Abstract
Bacteria play an important role in the initiation and progression of periodontal diseases and are part of a biofilm, which can contain over 100 different species. The aim of the present study was to show the potential of denaturing gradient gel electrophoresis (DGGE) as a tool for the detection of clinically relevant species and to compare the results of detection by DGGE with those by PCR and culturing. Hybridization of the bands from the DGGE profiles with species-specific probes was developed to confirm the band positions in the marker obtained with reference strains. The sensitivities of DGGE compared to those of cultivation for the detection of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, and Tannerella forsythensis were 100, 100, 88, and 100%, respectively; and the sensitivities of DGGE compared to those of PCR were 100, 90, 88, and 96%, respectively. DGGE as a diagnostic tool could easily be extended to other species, as shown for Treponema denticola, which could be detected in 48% of the samples. Three different groups of A. actinomycetemcomitans serotypes could be distinguished by DGGE (i.e., a group comprising serotypes a, d, e, and f; a group comprising serotype b; and a group comprising serotype c). Amplicons from P. gingivalis and T. denticola migrated to the same position in the gel, and P. intermedia produced multiple bands. In the present study we show that the DGGE profiles represent clinically relevant species which can be detected by hybridization with species-specific probes. With DGGE, large numbers of samples can be analyzed for different species simultaneously, and DGGE may be a good alternative in periodontal microbial diagnostics.
Periodontal diseases are infectious inflammatory disorders caused by bacteria that colonize the tooth surface. Upon bacterial plaque accumulation, gingivitis develops and causes redness, swelling, and gingival bleeding (11). As the dental plaque biofilm continues to accumulate, different bacterial species may colonize, develop, and cooperate in the biofilm. This results in a host response that includes lymphocyte infiltration and the subsequent secretion of cytokines, which may lead to the destruction of hard and soft periodontal tissues (32). The subgingival plaque comprises a complex microbiota that mainly consists of gram-negative anaerobic bacteria (12). The diversity of the subgingival microbiota was extensively explored by Moore and Moore (14), who isolated 509 species from plaque samples from 300 individuals by cultivation. With molecular techniques such as cloning and sequencing, this level of diversity was confirmed and unknown phylotypes were revealed (6, 10, 20). Socransky et al. (26) related the clinical parameters of periodontitis, such as bleeding on probing and pocket depth, with the subgingival microbiota. They proposed different complexes of bacteria which were related to the severity of periodontitis. The so-called red complex consists of Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythensis, and Treponema denticola. Actinobacillus actinomycetemcomitans is associated with aggressive forms of periodontitis, such as localized juvenile periodontitis and refractory periodontitis (21, 25). Although the subgingival bacterial diversity is widely appreciated, only a limited number of species have been recognized as clinically relevant. The complexity of the bacterial community, the presence of uncultivable or unknown species, and the costly and time-consuming cloning techniques hamper investigation of the relationship between the total microbial community and disease. As a consequence, little is known about, for example, the microbial changes that occur in the population during the transition of gingivitis into periodontitis. The potential of denaturing gradient gel electrophoresis (DGGE) for the study of total oral microbial populations has been shown previously (4, 23, 35). In DGGE, PCR-amplified DNA fragments of the same length but with different base-pair sequences can be separated. The fragments are loaded on a polyacrylamide gel containing a linear gradient of denaturants like formamide and urea. The two strands of DNA are denatured at a certain concentration of denaturant, depending on the G+C content and the composition of the fragment. Migration is retarded when a fragment reaches its first melting domain. Complete strand separation is prevented by the addition of a GC-rich fragment to one of the primers, a so-called GC clamp. When universal bacterial primers are used for PCR-DGGE, unknown and/or uncultivable species can be detected. PCR-DGGE of complex microbial populations results in a pattern of bands in which each band represents a different species. DGGE can be used to study microbial complexity and to monitor population dynamics (18).
DGGE has the potential advantage of detecting multiple species simultaneously on a large scale. However, there is no agreement that DGGE profiles are a representative fingerprint of the population under study (9, 16), and it is uncertain whether clinically relevant species are present in DGGE profiles. Consequently, the aim of the present study was to investigate the potential of DGGE as a tool for the detection of clinically relevant species and to compare the DGGE detection results to those obtained by PCR and anaerobic culture.
MATERIALS AND METHODS
Sampling protocol.
