Abstract
Community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) has emerged as a major public health problem in Australia, as in many other parts of the world. High rates of CA-MRSA skin and soft tissue infection have been reported from Aboriginal communities. We used a single-nucleotide polymorphism (SNP) genotyping typing system based on the multilocus sequence type (MLST) database to investigate the epidemiology of CA-MRSA and methicillin-sensitive S. aureus (MSSA) over a 12-month period in three remote Aboriginal communities of Northern Australia. This was supplemented by real-time PCR for Panton-Valentine leukocidin (PVL) genes, staphylococcal cassette chromosome mec (SCCmec) typing, and antimicrobial susceptibility testing. S. aureus was recovered from pyoderma lesions on 221 occasions and throat swabs on 44 occasions. The median monthly recovery rate of S. aureus from skin sores was 58% (interquartile range, 62 to 78%), and there was no seasonal variation. Twenty-three percent of isolates were CA-MRSA; the proportion was similar across the communities and did not vary over the study period. Erythromycin resistance was found in 47% of CA-MRSA and 21% of MSSA. SNP-based typing identified 14 different clonal complexes (cc); however, cc75 was predominant, accounting for 71% of CA-MRSA isolates. These were confirmed as ST75-like by using an additional SNP and MLST of selected isolates. All but one of the cc75 isolates had SSCmec type IV (one had type V), and all were PVL negative. Monthly tracking of SNP-based cc types showed a highly dynamic process. ST75-MRSA-IV appears to be unique to the region and probably evolved de novo in remote Aboriginal communities.
Methicillin-resistant Staphylococcus aureus (MRSA) was first recognized as an important nosocomial pathogen in Australia in the 1970s (34) and remains a major cause of healthcare-related infection worldwide (2). More recently, community-acquired MRSA (CA-MRSA) has emerged as a public health concern in Australia and elsewhere (9, 14). Classical healthcare-associated strains (HA-MRSA) tend to be multidrug resistant and have staphylococcal chromosomal mec (SCCmec) type I, II, or III, whereas CA-MRSA strains are usually not multidrug resistant and possess the smaller and potentially more mobile SCCmec type IV or V (1, 22). The genetic background of CA-MRSA usually reflects local genotypes (42), although some successful clones can rapidly spread across continents and even across the globe (27). Most strains of HA-MRSA fall into five broad lineages that are responsible for the majority of infections worldwide (38). Infections due to CA-MRSA can be seen in hospital settings, with occasional major outbreaks (4, 7), while HA-MRSA strains are also able to circulate within communities (32).
Methicillin resistance in the community setting probably arises de novo and is, for the most part, independent of the hospital reservoir. Methicillin-sensitive strains are likely to acquire the mecA gene from other sources, such as coagulase-negative staphylococci (38). Evidence suggests that this happens more frequently than previously thought (13) and is more likely in populations with substantial organism burdens associated with domestic crowding, often with frequent staphylococcal skin infection. High rates of CA-MRSA infection have been reported in people from Aboriginal communities in both North America and Australia (16, 24, 39, 43), with a reported spectrum of disease from mild to life-threatening (29). Many strains of CA-MRSA also have the virulence factor Panton-Valentine leukocidin (PVL), which has been associated with skin and soft tissue infection as well as necrotizing pneumonia and septic shock (8, 35, 42). In Australia, the presence of the PVL determinant is uncommon in Western Australian CA-MRSA clones but is found in those derived from the southwest Pacific and southern Queensland (28, 32).
The epidemiology of CA-MRSA and methicillin-sensitive Staphylococcus aureus (MSSA) in defined communities has been investigated to some extent, but most reports are hospital based or laboratory based and there are few community-based longitudinal studies. There has also been a lack of effective and standardized typing methods. For example, phenotyping using antimicrobial susceptibility patterns (antibiogram) has limitations, not the least of which is the mobility of resistance genes. Phage typing and pulsed-field gel electrophoresis (PFGE) are useful for investigating hospital outbreaks and have been used in longitudinal studies, but they do not lend themselves to the study of the genetic evolution of CA-MRSA. PFGE also has difficulties in achieving interlaboratory agreement. Multilocus sequence typing (MLST) uses recognized sequences in seven relatively stable housekeeping genes and is ideal for this purpose (12), especially when combined with typing of SCCmec. A major drawback is the workload and expense of MLST, making it available to only well-resourced laboratories.
An alternative approach is to use analysis of a specific set of single-nucleotide polymorphisms (SNPs) derived from an MLST database. This method has a high resolving power that can determine clonal complexes (cc) corresponding to the major staphylococcal lineages (40). When SNP-based typing by real-time PCR is combined with determination of specific virulence genes, such as those for PVL, it is both cost-effective and relatively rapid (37).
Previously, Robertson et al. (37) described a method that used the “Minimum SNPs” software to derive highly informative SNPs from DNA sequence alignments. We used SNP-based MLST clonal complex typing to explore the epidemiology of MRSA and MSSA over a 12-month period in remote Aboriginal communities of the Northern Territory (NT), Australia. This was combined with real-time PCR for PVL determinants, SCCmec typing of selected isolates, and antimicrobial susceptibility testing. We specifically set out to document the prevalence of CA-MRSA in these communities and to characterize changes in the S. aureus population over time. To our knowledge, it is the first time such a longitudinal study has been reported and the first time this technology has been used for community epidemiological investigation.
