Abstract
Characterization of nosocomial methicillin-susceptible Staphylococcus aureus isolates from Cape Verde showed that (i) Panton-Valentine leukocidin genes were present in 35% of the isolates and (ii) half of the collection had the same genetic background as methicillin-resistant pandemic clones. Introduction of the staphylococcal chromosome cassette mec (SCCmec) into virulent and epidemic isolates could pose serious threats to public health.
Staphylococcus aureus constitutes a major public health threat, and methicillin-resistant S. aureus (MRSA) is currently the most commonly identified antibiotic-resistant pathogen in many parts of the world. Concerning the African continent, although there is still a lack of epidemiologic data, MRSA was reported as a problem in different countries, including South Africa (6), Zimbabwe (16), Kenya (13), Ethiopia (29), Egypt (9), Senegal (27), and Ivory Coast (4). In contrast, the prevalence of MRSA was very low or nonexistent in Somalia (23), Nigeria (1), and Tanzania (30).
In a previous study we determined the rates of nosocomial colonization by staphylococci in Cape Verde islands and the antimicrobial susceptibility profiles (3). The S. aureus carriage rates were around 41%, and all isolates were susceptible to methicillin (3). The aim of the present study was to obtain insights into the methicillin-susceptible S. aureus (MSSA) genetic population, including the detection of virulence determinants, in a country where at least in a particular period the prevalence of MRSA in colonization samples seemed to be null.
The MSSA isolates included in this work were collected during a surveillance study in two hospitals on the two most populated islands of the Cape Verde archipelago (3). Hospital Dr. Agostinho Neto is located in Praia, on São Tiago, and Hospital Baptista de Sousa is in Mindelo, on São Vicente. The isolates represented colonization samples from patients or health care workers and were recovered from nasal or wound swabs. Five isolates from the previous study were not viable when cultured from the frozen stocks and could not be included in the present work.
Pulsed-field gel electrophoresis (PFGE) was performed after SmaI digestion as described by Chung et al. (7). The resulting band patterns were analyzed by visual inspection followed by analysis with BioNumerics software (version 4.0; Applied Maths, Ghent, Belgium) for relatedness evaluation. Dendrograms were generated from similarity matrixes calculated with the Jaccard coefficient, and patterns were clustered by the unweighted pair group method with arithmetic mean using an optimization of 1.5% and a tolerance of 1.3%. Profiles with more than 70% similarity were considered closely related.
spa typing was performed as previously described (17, 26). The spa types were assigned through the Egenomics and Ridom (http://www.ridom.de/spaserver/) (12) web servers. spa types with similar repeat profiles were grouped together as part of the same lineage (spa lineage); lineages are identified by the same capital letter.
Multilocus sequence typing (MLST) was carried out as described by Enright et al. (10), with the exception that primer arcCF2 (5′-CCT TTA TTT GAT TCA CCA GCG-3′) was used (8). spa typing and MLST PCR products were purified with a Wizard PCR Preps purification system (Promega, Madison, WI) and used as templates for sequencing of both strands at Macrogen (Seoul, South Korea). MLST alleles and sequence types (STs) were identified using the MLST database (http://www.mlst.net), which was also used to search for earlier reports on the STs identified during the present study.
The presence of the Panton-Valentine leukocidin (PVL) and the exfoliative toxin D (ETD) genes, i.e., lukS-PV and lukF-PV and etd, respectively, was determined by PCR as described previously (15, 19).