The study subjects were adult patients with chronic periodontitis with pockets of 6 mm or more showing bleeding on probing. Subgingival plaque samples from 25 patients were obtained from the deepest pocket in each quadrant of the dentition with sterile paper points. The samples were pooled in 1.5 ml reduced transport fluid (29); sent to the Department of Oral Microbiology, ACTA, Amsterdam, The Netherlands; and processed within 36 h. The samples were vortexed for 2 min, and 100 μl was used for anaerobic culture and 200 μl was used for molecular analysis.
Organisms and culture conditions.
A. actinomycetemcomitans serotypes a, b, c, d, e, and f were clinical isolates. P. gingivalis ATCC 33277, Actinomyces neaslundii DSM 43013, Fusobacterium magna DSM 20470, Actinomyces israellii DSM 43320, Streptococcus mutans DSM 20523, Peptostreptococcus micros DSM 20468, Escherichia coli ATCC 25922, and Fusobacterium nucleatum DSM 20482 were obtained from the American Type Culture Collection (ATCC strains; Manassas, Va.) or the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSM strains; Braunschweig, Germany). The strains from the Deutsche Sammlung von Mikroorganismen und Zellkulturen or the American Type Culture Collection were cultivated according to the instructions of the suppliers. A. actinomycetemcomitans was cultured as described below.
Cultivation.
Aliquots of 0.1 ml from 10-fold serial dilutions were plated onto 5% horse blood agar plates (no. 2; Oxoid, Basingstoke, United Kingdom) supplemented with hemin (5 mg/liter) and menadione (1 mg/liter) for the isolation and growth of obligate anaerobic bacteria and on tryptic soy-serum-bacitracin-vancomycin (TSBV) for the selective isolation and growth of A. actinomyctemcomitans (24). Blood agar plates were incubated anaerobically in 80% N2, 10% H2, and 10% CO2 for up to 14 days. TSBV plates were incubated in air supplemented with 5% CO2 for 5 days (30). The total number of CFU was determined and converted to the total number of CFU/ml. The total number of CFU/ml and the amounts of A. actinomycetemcomitans, P. gingivalis, P. intermedia, and T. forsythensis as a percentage of the total number of CFU/ml were determined. The limit of detection for A. actinomycetemcomitans was 20 cells/ml, and that for the other species was 104 cells/ml.
DNA extraction.
The 200-μl samples were incubated for 1 h at 58°C with 200 μl lysis buffer (10% sodium dodecyl sulfate [SDS], 0.2 mg/ml proteinase K). Proteinase K was inactivated by incubation at 80°C for 10 min. Subsequently, three cycles of freezing-thawing at −80°C for 15 min and 5 min at 80°C were performed. For DNA isolation, 200 μl phenol and 200 μl chloroform/iso-amylalcohol (24:1; vol/vol) were added to the samples. The samples were centrifuged at 14,000 × g for 5 min. A second phenol-chloroform extraction was performed; and after collection of the supernatant, the DNA was precipitated with 500 μl isopropanol at −20°C for 3 h. After centrifugation at 14,000 × g for 15 min, the supernatant was discarded and the pellet was washed twice in 100 μl 70% alcohol. After centrifugation at 14,000 × g for 15 min, the supernatant was removed. The pellet was dissolved in 100 μl sterile Milli-Q and stored at −20°C.
Species-specific PCR.
PCR was performed with a T-gradient thermocycler (Whatman Biometra, Germany) with primers targeting A. actinomycetemcomitans-, P. gingivalis-, P. intermedia-, or T. forsythensis-specific regions on the 16S rRNA gene (Table 1). The primers were designed by Ashimoto et al. (1). The PCR mixture contained 5.0 μl reaction buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl, 15 mM MgCl2), 4.0 μl deoxynucleoside triphosphates (dNTPs; 200 μM each dNTP), 35.5 μl Milli-Q, 2.0 μl (400 nM) of each primer, 0.5 μl (2.5 U) Taq, and 1.0 μl template. The temperature profile included an additional denaturation step of 10 min at 96°C, followed by 34 cycles of a denaturation step at 96°C for 1 min, a primer annealing step at 61°C for 45 s, and an extension step at 72°C for 1 min, with a final extension step of 72°C for 5 min. The PCR products were analyzed by electrophoresis on a 2.0% agarose gel. The gel contained 0.5 μg/ml ethidium bromide. The limit of detection of the PCRs for all four species was less than 50 cells.