MATERIALS AND METHODS
Study sites.
The study was conducted in three large remote communities in the tropical Top End of the NT and was part of a larger prospective study of skin and throat infection in relation to acute rheumatic fever (25). The approximate community populations were 2,500 (community 1), 800 (community 2), and 1,800 (community 3). Approval was granted by the regional ethics committee prior to community consultation. Informed consent was obtained from the occupants of 49 study households. We then visited households on a monthly basis to conduct surveillance for pyoderma, pharyngitis, and acute rheumatic fever. Throat swabs were taken from every person, those with and without symptoms of pharyngitis, and skin sore swabs (maximum of three) were taken from all adults and children with pyoderma. Additional demographic information was collected, including past history of acute rheumatic fever/rheumatic heart disease, general skin health, and household size (number of people per bedroom); a detailed analysis of this information is contained in another paper (26).
Recovering S. aureus isolates.
During the period May 2004 to June 2005, S. aureus isolates were specifically identified and characterized from swabs of all people with pyoderma. However, S. aureus isolates were only identified and characterized from throat swabs taken over a 2-month period (June and July 2004). Swabs were struck out on horse blood agar culture plates (Oxoid) in the community health centers or sealed in airtight bags with desiccant for transportation back to the laboratory in Darwin, Australia, by aircraft (25). Plates were incubated at 37°C and inspected at 24 h and 48 h. S. aureus was identified by colonial morphology and a latex slide agglutination test for detection of clumping factor, protein A, and specific capsular polysaccharides (Staphytect Plus; Oxoid). One representative colony per plate was stored in glycerol for further analysis.
Antimicrobial susceptibility testing.
Susceptibility testing was performed by disk diffusion using CLSI (formerly NCCLS) methodology (31). Resistance was determined by CLSI susceptibility breakpoints (3), and plates were read with an optical analyzer system (Biomic vision; Giles Scientific, Inc., California). A disk approximation test was used to detect inducible clindamycin resistance (3). S. aureus isolates that were resistant to cefoxitin by disk diffusion were tested for the presence of specific penicillin binding protein (PBP2) using a latex agglutination test (Oxoid).
DNA extraction.
Isolates were plated from glycerol stocks onto horse blood agar plates and incubated at 37°C overnight. A single colony was picked into 5 ml of Todd-Hewitt broth (Oxoid) supplemented with yeast and grown at 37°C overnight with agitation. DNA was extracted from 1 ml of pelleted bacterial cells using a QIAGEN QIAamp DNA mini kit according to the manufacturer's instructions, using 100 μg/ml lysostaphin (Sigma). The DNA was eluted in 200 μl of 10 mM Tris, 0.5 mM EDTA, pH 9, and stored at −20°C.
Clonal complex, nucA, and PVL determination.
MLST-derived clonal complexes using SNP sets were determined using a single-tube kinetic PCR method as described by Huygens et al. (17). The primers used in this study are shown in Table 1. A kinetic PCR uses the SNP at the 3′ end of the primer and ascertains the rate of appearance of the reaction product. SNP tpi36 was included as an internal control and used to calculate the change in cycle threshold.
TABLE 1.
Primers used for SNP typing, PVL and NucA gene detection, and for SCCmec typing
| Description | Primer name | Nucleotide sequence (5′-3′) | Reference |
|---|---|---|---|
| SNP primers | |||
| arc162 | arcC162_99FL | CAGGGTATGATAGGCTATTGGTTG | 17 |
| arcC162cAR | GATCATCTTTATCTACTTCCACACGTGCT | 17 | |
| arcC210 | arcC210_99FL | CAGGGTATGATAGGCTATTGGTTG | 17 |
| arcC210RCC | CGTATAAAAAGGACCAATTGGTCTG | 37 | |
| gmk318 | gmk318F | TGCGTGAAGGTGAAGTTGATG | 17 |
| gmk318aTR | ACCTACTAATCGCTCTCTCAAGTAA | 17 | |
| pta294 | pta294_411RL | CCTTGTGAATCAAGTTCTGGATTG | 17 |
| pta294aAF | TGCAGCACATTCAACAGAA | 17 | |
| pta383 | pta383_142FL | CTGCGACAAGTGAATTGAAAGCTG | 17 |
| pta383CT | TTGCACAATCACCAAAGATGTATTGCA | 17 | |
| tpi36 | tpi36_206RL | TTGATGATTTACCAGTTCCGATTG | 17 |
| tpi36cCF | GATGAAGAAATTAACAAAAAAGCGCCC | 17 | |
| tpi36cTF | GATGAAGAAATTAACAAAAAAGCGCCT | 17 | |
| tpi241/3 | tpi_365RL | GCCCCATCAATATCAGTTTGTG | 17 |
| tpi241 + 243GTA | GTAAATCATCAACATCTGAAGATGTA | 17 | |
| aroE252 | aroE 252GF | GGTATAATACAGATGGTATCGGTTATGTG | 17 |
| aroE 252GR | ACCTGCGCCCAAAATTAAAA | 17 | |
