, and J. Siegel. 2004. Real time TaqMan PCR detection and quantitation of HBV genotypes A-G with the use of an internal quantitation standard. J. Clin. Virol. 30: 86-93. [DOI] [PubMed] [Google Scholar]
- 59.Yuen, M. F., E. Sablon, Y. Tanaka, T. Kato, M. Mizokami, J. Doutreloigne, H. J. Yuan, D. K. Wong, S. M. Sum, and C. L. Lai. 2004. Epidemiological study of hepatitis B virus genotypes, core promoter and precore mutations of chronic hepatitis B infection in Hong Kong. J. Hepatol. 41:119-125. [DOI] [PubMed] [Google Scholar]