TABLE 1.
Primer and probe sequences targeting the 16S rRNA gene used in this study for PCR amplification and hybridization analysis
| Primer or probe | Sequence (5′→3′) | Reference |
|---|---|---|
| Aa1034 | AAA CCC ATC TCT GAG TTC TTC TTC | 1 |
| Aa478 | ATG CCA ACT TGA CGT TAA AT | 1 |
| Pg1132 | ACT GTT AGC AAC TAC CGA TGT | 1 |
| Pg729 | AGG CAG CTT GCC ATA CTG CG | 1 |
| Bf760 | TGC TTC AGT GTC AGT TAT ACC T | 1 |
| Bf120 | GCG TAT GTA ACC TGC CCG CA | 1 |
| Pi1032 | TTT GTT GGG GAG TAA AGC GGG | 1 |
| Pi458 | TCA ACA TCT CTG TAT CCT GCG T | 1 |
| I-341fGC | GC clamp-CCTACGGGIGGCIGCA | 31 |
| I-533r | TIACCGIIICTICTGGCAC | 31 |
| GC clamp | CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG | 16 |
| Aa502 | AAC GTC AAK TTG GCA TGC | This study |
| Pg477 | CGT ATC GCC CGT TAT TCC C | This study |
| Pi425 | CTT TAC TCC CCA ACA AAA GCA GTT TAC AA | 28 |
| Tf440 | CGT ATC TCA TTT TAT TCC CCT GTA | 28 |
| Td469 | CAT GAC TAC CGT CAT CAA AGA AGC | 15 |
PCR-DGGE.
PCR amplification of the 16S rRNA gene fragment region at positions ∼341 and ∼533 (E. coli numbering) was performed on a T-gradient thermocycler (Whatman Biometra) with universal bacterial primers I-341fGC and I-533r (Table 1) (31). The PCR mixture contained 5.0 μl reaction buffer (100 mM Tris-HCl, pH 8.3, 500 mM KCl, 15 mM MgCl2), 4.0 μl dNTPs (200 μM each dNTP), 35.75 μl Milli-Q, 2.0 μl (400 nM) of each primer, 0.25 μl (1.25 U) Taq, and 1.0 μl template. The temperature profile included an additional denaturation step of 1 min at 94°C, followed by 35 cycles of a denaturation step at 94°C for 45 s, a primer annealing step at 49°C for 30 s, an extension step at 72°C for 1 min, with a final extension step of 72°C for 5 min. The PCR products were analyzed by electrophoresis on a 2.0% agarose gel containing 0.5 μg/ml ethidium bromide.
DGGE analysis of PCR amplicons.
DGGE of the PCR products generated with the I-341fGC/I-533r primer set was performed as described by Muyzer et al. (17) with the use of a PhorU system (Ingeny, The Netherlands). The PCR products were loaded on a 6% (wt/vol) polyacrylamide gel in 0.5× TAE (1× TAE is 0.04 M Tris base, 0.02 M acetic acid, and 1.0 mM EDTA [pH 7.5]). The denaturing gradient consisted of 30% to 70% denaturant (100% denaturant equals 7 M urea and 40% formamide). The gels were poured by use of a gradient mixer. A 10-ml stacking gel without denaturant was layered on top. Electrophoresis was performed for 16 h at 100 V at 60°C. The gels were stained with silver nitrate.
Southern blotting of DGGE gels.
After DGGE, the DNA from a nonstained gel was transferred to nylon hybridization membranes (Hybond N+; Amersham International plc) by use of a semidry electroblotter, according to the manufacturer's instructions, with the transfer medium consisting of 0.1× TAE buffer; and the blot was run at 252 mA for 3 h. Following transfer, the DNA was simultaneously denatured and covalently cross-linked to the hybridization membrane by incubation on a pad of 3MM paper (Whatman International, Kent, United Kingdom) soaked in 0.4 M NaOH for 30 min, followed by neutralization on two pads of 1 M Tris-HCl, pH 7.0, for 2 min each. The filters were then baked for 2 h at 80°C and stored at −20°C until further processing.
Hybridization of blots.