| gmk159G | gmk159GF | GATGATCAATTTATAGAATATGCCGAG | This study |
| gmk159GR | TCGTCCATAGTATCTTTAACATATTGAACTG | This study | |
| PVL gene | PVLlukFor | TATCTCTAACGGCTTGTCAGGTG | 17 |
| PVLlukRev | TGCTTCAACATCCCAACCAA | 17 | |
| NucA gene | nucAFor | GCGATTGATGGTGATACGGTT | 23 |
| nucARev | AGCCAAGCCTTGACGAACTAAAGC | 23 | |
| CCR gene complex | |||
| Types 1, 2, 3 | βc | ATTGCCTTGATAATAGCCITCT | 33 |
| αc | ATCTATTTCAAAAATGAACCA | 33 | |
| Type 2 | α2 (used with βc) | TAAAGGCATCAATGCACAAACACT | 33 |
| ccrC gene | γF | CGTCTATTACAAGATGTTAAGGATAAT | 19 |
| γR | CCTTTATAGACTGGATTATTCAAAATAT | 19 | |
| mec gene complex | |||
| mecR1 (MS domain) | mcR4 | GTCGTTCATTAAGATATGACG | 33 |
| mcR3 | GTCTCCACGTTAATTCCATT | 33 | |
| mecA | mA1 | TGCTATCCACCCTCAAACAGG | 33 |
| mA2 | AACGTTGTAACCACCCCAAGA | 33 | |
| Class B | IS5 | AACGCCACTCATAACATATGGAA | 33 |
| mA6 | TATACCAAACCCGACAAC | 33 | |
| Class C | IS2 (used with mA2) | TGAGGTTATTCAGATATTTCGATGT | 33 |
Real-time PCR was carried out using a Corbett Rotorgene 2000. Reaction mixtures contained 1 μl of a 1:10 dilution of extracted DNA, 1× SYBR green PCR MasterMix (Applied Biosystems), and 0.4 μM concentrations of each primer in a total volume of 10 μl. Cycling conditions consisted of 50°C for 1 min and 95°C for 2 min, followed by 40 cycles of 95°C for 5 s, 56°C for 20 s, and 72°C for 35 s. Isolates assigned to cc93 were typed with an additional SNP (aroE252) to determine if they were of sequence type 93 (ST93). In addition, all of the isolates assigned to cc75 were typed with an additional SNP, gmk159G, to determine their relatedness to ST75. Real-time PCR was also used to verify the identity of S. aureus isolates by confirming the presence of the nucA gene (23) and to check for the presence of PVL determinants (40).
Identification of SCCmec type IV and V.
The presence of the MS domain of the mecR1 gene, present in mec gene complexes of type A, B, C1, or D, was determined using primers mcR3 and mcR4. Isolates positive for this region were tested for a class B mec gene complex using the primers IS5 and mA6. Isolates negative for this region were tested with the primers mA1 and mA2 to confirm the presence of the mecA gene and primers mA2 and IS2 for detection of a class C2 mec gene complex. Isolates were also tested for the presence of a type 1, 2, or 3 ccr gene complex using the primers βc and αc and then specifically for a type 2 ccr gene complex using primers βc and α2. Isolates that did not have a type 1, 2, or 3 ccr gene complex were tested with primers specific for the ccrC gene using primers γF and γR (19).
SCCmec typing amplification was performed using 1 μl of a 1:10 dilution of template DNA in a 25-μl reaction mixture containing 1× PCR buffer, 1 μl of each primer, 200 μM deoxynucleoside triphosphates, and 1 U QIAGEN Taq polymerase. Cycling for all primers consisted of an initial denaturation of 94°C for 1 min followed by 35 cycles at 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min.
MLST of specific isolates.
MLST was performed on five selected isolates as specified by Enright et al. (11). The sequences obtained were compared with sequences at the MLST web site at http://www.mlst.net to assign an ST.
Analysis.
Laboratory data were compared to the MLST database (http://saureus.mlst.net) using a specific computer program (18, 40). Epidemiological data were analyzed using Stata 9 (Stata Corporation, College Station, Texas). Differences in proportions were compared with chi-square analysis.
RESULTS
S. aureus and pyoderma.
Over the study period, there were 429 household visits, mostly in community 1 and community 3. There were logistic problems maintaining the project in community 2, and surveillance was ceased after only 28 household visits. Skin infection was common, and 38% of children <15 years had pyoderma on at least one occasion. S. aureus was recovered from skin sores in 221 of 375 children with pyoderma (59%) and was found in company with group A beta-hemolytic streptococcus on 110 occasions (29%). The median monthly recovery rate of S. aureus from skin sore swabs was 58% (interquartile range, 62 to 78%), and there was no significant difference in recovery rates between the wet season (November to April) and dry season (May to October). On 21 occasions, S. aureus was recovered from multiple pyoderma lesions from a child on the same day. There were 44 additional isolates of S. aureus from 513 throat swabs (8.6%) taken over a 2-month period; these were presumably related to nasal carriage.
Antibiotic resistance patterns.