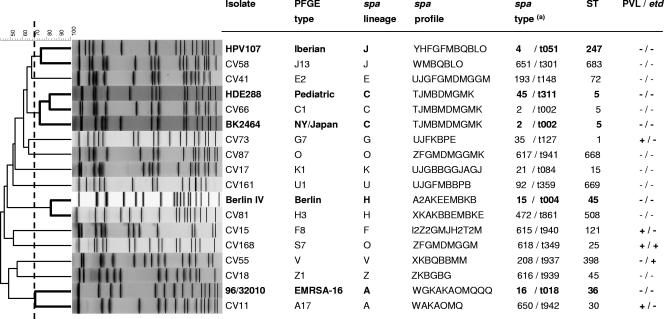
The characterization of 63 MSSA strains by PFGE resulted in the definition of 13 different genotypes (Fig. 1). Representatives of these 13 PFGE patterns (17 isolates) were characterized by spa typing and classified into 17 spa types and 12 spa lineages. MLST was performed on single isolates of each PFGE pattern and/or spa lineage identified (13 isolates). All PFGE types belonged to a different ST, confirming the existence of 13 distinct clones among the 63 MSSA isolates, whose main characteristics are shown in Fig. 1.
FIG. 1.
Molecular characterization of MSSA strains and comparison with MRSA pandemic clones. On the left is a dendrogram showing the estimated relationships of PFGE types based on Bionumerics analysis, including representatives of five international pandemic MRSA clones (in bold). (a) The spa type was assigned through both the Egenomics and the Ridom web servers.
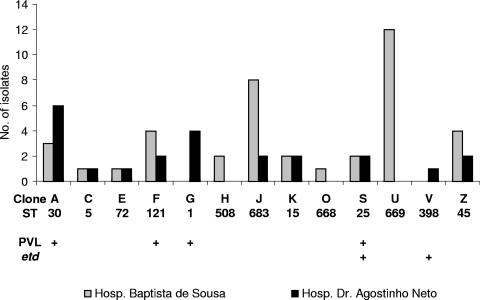
The genetic studies on the clonal population of the MSSA isolates in Cape Verde demonstrated that some clones were very successful. Clone U (spa type 92/t359 or related, ST 669) was represented by 19% of the isolates (n = 12), whereas the second and third most prevalent clones, clone J (spa type 651/t301 or related, ST 683) and clone A (spa type 650/t942 or related, ST 30), had incidences of 16% (n = 10) and 14% (n = 9), respectively. In addition, some of these clones have the capacity to disseminate between islands: 8 of the 13 clones occurred in both hospitals (Fig. 2). However, the most successful clone, clone U, was identified in HBS only.
FIG. 2.
Clonal distribution between Hospital Baptista de Sousa (Praia, São Tiago) and Hospital Dr. Agostinho Neto (Mindelo, São Vicente).
Although no MRSA strains have been found in Cape Verde, about half of the isolates (46%, n = 29) showed a genetic background identical (same ST) or very similar (single-locus variant [SLV] or double-locus variant [DLV]) to the genotypes of pandemic MRSA clones. For instance, clone J (ST 683) is a DLV of the Iberian MRSA (ST 247), clone A (ST 30) is an SLV of EMRSA-16 (ST 36), and clone C (ST 5) has a background identical to the New York/Japan and Pediatric (ST 5) clones. Moreover, clone H (ST 508) is an SLV of and clone Z (ST 45) shares the same ST as the Berlin clone (ST 45) (Fig. 1). The fact that some of the major MSSA clones established in Cape Verde correspond to internationally successful MRSA clones provides evidence that the emergence of MRSA isolates in an environment is not exclusively dependent on the presence of successful MSSA lineages. A similar observation was recently published from Portugal (2).
Although most MSSA clones in Cape Verde are recognized as international clonal types, three STs found in the present study have not been described before (STs 668, 669, and 683). Interestingly, ST 398, which was detected in a single isolate in the present study, was recently reported to be resistant to digestion with SmaI restriction endonuclease (33) and seems to be associated with pigs and pig farmers (5, 33). Whether the isolate from Cape Verde was pig related is unknown. However, there were no signs of resistance to SmaI digestion, and the isolate was classified as PFGE type V.