The optimal hybridization temperatures for probes Aa502, Pg477, Pi425, Tf440, and Td469 were calculated with the web-based application OligoAnalyzer 3.0 (http://207.32.43.70/biotools/oligocalc/oligocalc.asp) and were determined empirically. Tenfold serial dilutions of the PCR amplicons were blotted onto a nylon membrane and hybridized. The calculated hybridization temperatures were lower than the empirically determined temperatures. For increased specificity, we chose to use the empirically determined hybridization temperatures. Prehybridization and hybridization were carried out in hybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate-0.1% sodium lauryl sulfate], 0.02% [wt/vol] SDS, 0.1% N-lauroylsarcosine) at 42°C for probes Aa502, Tf440, and Td469 or 50°C for probes Pg477 and Pi425. At the hybridization temperature, two stringency washes of 15 min each with 2× SSC-0.1% SDS were followed by two stringency washes of 15 min each with 0.1× SSC-0.1% SDS. According to the manufacturer's instructions, the hybridized blots were incubated for 1 h at room temperature with alkaline phosphatase conjugate in buffer A (0.3 M NaOH, 0.1 M Tris-HCl, pH 7.5). The membranes were washed free of unbound conjugate with three washes of 10 min each with 0.3% Tween 20 in buffer A. For signal generation, the membranes were incubated overnight with 1.3 ml enhanced chemifluorescence substrate (Amersham Biosciences, Little Chalfont, United Kingdom). Fluorescent signals were detected with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and analyzed with image analysis software.
RESULTS
Probe design and determination of optimal hybridization conditions.
Probes Aa502 and Pg477 have been designed by using the ARB software package (13) and tested for specificity against reference strains. Both probes were specific for their target organisms. The optimal hybridization temperatures were 42°C for probes Aa502, Tf440, and Td469 and 50°C for probes Pg477 and Pi425. Probe Aa502 is complementary to a conserved region of the 16S rRNA where intrahelix secondary base interactions may interfere during hybridization (2). It is therefore not possible to use probe Aa502 in fluorescent in situ hybridization experiments. Probe Aa502 hybridized to PCR amplicons of the six A. actinomycetemcomitans serotypes baked on a nylon membrane but not to amplicons of the P. gingivalis, P. intermedia, or T. forsythensis strains.
Cultivation.
Twenty-five patient samples were analyzed by cultivation. The average total number of CFU/ml was 2.1 × 108 (range, 6.0 × 106 to 2.1 × 109 CFU/ml; standard deviation, 4.7 × 108 CFU/ml). Three of the 25 samples were positive for A. actinomycetemcomitans. A. actinomycetemcomitans contributed 0.009 to 2.0% of the total number of CFU/ml. Nine of the 25 samples were positive for P. gingivalis. P. gingivalis contributed 2 to 38% to the total number of CFU/ml. Sixteen of the 25 samples were positive for P. intermedia. P. intermedia contributed 0.4 to 18% to the total number of CFU/ml. Twenty-three of the 25 samples were positive for T. forsythensis. T. forsythensis contributed 0.008 to 21% to the total number of CFU/ml.
Cultivation versus species-specific PCR.
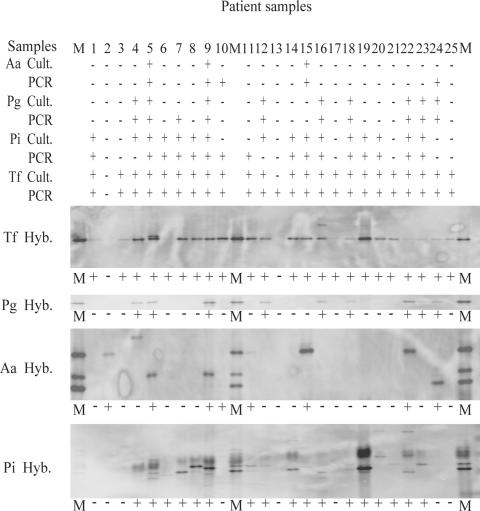
DNA extracts from 25 samples were analyzed for the presence of A. actinomycetemcomitans, P. gingivalis, P. intermedia, and T. forsythensis. The results are shown in Fig. 1 and Table 2. The three samples that were positive for A. actinomycetemcomitans by culture were also positive by PCR. An additional two samples were positive for A. actinomycetemcomitans by PCR. Next to the nine samples positive for P. gingivalis by cultivation, one additional sample was positive for P. gingivalis by PCR. For P. intermedia, 13 samples were positive by both cultivation and PCR, 3 samples were positive by culture but negative by PCR, and 3 samples were positive by PCR and negative by culture. Twenty-four samples were positive for T. forsythensis by PCR, whereas 23 were positive for T. forsythensis by culture.