PBP2 (methicillin resistance) was detected in 23% (66 of 283) of isolates (Table 2): 22% (40/184) in community 1, 30% (3/10) in community 2, and 26% (23/89) in community 3. The rates did not vary substantially over the period of the study. PBP2 was detected in 21% (51 of 239) of skin isolates and 34% (15 of 44) of throat isolates. Six isolates (2%) were susceptible to all the antibiotics in the panel, including penicillin. A further 164 isolates (58%) were resistant to penicillin alone, and the remaining 113 (40%) were resistant to multiple agents (Fig. 1). Erythromycin resistance was detected in 47% (31/66) of methicillin-resistant isolates and 21% (46/217) of methicillin-sensitive isolates. Likewise, 44% (29/66) of methicillin-resistant isolates were resistant to clindamycin, as were 19% (41/217) of methicillin-sensitive isolates. Thirty-three of the MRSA isolates (50%) were resistant only to beta-lactam antibiotics. The antibiogram showed no cotrimoxazole-, tetracycline-, gentamicin-, or rifampin-resistant isolates, a pattern typical of CA-MRSA. The throat MRSA carriage rate of 3% (15 of 513) in this study probably reflected a substantially higher nasal carriage rate.
TABLE 2.
Antibiotic susceptibility profile of 283 community isolates of S. aureus
| Antimicrobial agent | No. (%) of isolates resistant |
|---|---|
| Penicillin | 277 (98) |
| Methicillin (PBP2 positive) | 66 (23) |
| Erythromycin | 77 (27) |
| Clindamycin | 70 (25) |
| Cotrimoxazole | 0 (0) |
| Chloramphenicol | 6 (2) |
| Gentamicin | 0 (0) |
| Ciprofloxacin | 2 (1) |
| Rifampin | 0 (0) |
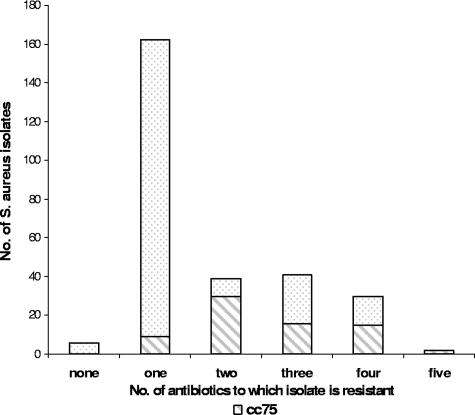
FIG. 1.
Antibiotic resistance profile of 283 S. aureus isolates showing that isolates of cc75 tend to be resistant to more antibiotics than the total pool.
SNP typing and presence of PVL.
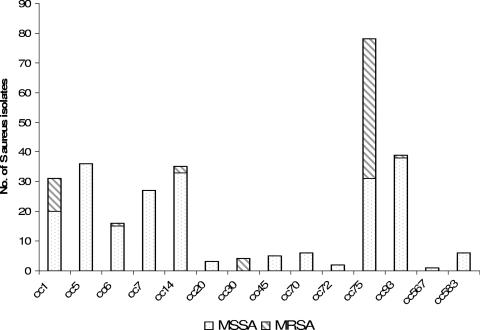
A total of 283 isolates were SNP typed and assigned to 14 different clonal complexes (Fig. 2). A unique and dominant clonal complex was initially identified as cc49 because of highly aberrant alleles. These isolates were subsequently confirmed as belonging to ST75 using data from an additional SNP (gmk159G) and 5 of the isolates that were typed by MLST. This clonal complex accounted for 25% (72 of 283) of all S. aureus isolates and 71% of CA-MRSA (47 of 66) isolates. The cc75 isolates were found in communities 1 (20% of isolates) and 3 (38% of isolates) but were not concentrated in particular households. We demonstrated that cc75 was resistant to more antibiotics on the panel than other clonal complexes (Fig. 1). Thirty-four of 72 cc75 isolates (47%) were resistant to erythromycin, compared to 43 of the remaining 211 staphylococcal isolates (20%) (P = <0.0001).
FIG. 2.
Distribution of SNP-determined clonal complexes and methicillin resistance in 283 isolates of S. aureus from remote communities.
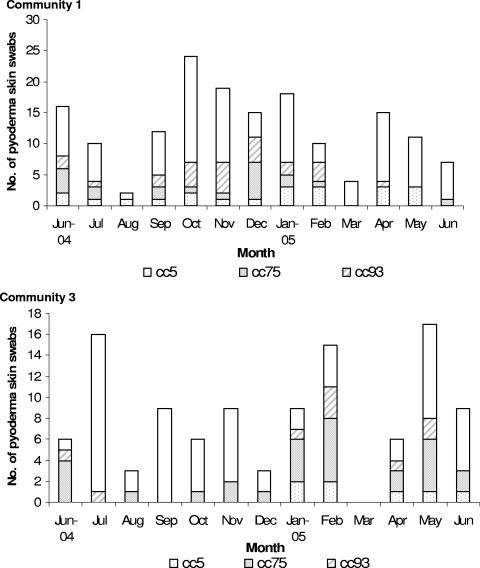
Thirty-nine isolates (14%) belonged to cc93, and typing with an additional SNP showed them to be ST93. A single cc93 isolate was methicillin resistant, indicating that it was ST93-MRSA-IV, also known as the Queensland clone. Most of the remaining CA-MRSA isolates belonged to cc1, the most important member being ST1-MRSA-IV or WA MRSA-1, or to cc30. ST30-MRSA-IV is the southwest Pacific clone (1). Monthly fluctuations of the three most common clonal complexes recovered from pyoderma lesions in communities 1 and 3 are shown in Fig. 3. It can be seen that the month-to-month prevalence of pyoderma is variable and organisms of specific lineages ebb and flow in each community as part of a highly dynamic process. Using the SNP typing data, it was also shown that on only 4 occasions, organisms of the same clonal complex were recovered from two or more different lesions on the same person; while on 15 occasions, they were from different clonal complexes. One child had isolates of cc6 (MSSA), cc45 (MSSA), and cc75 (MRSA) on three separate, but concurrent, skin sores.