A total of 22 (35%) and 5 (8%) isolates tested positive by PCR amplification for the PVL and ETD genes, respectively. All but one isolate belonging to STs 30 (n = 8), 121 (n = 6), 1 (n = 4), and 25 (n = 4) were PVL positive, whereas the five ETD-positive isolates belonged to ST 25 (n = 4) and ST 398 (n = 1). Out of the 22 PVL-positive isolates that were almost equally distributed among patients and health care workers, four were recovered from wound infections, whereas 18 were obtained from nasal samples. We found a proportion of PVL-positive MSSA isolates that was fairly high (35%) compared with data in studies from other countries. Melles et al. (21) found a PVL prevalence of only 0.6% in a large (n = 829) Dutch collection of carriage isolates, whereas Kuehnert et al. (18) reported the presence of PVL in 1% of 297 American MSSA nasal isolates, and Holmes et al. (14) found only 8 PVL-positive isolates (1.6%) among 515 isolates of S. aureus from different sites of infection. An additional study demonstrated that PVL genes were very rare in Germany in both nasal and blood isolates (32). Moreover, the PVL gene has been reported to be more common in MRSA isolates than in MSSA isolates (18, 20), particularly among community-acquired MRSA strains of STs 1, 8, 30, 59, 80, and 93 (31).
In the present study, the PVL genes were detected in all isolates belonging to clones A-ST 30 (with the exception of one isolate), F-ST 121, G-ST 1, and S-ST 25, whereas etd was present in all isolates of clones S and V-ST 398. A previous study comparing the genetic backgrounds of MSSA and MRSA in Portugal showed some MSSA clonal overlapping between Portugal and Cape Verde. Although the four PVL-positive clonal types detected in Cape Verde had been identified among the Portuguese MSSA nosocomial and/or community population, the cytotoxin was found exclusively in clone F-ST 121 and in a very restricted number of Portuguese isolates (2 out of 19 isolates) (2). Clones ST 1 and ST 30 PVL+ have been described as the most frequent community-acquired MRSA clones in the United States and Oceania, respectively (31). ST 121 has been detected among isolates carrying PVL genes in England and Wales (14), and ST 25 was recently reported as a highly transmissible clone associated with colonization and infection in the niche of AIDS-infected patients with a history of drug use (11). It is known that the genes that encode PVL are carried by different temperate phages and that S. aureus strains are usually lysogenized by a phage(s) (22). Our results suggest that hospitals in Cape Verde may represent a reservoir of PVL-positive clones. However, the reason why MSSA strains from Cape Verde seem to be more prone to PVL-converting phage infection is currently unclear. This study indicates that the acquisition of virulence might confer a greater advantage to S. aureus than antibiotic resistance genes in a country and period in which the use of antibiotics is very limited.
Interestingly, clone A is closely related to phage type 80/81 strains, which became pandemic during the 1950s, causing a high frequency of skin lesions, sepsis, and pneumonia in children and young adults in hospitals and the community (25). The phage type 80/81 clone is ST 30, carries the PVL genes, and has been suggested to be the ancestor of the community-acquired PVL-positive southwest Pacific MRSA clone (ST 30, SCCmec IV) (24). On the other hand, Taneike et al. recently reported that the MRSA isolates that caused nosocomial outbreaks in Japan in the 1980s were ST 30 and PVL positive (28). This clone was replaced by the PVL-negative New York/Japan MRSA ST 5 clone in the 1990s and now seems to be re-emerging in the community after having lost some virulence genes (28).
In summary, the genetic backgrounds corresponding to five pandemic MRSA clones were represented among the MSSA isolates from Cape Verde, and the genes that code for PVL were detected in a high proportion of strains with different genetic backgrounds. Although a fitness cost associated with the maintenance of both SCCmec and the PVL genes may exist, we should be aware of the eventual emergence of MRSA strains by the introduction of SCCmec into highly virulent and epidemic MSSA backgrounds. This could pose an additional threat to public health, warranting continuing surveillance studies, namely in African countries.
Acknowledgments
This work was supported by project FCG 61052 from Fundação Calouste Gulbenkian and project POCTI/SAU-ESP/57841/2004 from Fundação para a Ciência e Tecnologia, Portugal, awarded to H. de Lencastre. T. Conceição was supported by grant SFRH/BD/21424/2005 from Fundação para a Ciência e Tecnologia.