FIG. 1.
Cultivation (Cult.), PCR, and hybridization (Hyb.) results for A. actinomycetemcomitans (Aa) P. gingivalis (Pg), P. intermedia (Pi), and T. forsythensis (Tf) presented in line. Lanes M, markers containing a mixture of PCR amplicons of reference species.
TABLE 2.
Detection results from cultivation, species-specific PCR, and DGGE for the four pathogens associated with periodontitis in 25 subgingival plaque samples
| Species | No. of samples with positive results (sensitivitya [%])
|
||
|---|---|---|---|
| Cultivation | PCR | DGGE | |
| Actinobacillus actinomycetemcomitans | 3 (38) | 5 (100) | 8 (100/100) |
| Porphyromonas gingivalis | 9 (100) | 10 (100) | 9 (100/90) |
| Prevotella intermedia | 16 (88) | 16 (88) | 17 (94/88) |
| Tannerella forsythensis | 23 (100) | 24 (100) | 23 (100/96) |
| Treponema denticola | ND | ND | 12 |
The sensitivity of cultivation was calculated with DGGE as the reference standard. The sensitivity of PCR was calculated with cultivation as the reference standard. The sensitivity of DGGE was calculated with both cultivation and PCR as the reference standards, as indicated by the two sensitivity values, respectively, in parentheses. ND, not determined.
Cultivation versus hybridization analysis of DGGE blots.
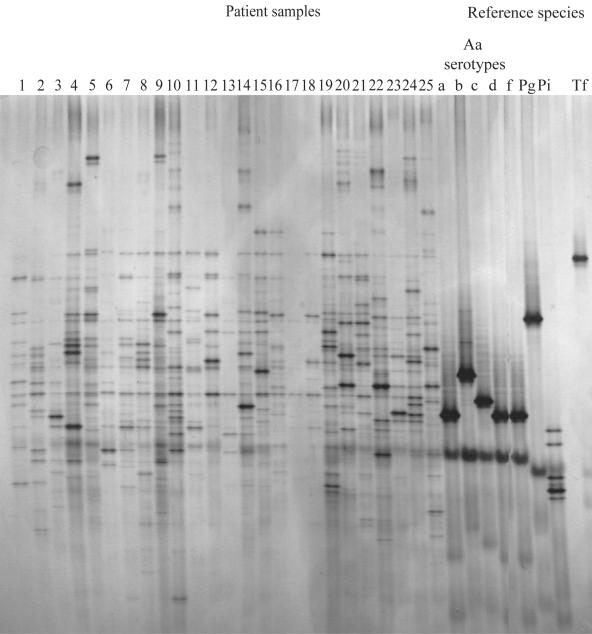
The DGGE profiles for the strains from the 25 patient samples and the reference strains are shown in Fig. 2. On average, the profiles for strains from the patient samples show a high number of bands, and between patients, a variation in the banding patterns can be observed (Fig. 2). The identification of pathogens by comparison of their migration patterns with those of reference strains is therefore difficult. Identification is even more complicated for P. gingivalis and T. denticola, which migrate to the same position in the gel. Even when a mixture of the PCR products of P. gingivalis and T. denticola is loaded on the gel, the fragments migrate to the same position in the gel, resulting in a single band. DNA fragments from A. actinomycetemcomitans serotypes a, d, e, and f migrate to the same position in the gel, whereas fragments from serotypes b and c migrate to different positions. The DGGE profiles were transferred to nylon membranes and analyzed for the presence of A. actinomycetemcomitans, P. gingivalis, P. intermedia, T. forsythensis, and T. denticola with species-specific probes. The results are presented in Fig. 1 and Table 2. The three samples positive for A. actinomycetemcomitans by culture were also positive by DGGE. In addition, five more samples appeared to be positive by DGGE. Four patients harbored serotype c; three patients harbored serotype b; and three patients harbored serotype a, d, e, or f. Furthermore, two patients harbored a combination of a serotype b strain with a serotype a, d, e, or f strain. The nine samples positive for P. gingivalis by culture were positive by DGGE. For P. intermedia, 15 of the 16 culture-positive samples were positive by DGGE. In addition, two other samples were positive. The 23 samples positive for T. forsythensis by cultivation were positive by DGGE. Although cultivation of T. denticola was not performed, the profiles were screened for the presence of T. denticola to show the ease and the possibility of extension of the set of species that can be detected by DGGE. Twelve (48%) patients were positive for T. denticola (Table 2).