FIG. 3.
Monthly fluctuations of pyoderma and skin swabs in communities 1 and 3 showing the three most common S aureus clonal complexes, as determined by SNP sets. Community visits were restricted in August 2004 and March 2005 for ceremonial reasons.
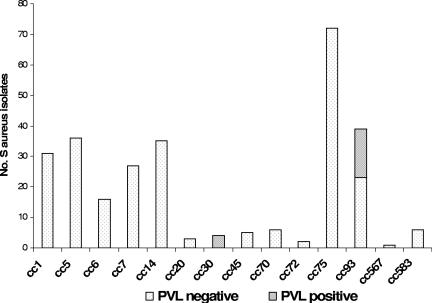
The genes for PVL were found in organisms of only two clonal complexes: the four cc30 isolates and 14 (41%) of the cc93 isolates. None of the cc75 isolates were positive for PVL (Fig. 4). Of the skin isolates, 19 (8%) were PVL positive, compared to 1 of 44 throat isolates (2%), although the difference was not statistically significant (P = 0.16).
FIG. 4.
Distribution of PVL genes within the clonal complexes.
SCCmec allotyping.
CA-MRSA usually carries type IV SCCmec that consists of a class B mec gene complex and a type 2 ccr gene complex (33). CA-MRSA isolates assigned to cc75 were tested by PCR for products indicative of a class B mec gene complex and a type 2 ccr gene complex. All but one was found to have SCCmec type IV. No product was obtained for the MS domain of the mecR1 gene in the remaining isolate, indicating that it was likely to contain a class C2 mec gene complex, typical of type V SCCmec. This was established with primers specific for class C2. The isolate also failed to provide a product indicative of a ccr gene complex of types 1, 2, and 3. However, it was shown to contain a ccrC gene, using the primers γF and γR, confirming it as having SCCmec type V.
MLST results.
Five isolates were selected from the most common SNP profile group. These isolates were found to closely match ST75 on the MLST database, with variations in 2 of the 7 genes. Variations in allele sequences were found for arcC and pta loci in four of five isolates. MLST sequencing of these isolates proved to be problematic because alleles at the aroE, glpF, and yqiL loci were far removed from other known alleles at these loci. ST-75 appears to be phylogenetically remote from other S. aureus isolates. The great majority of S. aureus MLST alleles are 97% similar or greater. However, the similarity of ST-75 alleles with other alleles ranges from 90 to 95%. The only exception to this is the aroE3 allele. This is common in the database and very similar to other alleles. Our current model is that ST-75 diverged from other S. aureus clones much earlier than the major clones diverged from each other, but there has been one recent horizontal gene transfer event that resulted in the common aroE3 allele being introduced into the clone.
DISCUSSION
Most CA-MRSA infections that occur in communities are skin and soft tissue infections; occasionally they can be severe, resulting in necrotizing pneumonia and septicemia (29, 35). Endemic CA-MRSA also has the potential for repeated introduction into the healthcare setting (7, 21). To our knowledge, there have been no previously published longitudinal studies of methicillin resistance in CA-MSSA infections in Australian Aboriginal or even non-Aboriginal communities. The findings are disquieting, with 23% of the isolates being methicillin resistant, and have important implications for future antibiotic prescribing guidelines in the region and across tropical Australia. Likewise, the high rates of resistance to erythromycin and clindamycin give little cause for comfort.
Reports from Western Australian more than 20 years ago were among the first in the world to raise the alarm about CA-MRSA (41). These Western Australian strains have subsequently spread to other parts of the country and beyond (14). Important clonal groups of MRSA in Australia now include the Western Australian group (primarily cc1 also called WA MRSA), the Southwest Pacific group (cc30), the Queensland group (cc93), the United Kingdom EMRSA-15 (cc22), and the so-called East Coast clonal group, AUS-2/AUS-3 (cc239) (1, 14).
The predominant CA-MRSA cc type found in the Aboriginal communities of the Top End was cc75, accounting for 71% if isolates. This clonal complex does not belong to any of the previously described clonal groups and appears to stand alone. It also lacks the genes for PVL. The additional specific SNP (gmk159G) and MLST sequencing confirmed relatedness to ST75. ST75-MRSA-IV has appeared in a previous report from Western Australia (5), and two of these isolates are represented on the current MLST database. However, on further investigation, it appears the isolates in the report, and on the database, actually came from patients in communities of the Top End of the Northern Territory (Frances O'Brien, personal communication). It is likely that ST75-MRSA-IV is unique to the region with CA-MRSA evolving de novo in these communities, just as it has done in Western Australia.