We thank Barry N. Kreiswirth and Steve Naidich for the new spa type assignments (spa types 615 to 619, 650, and 651). This study made use of the Multi Locus Sequence Typing website (http://www.mlst.net), which is hosted at Imperial College, London, United Kingdom.
REFERENCES
- 1.Adesida, S., H. Boelens, B. Babajide, A. Kehinde, S. Snijders, W. van Leeuwen, A. Coker, H. Verbrugh, and A. van Belkum. 2005. Major epidemic clones of Staphylococcus aureus in Nigeria. Microb. Drug Resist. 11:115-121. [DOI] [PubMed] [Google Scholar]
- 2.Aires de Sousa, M., T. Conceição, C. Simas, and H. de Lencastre. 2005. Comparison of genetic backgrounds of methicillin-resistant and -susceptible Staphylococcus aureus isolates from Portuguese hospitals and the community. J. Clin. Microbiol. 43:5150-5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires de Sousa, M., I. Santos Sanches, M. L. Ferro, and H. de Lencastre. 2000. Epidemiological study of staphylococcal colonization and cross-infection in two West African Hospitals. Microb. Drug Resist. 6:133-141. [DOI] [PubMed] [Google Scholar]
- 4.Akoua-Koffi, C., N. Guessennd, V. Gbonon, H. Faye-Kette, and M. Dosso. 2004. Methicillin-resistance of Staphylococcus aureus in Abidjan (1998-2001): a new hospital problem. Med. Mal. Infect. 34:132-136. [DOI] [PubMed] [Google Scholar]
- 5.Armand-Lefevre, L., R. Ruimy, and A. Andremont. 2005. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg. Infect. Dis. 11:711-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bell, J. M., and J. D. Turnidge. 2002. High prevalence of oxacillin-resistant Staphylococcus aureus isolates from hospitalized patients in Asia-Pacific and South Africa: results from SENTRY antimicrobial surveillance program, 1998-1999. Antimicrob. Agents Chemother. 46:879-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, M., H. de Lencastre, P. Matthews, A. Tomasz, I. Adamsson, M. Aires de Sousa, T. Camou, C. Cocuzza, A. Corso, I. Couto, A. Dominguez, M. Gniadkowski, R. Goering, A. Gomes, K. Kikuchi, A. Marchese, R. Mato, O. Melter, D. Oliveira, R. Palacio, R. Sá-Leão, I. Santos Sanches, J. H. Song, P. T. Tassios, and P. Villari. 2000. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb. Drug Resist. 6:189-198. [DOI] [PubMed] [Google Scholar]
- 8.Crisóstomo, M. I., H. Westh, A. Tomasz, M. Chung, D. C. Oliveira, and H. de Lencastre. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. USA 98:9865-9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Kholy, A., H. Baseem, G. S. Hall, G. W. Procop, and D. L. Longworth. 2003. Antimicrobial resistance in Cairo, Egypt 1999-2000: a survey of five hospitals. J. Antimicrob. Chemother. 51:625-630. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon, R. J., B. Quagliarello, C. Cespedes, M. Chung, H. de Lencastre, P. Vavagiakis, M. Miller, B. Zeller, and F. D. Lowy. 2005. A molecular epidemiological analysis of 2 Staphylococcus aureus clonal types colonizing and infecting patients with AIDS. Clin. Infect. Dis. 40:1028-1036. [DOI] [PubMed] [Google Scholar]
- 12.Harmsen, D., H. Claus, W. Witte, J. Rothganger, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayanga, A., A. Okello, R. Hussein, and A. Nyong'o. 1997. Experience with methicillin resistant Staphylococcus aureus at the Nairobi Hospital. East Afr. Med. J. 74:203-204. [PubMed] [Google Scholar]