FIG. 2.
DGGE profiles for 25 samples and reference species. Aa, A. actinomycetemcomitans; Pg, P. gingivalis; Pi, P. intermedia; Tf, T. forsythensis.
DISCUSSION
The present study examined the use of DGGE as a diagnostic tool. Samples from 25 patients with periodontal disease were analyzed for the detection of A. actinomycetemcomitans, P. gingivalis, P. intermedia, and T. forsythensis by cultivation, species-specific PCR, and DGGE. The results showed that there was good agreement in the results for the detection of A. actinomycetemcomitans, P. gingivalis, P. intermedia, and T. forsythensis by either cultivation, PCR, or DGGE. The sensitivity of DGGE for P. gingivalis and T. forsythensis was lower by use of PCR as a reference than by use of cultivation as a reference. This can be explained by the lower limit of detection of the species-specific PCR. For A. actinomycetemcomitans, an additional two samples were positive by species-specific PCR than by cultivation and five more samples were positive by DGGE than by cultivation. This may be the result of the lower limit of detection of PCR-based techniques and the fact that dead cells are also detected by PCR. Interestingly, three samples were PCR negative and DGGE positive for A. actinomycetemcomitans. This may be due to the higher specificity of probe Aa502 compared to those of the primers used. Online analysis at the RDPII database revealed 42 positive matches for probe Aa502, compared to only 8 positive matches for the reverse primer and 15 positive matches for the forward primer. Except for nine sequences, the available sequences are partial and do not include the V6-V8 region where the sequence of the A. actinomycetemcomitans reverse primer used is positioned. Kaplan et al. (8) showed that differences in 16S rRNA gene sequences could be related to different serotypes of A. actinomycetemcomitans. Ihalin and Asikainen (7) noted different unique migration patterns within serotype e strains when they used the V6-V8 region to study the migration patterns of different serotypes by DGGE. The available data show that variable regions in the 16S rRNA gene are present between and within serotypes. These findings may explain the PCR-negative, DGGE-positive results. Boutaga et al. (3) and Siqueira et al. (22) also used the V6-V8 region for A. actinomycetemcomitans-specific primer design. Those two groups of investigators found five culture-positive, quantitative PCR-negative samples and three checkerboard DNA-DNA hybridization assay-positive, PCR-negative samples, respectively. The primers that were designed may not cover some variants or serotypes, especially serotype e. On the basis of these findings, we conclude that the V6-V8 region is not suitable for A. actinomycetemcomitans-specific primer or probe design until complete 16S rRNA gene sequences covering all six serotypes are available. Probe Aa502 was specific for A. actinomycetemcomitans, and hybridization of DGGE profiles generated with primer set I-341fGC/I-533r with probe Aa502 could distinguish between serotypes a, d, e, and f and serotype b or c. According to Yang et al. (34) and Paju et al. (19), serotype b is the predominant serotype in periodontitis, endocarditis, and bacteremia; serotype c is predominant in periodontally healthy subjects; and serotypes d and e should be considered rare.
The culture-positive, PCR-negative results for P. intermedia may be due to the discriminating capacity of PCR between P. intermedia and P. nigrescens, which cannot be obtained with standard culture techniques, resulting in false-positive results (5). This resulted in low sensitivities for DGGE and PCR. When PCR was used as the reference, the sensitivity of DGGE decreased to 88% for P. intermedia. This may be due to the specificity of probe Pi425, the lower detection limit of PCR, and the presence of multiple bands for P. intermedia in a DGGE profile. Multiple bands can be caused by microvariations introduced during PCR (27), the application of degenerated primers, and the presence of four open reading frames for the 16S rRNA gene (The Institute of Genomic Research [www.tigr.org]). The presence of multiple bands complicates the interpretation of the profile.