Increasing rates of CA-MRSA were recognized in Darwin in the early 1990s (24), and the trend has continued (29), with most of the infections occurring in the Aboriginal population. At the Royal Darwin Hospital, the rate of CA-MRSA as a proportion of S. aureus sepsis increased from 9% in 1998 to 20% in 2001 (10). The majority are typical CA-MRSA strains that are not multiresistant and have SCCmec type IV (29); this corresponds with the findings of this study where multiresistant strains were not encountered. The Top End of the Northern Territory has about the same area as the US state of Texas, and the communities in this study are truly remote. Yet, there is a relatively frequent movement of people between the communities and the capital city, Darwin, and from community to community. This includes people traveling for health reasons, many of whom are admitted to Darwin's only large public hospital. There is potential for taking HA-MRSA back to communities, just as CA-MRSA is frequently transmitted to the hospital environment (24). Yet, in this study, typical HA-MRSA strains were not found in the Aboriginal communities.
The finding that 21% of S. aureus isolates from skin infection were CA-MRSA is one of the highest rates reported. The proportion of CA-MRSA associated with skin infection rose fourfold, from 4% to 17%, over 3 years in Frejus, France (8), and accounted for 11% of pyoderma in a recent study from Southern India (30). Extremely high rates, 55% of all S. aureus infection, have been reported in one Native American community (16), and an extended community outbreak has been reported in South Texas, with frequent cases of cellulitis and abscesses (36). A recent large study in Atlanta, Georgia, found that the CA-MRSA clone USA 300 had become the predominant cause of skin and soft tissue infection in the region (20). There are more than a dozen published community MRSA nasal carriage studies with rates varying from <1% in Italy (44) to 9.2% in the southern United States (6). A recent national study in the United States found an MRSA carriage rate of 0.84% (15). In our study of remote communities, the MRSA throat carriage rate of 3% (15 of 513) probably reflects a much higher nasal carriage rate.
The surprise finding of cc75 (ST75) in communities 1 and 3 highlights the regional nature of CA-MRSA. However, the communities are more than 400 km apart and in opposite directions from the regional capital of Darwin. They have markedly different languages and cultural backgrounds, and there is limited interaction between them. As distinct from HA-MRSA, CA-MRSA strains are more likely to reflect preexisting regional S. aureus lineages with local acquisition of SCCmec type IV or V (38). Indeed both MRSA and MSSA cc75 were shown to be widely circulating in these communities. Our study shows that the community profile of clonal groups was highly dynamic; moreover, different strains were often present on adjacent pyoderma lesions. One of the study limitations was that only one colony was chosen for analysis from each culture plate, where there may have been multiple types infecting the same wound. The SCCmec gene was restricted to but a few clonal groups, and the overall proportion of MRSA isolates was stable over the study period. Could SCCmec become more mobile across clonal groups with increased community antibiotic prescribing pressure? This is a question that requires urgent investigation.
MLST itself is based upon arbitrary gene fragments and would give significantly different results if different gene fragments were used. The SNP method gives a portable and unambiguous genotype that is of sufficient resolution for an initial screen of a large number of isolates and also is compatible with MLST data. The resolution can easily be increased by interrogating binary markers (17, 40) and adding an additional SNP. Isolates that appear interesting can be further characterized by full MLST, as was done in this study. This approach led to the identification of a new numerically dominant clone, an important epidemiological finding. The SNP genotyping method is simple and relatively inexpensive when compared to other typing methods such as MLST and PFGE. It can also be used to study the epidemiology of MRSA in other settings and even other microorganisms, such as Streptococcus pyogenes (37). An understanding of community acquisition and transmission should lead to more enlightened interventions aimed at treatment and prevention of CA-MRSA infection.
Acknowledgments
We thank the families in the communities, the Aboriginal research officers, the community councils, and community health center staff for their participation and support. Murin Air (NT) and Western Pathology provided assistance with transportation of specimens. Christopher Pearce, Department of Microbiology, Royal Children's Hospital performed the antibiotic susceptibility testing.
The study was supported by grants from the National Heart Foundation of Australia (no. PB 02 M 0996), the National Health and Medical Research Council (no. ID 251690), and the Cooperative Research Centre for Aboriginal Health.
F.H., P.M.G., J.I.-B., and A.J.S. are inventors of a patent describing bacterial genotyping (assessing data sets PCT/AU03/00320) and may be eligible for royalties resulting from the sale of the patent. Otherwise, the authors have no conflict of interest.
REFERENCES
- 1.The Australian Group on Antimicrobial Resistance. 2005. MRSA epidemiology and typing report. Staphylococcus aureus Programme 2004. Australian Group on Antimicrobial Resistance, Perth, Western Australia, Australia.
- 2.Boyce, J., B. Cookson, K. Christainsen, S. Hori, J. Vuopio-Varkila, S. Kocagoz, A. Oztop, C. Vandenbroucke-Grauls, S. Harbarth, and D. Pittet. 2005. Forum: methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 5:653-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing. 15th informational supplement. M2-A8. Wayne, Pa.