- 14.Holmes, A., M. Ganner, S. McGuane, T. L. Pitt, B. D. Cookson, and A. M. Kearns. 2005. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J. Clin. Microbiol. 43:2384-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klugman, K. P. 1998. Emerging infectious diseases—South Africa. Emerg. Infect. Dis. 4:517-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuehnert, M. J., D. Kruszon-Moran, H. A. Hill, G. McQuillan, S. K. McAllister, G. Fosheim, L. K. McDougal, J. Chaitram, B. Jensen, S. K. Fridkin, G. Killgore, and F. C. Tenover. 2006. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. J. Infect. Dis. 193:172-179. [DOI] [PubMed] [Google Scholar]
- 19.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Aguilar, G., A. Avalos-Mishaan, K. Hulten, W. Hammerman, E. O. Mason, Jr., and S. L. Kaplan. 2004. Community-acquired, methicillin-resistant and methicillin-susceptible Staphylococcus aureus musculoskeletal infections in children. Pediatr. Infect. Dis. J. 23:701-706. [DOI] [PubMed] [Google Scholar]
- 21.Melles, D. C., R. F. Gorkink, H. A. Boelens, S. V. Snijders, J. K. Peeters, M. J. Moorhouse, P. J. van der Spek, W. B. van Leeuwen, G. Simons, H. A. Verbrugh, and A. van Belkum. 2004. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J. Clin. Investig. 114:1732-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narita, S., J. Kaneko, J. Chiba, Y. Piemont, S. Jarraud, J. Etienne, and Y. Kamio. 2001. Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, phiSLT. Gene 268:195-206. [DOI] [PubMed] [Google Scholar]
- 23.Nur, Y. A., M. Vandenberg, M. Yussuf, A. van Belkum, and H. Verbrugh. 1996. Nasal carriage of multiresistant Staphylococcus aureus among healthcare workers and pediatric patients in two hospitals in Mogadishu, Somalia. Int. J. Infect. Dis. 1:186-191. [Google Scholar]
- 24.Robinson, D. A., A. M. Kearns, A. Holmes, D. Morrison, H. Grundmann, G. Edwards, F. G. O'Brien, F. C. Tenover, L. K. McDougal, A. B. Monk, and M. C. Enright. 2005. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired methicillin-resistant clone. Lancet 365:1256-1258. [DOI] [PubMed] [Google Scholar]
- 25.Rountree, P. M., and M. A. Beard. 1958. Further observations on infection with phage type 80 staphylococci in Australia. Med. J. Aust. 45:789-795. [PubMed] [Google Scholar]
- 26.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sow, A. I., A. Wade, M. A. Faye-Niang, M. Seydi, C. S. Boye, M. Soumare, M. Gaye, N. M. Dia, and M. F. Cisse. 1998. Methicillin-resistant Staphylococcus aureus in Dakar. Med. Trop. 58:155-157. [PubMed] [Google Scholar]
- 28.Taneike, I., T. Otsuka, S. Dohmae, K. Saito, K. Ozaki, M. Takano, W. Higuchi, T. Takano, and T. Yamamoto. 2006. Molecular nature of methicillin-resistant Staphylococcus aureus derived from explosive nosocomial outbreaks of the 1980s in Japan. FEBS Lett. 580:2323-2334. [DOI] [PubMed] [Google Scholar]
- 29.Tenssay, Z. W. 2000. Staphylococci: frequency of isolation and antibiotic susceptibility patterns in Jimma Hospital, south-west Ethiopia. Ethiopian Med. J. 38:175-184. [PubMed] [Google Scholar]
- 30.Urassa, W. K., E. A. Haule, C. Kagoma, and N. Langeland. 1999. Antimicrobial susceptibility of Staphylococcus aureus strains at Muhimbili Medical Centre, Tanzania. East Afr. Med. J. 76:693-695. [PubMed] [Google Scholar]
- 31.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Eiff, C., A. W. Friedrich, G. Peters, and K. Becker. 2004. Prevalence of genes encoding for members of the staphylococcal leukotoxin family among clinical isolates of Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 49:157-162. [DOI] [PubMed] [Google Scholar]
- 33.Voss, A., F. Loeffen, J. Bakker, C. Klaassen, and M. Wulf. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11:1965-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]