Screening for T. denticola shows the possibility of the extension of detection by DGGE to fastidious or noncultivable bacteria. Differences in hybridization temperatures offer the possibility of detection of P. gingivalis and T. denticola during subsequent hybridization sessions. By hybridization with the bands from the DGGE profiles, T. denticola was detected in 12 (48%) samples. This finding is in agreement with those of a previous report (33), showing a T. denticola prevalence of 40 to 50% in subgingival samples.
DGGE has mostly been used to study population dynamics and bacterial diversity and to monitor bacterial colonization and succession (17). Besides the benefits, DGGE has potential pitfalls. First, only bacterial populations making up more than 1% of the total community can be detected by DGGE (18). In the present study we demonstrated that clinically relevant species are present and can be detected from their DGGE profiles, even when they comprise less than 1% of the cultivable population. Second, amplified fragments from different species might migrate to the same location in the gel (9) or multiple bands are observed from a single species. Subgingival diversity measurements are also biased by multiple bands from single species (A. actinomycetemcomitans and P. intermedia) and by fragments from different species that migrate to the same position in the gel (P. gingivalis and T. denticola). These limitations of DGGE can be overcome by the application of species-specific probes during different hybridization sessions. Detailed information about species that produce multiple bands or the migration of fragments to similar locations will provide more insight into species diversity and can be used to refine statistical analyses of DGGE profiles.
In conclusion, the present study shows that the results of DGGE and hybridization with species-specific probes correlate with those of cultivation and PCR for the detection of clinically relevant periodontal pathogens. DGGE outcompetes cultivation and PCR in its sensitivity for the detection of A. actinomycetemcomitans. DGGE offers the opportunity to detect multiple species simultaneously and to distinguish between A. actinomycetemcomitans serotypes and can easily be extended to other species of interest. Moreover, the use of DGGE and hybridization offers the opportunity to study the presence of these pathogens in relation to the presence of other species. Therefore, DGGE may be the next alternative in clinical microbiological diagnostics.
Acknowledgments
We thank Erwin Raangs for technical assistance and the technicians from the Oral Microbiology Department at ACTA for culturing the reference strains.
REFERENCES
- 1.Ashimoto, A., C. Chen, I. Bakker, and J. Slots. 1996. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol. Immunol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 2.Behrens, S., C. Ruhland, J. Inacio, H. Huber, A. Fonseca, I. Spencer-Martins, B. M. Fuchs, and R. Amann. 2003. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boutaga, K., A. J. van Winkelhoff, C. M. Vandenbroucke-Grauls, and P. H. Savelkoul. 2005. Periodontal pathogens: a quantitative comparison of anaerobic culture and real-time PCR. FEMS Immunol. Med. Microbiol. 45:191-199. [DOI] [PubMed] [Google Scholar]
- 4.Fujimoto, C., H. Maeda, S. Kokeguchi, S. Takashiba, F. Nishimura, H. Arai, K. Fukui, and Y. Murayama. 2003. Application of denaturing gradient gel electrophoresis (DGGE) to the analysis of microbial communities of subgingival plaque. J. Periodontal Res. 38:440-445. [DOI] [PubMed] [Google Scholar]
- 5.Gmür, R., and T. Thurnheer. 2002. Direct quantitative differentiation between Prevotella intermedia and Prevotella nigrescens in clinical specimens. Microbiology 148:1379-1387. [DOI] [PubMed] [Google Scholar]
- 6.Hutter, G., U. Schlagenhauf, G. Valenza, M. Horn, S. Burgemeister, H. Claus, and U. Vogel. 2003. Molecular analysis of bacteria in periodontitis: evaluation of clone libraries, novel phylotypes and putative pathogens. Microbiology 149:67-75. [DOI] [PubMed] [Google Scholar]
- 7.Ihalin, R., and S. Asikainen. 2006. 16S rDNA PCR-denaturing gradient gel electrophoresis in determining proportions of coexisting Actinobacillus actinomycetemcomitans strains. J. Microbiol. Methods 65:417-424. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan, J. B., H. C. Schreiner, D. Furgan, and D. H. Fine. 2002. Population structure and genetic diversity of Actinobacillus actinomycetemcomitans strains isolated from localized juvenile periodontitis patients. J. Clin. Microbiol. 40:1181-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kisand, V., and J. Wikner. 2003. Limited resolution of 16S rDNA DGGE caused by melting properties and closely related DNA sequences. J. Microbiol. Methods 54:183-191. [DOI] [PubMed] [Google Scholar]