- 4.Coombs, G. W., G. R. Nimmo, J. M. Bell, F. Huygens, F. G. O'Brien, M. J. Malkowski, J. C. Pearson, A. J. Stephens, and P. M. Giffard. 2004. Genetic diversity among community methicillin-resistant Staphylococcus aureus strains causing outpatient infections in Australia. J. Clin. Microbiol. 42:4735-4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coombs, G. W., J. C. Pearson, F. G. O'Brien, R. J. Murray, W. B. Grubb, and K. J. Christiansen. 2006. Methicillin-resistant Staphylococcus aureus clones, Western Australia. Emerg. Infect. Dis. 12:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creech, C. B., II, D. S. Kernodle, A. Alsentzer, C. Wilson, and K. M. Edwards. 2005. Increasing rates of nasal carriage of methicillin-resistant Staphylococcus aureus in healthy children. Pediatr. Infect. Dis. J. 24:617-621. [DOI] [PubMed] [Google Scholar]
- 7.Dailey, L., G. W. Coombs, F. G. O'Brien, J. W. Pearman, K. Christiansen, W. B. Grubb, and T. V. Riley. 2005. Methicillin-resistant Staphylococcus aureus, Western Australia. Emerg. Infect. Dis. 11:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Giudice, P., V. Blanc, F. Durupt, M. Bes, J. P. Martinez, E. Counillon, G. Lina, F. Vandenesch, and J. Etienne. 2006. Emergence of two populations of methicillin-resistant Staphylococcus aureus with distinct epidemiological, clinical and biological features, isolated from patients with community-acquired skin infections. Br. J. Dermatol. 154:118-124. [DOI] [PubMed] [Google Scholar]
- 9.Deresinski, S. 2005. Methicillin-resistant Staphylococcus aureus: an evolutionary, epidemiologic, and therapeutic odyssey. Clin. Infect. Dis. 40:562-573. [DOI] [PubMed] [Google Scholar]
- 10.Douglas, M. W., G. Lum, J. Roy, D. A. Fisher, N. M. Anstey, and B. J. Currie. 2004. Epidemiology of community-acquired and nosocomial bloodstream infections in tropical Australia: a 12-month prospective study. Trop. Med. Int. Health 9:795-804. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright, M. C., and B. G. Spratt. 1999. Multilocus sequence typing. Trends Microbiol. 7:482-487. [DOI] [PubMed] [Google Scholar]
- 13.Feil, E. J., and M. C. Enright. 2004. Analyses of clonality and the evolution of bacterial pathogens. Curr. Opin. Microbiol. 7:308-313. [DOI] [PubMed] [Google Scholar]
- 14.Gosbell, I. B. 2005. Epidemiology, clinical features and management of infections due to community methicillin-resistant Staphylococcus aureus (cMRSA). Intern. Med. J. 35(Suppl. 2):S120-S135. [DOI] [PubMed] [Google Scholar]
- 15.Graham, P. L., S. X. Lin, and E. L. Larson. 2006. A US population-based survey of Staphylococcus aureus colonization. Ann. Intern. Med. 144:318-325. [DOI] [PubMed] [Google Scholar]
- 16.Groom, A., D. Wolsey, T. Naimi, K. Smith, S. Johnson, D. Boxrud, et al. 2001. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA 286:1201-1205. [DOI] [PubMed] [Google Scholar]
- 17.Huygens, F., J. Inman-Bamber, G. R. Nimmo, W. Munckhof, J. Schooneveldt, B. Harrison, J. A. McMahon, and P. M. Giffard. 2006. Staphylococcus aureus genotyping using novel real-time PCR formats. J. Clin. Microbiol. 44:3712-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huygens, F., A. J. Stephens, G. R. Nimmo, and P. M. Giffard. 2004. mecA locus diversity in methicillin-resistant Staphylococcus aureus isolates in Brisbane, Australia, and the development of a novel diagnostic procedure for the Western Samoan phage pattern clone. J. Clin. Microbiol. 42:1947-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King, M. D., B. J. Humphrey, E. V. Kourbatova, S. M. Ray, and H. M. Blumberg. 2006. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann. Intern. Med. 144:309-317. [DOI] [PubMed] [Google Scholar]
- 21.Kluytmans-Vandenbergh, M. F., and J. A. Kluytmans. 2006. Community-acquired methicillin-resistant Staphylococcus aureus: current perspectives. Clin. Microbiol. Infect. 12(Suppl. 1):9-15. [DOI] [PubMed] [Google Scholar]
- 22.Lim, T. T., F. N. Chong, F. G. O'Brien, and W. B. Grubb. 2003. Are all community methicillin-resistant Staphylococcus aureus related? A comparison of their mec regions. Pathology 35:336-343. [PubMed] [Google Scholar]
- 23.Louie, L., J. Goodfellow, P. Mathieu, A. Glatt, M. Louie, and A. E. Simor. 2002. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J. Clin. Microbiol. 40:2786-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maguire, G., A. Arthur, P. Boustead, B. Dwyer, and B. Currie. 1996. Emerging epidemic of community-acquired methicillin-resistant Staphylococcus aureus infection in the Northern Territory. Med. J. Aust. 164:721-723. [DOI] [PubMed] [Google Scholar]