- 10.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loë, H., E. Theilade, and S. B. Jensen. 1965. Experimental gingivitis in man. J. Periodontol. 36:177-187. [DOI] [PubMed] [Google Scholar]
- 12.Loesche, W. J., and N. S. Grosman. 2001. Periodontal disease as a specific albeit chronic infection: diagnosis and treatment. Clin. Microbiol. Rev. 14:727-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 14.Moore, W. E., and L. V. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 15.Moter, A., C. Hoenig, B. K. Choi, B. Riep, and U. B. Gobel. 1998. Molecular epidemiology of oral treponemes associated with periodontal disease. J. Clin. Microbiol. 36:1399-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 3.4.4.1-3.4.4.27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, 3rd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 18.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 19.Paju, S., P. Carlson, H. Jousimies-Somer, and S. Asikainen. 2000. Heterogeneity of Actinobacillus actinomycetemcomitans strains in various human infections and relationships between serotype, genotype, and antimicrobial susceptibility. J. Clin. Microbiol. 38:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodenburg, J. P., A. J. van Winkelhoff, E. G. Winkel, R. J. Goene, F. Abbas, and J. de Graaff. 1990. Occurrence of Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in severe periodontitis in relation to age and treatment history. J. Clin. Periodontol. 17:392-399. [DOI] [PubMed] [Google Scholar]
- 22.Siqueira, J. F., I. N. Rocas, M. De Uzeda, A. P. Colombo, and K. R. Santos. 2002. Comparison of 16S rDNA-based PCR and checkerboard DNA-DNA hybridization for detection of selected endodontic pathogens. J. Med. Microbiol. 51:1090-1096. [DOI] [PubMed] [Google Scholar]
- 23.Siqueira, J. F., Jr., I. N. Rocas, and A. S. Rosado. 2004. Investigation of bacterial communities associated with asymptomatic and symptomatic endodontic infections by denaturing gradient gel electrophoresis fingerprinting approach. Oral Microbiol. Immunol. 19:363-370. [DOI] [PubMed] [Google Scholar]
- 24.Slots, J. 1982. Selective medium for isolation of Actinobacillus actinomycetemcomitans. J. Clin. Microbiol. 15:606-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slots, J., and M. Ting. 1999. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol. 2000 20:82-121. [DOI] [PubMed] [Google Scholar]
- 26.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 27.Speksnijder, A. G., G. A. Kowalchuk, S. De Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sunde, P. T., I. Olsen, U. B. Gobel, D. Theegarten, S. Winter, G. J. Debelian, L. Tronstad, and A. Moter. 2003. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. Microbiology 149:1095-1102. [DOI] [PubMed] [Google Scholar]
- 29.Syed, S. A., and W. J. Loesche. 1972. Survival of human dental plaque flora in various transport media. Appl. Microbiol. 24:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Steenbergen, T. J., A. J. van Winkelhoff, L. van der Mispel, U. van der Velden, F. Abbas, and J. de Graaff. 1986. Comparison of two selective media for Actinobacillus actinomycetemcomitans. J. Clin. Microbiol. 24:636-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe, K., Y. Kodama, and S. Harayama. 2001. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J. Microbiol. Methods 44:253-262. [DOI] [PubMed] [Google Scholar]
- 32.Williams, R. C. 1993. Periodontal diseases: gingivitis, juvenile periodontitis, adult periodontitis. Curr. Clin. Top. Infect. Dis. 13:146-163. [PubMed] [Google Scholar]
- 33.Ximenez-Fyvie, L. A., A. D. Haffajee, and S. S. Socransky. 2000. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27:648-657. [DOI] [PubMed] [Google Scholar]
- 34.Yang, H. W., Y. F. Huang, Y. Chan, and M. Y. Chou. 2005. Relationship of Actinobacillus actinomycetemcomitans serotypes to periodontal condition: prevalence and proportions in subgingival plaque. Eur. J. Oral Sci. 113:28-33. [DOI] [PubMed] [Google Scholar]
- 35.Zijnge, V., H. J. M. Harmsen, J. Kleinfelder, M. E. van der Rest, J. E. Degener, and G. W. Welling. 2003. Denaturing gradient gel electrophoresis analysis to study bacterial community structure in pockets of periodontitis patients. Oral Microbiol. Immunol. 18:59-65. [DOI] [PubMed] [Google Scholar]