- 25.McDonald, M., R. Towers, P. Fagan, M. McKinnon, M. Benger, R. Andrews, B. Currie, and J. Carapetis. 2006. Recovering streptococci from the throat in remote tropical communities: a practical alternative to direct plating. J. Clin. Microbiol. 44:547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald, M., R. J. Towers, R. Andrews, N. Benger, B. J. Currie, and J. R. Carapetis. 2006. Low rates of streptococcal pharyngitis and high rates of pyoderma in communities where rheumatic fever is hyperendemic. Clin. Infect. Dis. 43:683-689. [DOI] [PubMed]
- 27.Moellering, R. C. 2006. The growing menace of community-acquired methicillin-resistant Staphylococcus aureus. Ann. Intern. Med. 144:368-370. [DOI] [PubMed] [Google Scholar]
- 28.Munckhof, W. J., J. Schooneveldt, G. W. Coombs, J. Hoare, and G. R. Nimmo. 2003. Emergence of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) infection in Queensland, Australia. Int. J. Infect. Dis. 7:259-264. [DOI] [PubMed] [Google Scholar]
- 29.Murray, R. J., T. T. Lim, J. C. Pearson, W. B. Grubb, and G. D. Lum. 2004. Community-onset methicillin-resistant Staphylococcus aureus bacteremia in Northern Australia. Int. J. Infect. Dis. 8:275-283. [DOI] [PubMed] [Google Scholar]
- 30.Nagaraju, U., G. Bhat, M. Kuruvila, G. S. Pai, Jayalakshmi, and R. P. Babu. 2004. Methicillin-resistant Staphylococcus aureus in community-acquired pyoderma. Int. J. Dermatol. 43:412-414. [DOI] [PubMed] [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial disc susceptibility testing; approved standards, 8th ed. M2-A8. Wayne, Pa.
- 32.O'Brien, F. G., T. T. Lim, F. N. Chong, G. W. Coombs, M. C. Enright, D. A. Robinson, A. Monk, B. Said-Salim, B. N. Kreiswirth, and W. B. Grubb. 2004. Diversity among community isolates of methicillin-resistant Staphylococcus aureus in Australia. J. Clin. Microbiol. 42:3185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavillard, R., K. Harvey, D. Douglas, A. Hewstone, J. Andrew, B. Collopy, V. Asche, P. Carson, A. Davidson, G. Gilbert, J. Spicer, and F. Tosolini. 1982. Epidemic of hospital-acquired infection due to methicillin-resistant Staphylococcus aureus in major Victorian hospitals. Med. J. Aust. 1:451-454. [PubMed] [Google Scholar]
- 35.Peleg, A. Y., W. J. Munckhof, S. L. Kleinschmidt, A. J. Stephens, and F. Huygens. 2005. Life-threatening community-acquired methicillin-resistant Staphylococcus aureus infection in Australia. Eur. J. Clin. Microbiol. Infect. Dis. 24:384-387. [DOI] [PubMed] [Google Scholar]
- 36.Purcell, K., and J. Fergie. 2005. Epidemic of community-acquired methicillin-resistant Staphylococcus aureus infections: a 14-year study at Driscoll Children's Hospital. Arch. Pediatr. Adolesc. Med. 159:980-985. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, G. A., V. Thiruvenkataswamy, H. Shilling, E. P. Price, F. Huygens, F. A. Henskens, and P. M. Giffard. 2004. Identification and interrogation of highly informative single nucleotide polymorphism sets defined by bacterial multilocus sequence typing databases. J. Med. Microbiol. 53:35-45. [DOI] [PubMed] [Google Scholar]
- 38.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shukla, S. K., M. E. Stemper, S. V. Ramaswamy, J. M. Conradt, R. Reich, E. A. Graviss, and K. D. Reed. 2004. Molecular characteristics of nosocomial and Native American community-associated methicillin-resistant Staphylococcus aureus clones from rural Wisconsin. J. Clin. Microbiol. 42:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stephens, A. J., F. Huygens, J. Inman-Bamber, E. P. Price, G. R. Nimmo, J. Schooneveldt, W. Munckhof, and P. M. Giffard. 2006. Methicillin-resistant Staphylococcus aureus genotyping using a small set of polymorphisms. J. Med. Microbiol. 55:43-51. [DOI] [PubMed] [Google Scholar]
- 41.Townsend, D. E., N. Ashdown, J. W. Pearman, D. L. Annear, and W. B. Grubb. 1985. Genetics and epidemiology of methicillin-resistant Staphylococcus aureus in a Western Australian hospital. Med. J. Aust. 142:108-111. [DOI] [PubMed] [Google Scholar]
- 42.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vlack, S., L. Cox, A. Y. Peleg, C. Canuto, C. Stewart, A. Conlon, A. Stephens, P. Giffard, F. Huygens, A. Mollinger, R. Vohra, and J. S. McCarthy. 2006. Carriage of methicillin-resistant Staphylococcus aureus in a Queensland indigenous community. Med. J. Aust. 184:556-559. [DOI] [PubMed] [Google Scholar]
- 44.Zanelli, G., A. Sansoni, A. Zanchi, S. Cresti, S. Pollini, G. M. Rossolini, and C. Cellesi. 2002. Staphylococcus aureus nasal carriage in the community: a survey from central Italy. Epidemiol. Infect. 129:417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]