Abstract
The aim of this study was to develop a PCR and reverse line blot hybridization (PCR-RLB) macroarray assay based on 16S-23S rRNA gene internal transcribed spacer sequences for the identification and differentiation of 34 mycobacterial species or subspecies. The performance of the PCR-RLB assay was assessed and validated by using 78 reference strains belonging to 55 Mycobacterium species, 219 clinical isolates which had been identified as mycobacteria by high-performance liquid chromatography or gas chromatography, three skin biopsy specimens from patients with suspected leprosy which had been shown to contain acid-fast bacilli, and isolates of 14 nonmycobacterial species. All mycobacteria were amplified in the PCR and hybridized with a genus-specific probe (probe MYC). The 34 species-specific probes designed in this study hybridized only with the corresponding Mycobacterium species. The mycobacterial PCR-RLB assay is an efficient tool for the identification of clinical isolates of mycobacteria; it can reliably identify mixed mycobacterial cultures and M. leprae in skin biopsy specimens.
Mycobacterium tuberculosis infection continues to increase worldwide at an estimated annual rate of 3.5% (http://www.who.int/tb/publications/global_report/2005/pdf/Full.pdf). The rapidly spreading human immunodeficiency virus (HIV) epidemic in many parts of the world will further increase the number of HIV-related cases of tuberculosis as well as the number of nontuberculous mycobacterial infections. The rapid and accurate identification of clinically significant mycobacterial species is necessary for optimal medical and public health interventions (3, 21).
The results of traditional methods for the identification of closely related Mycobacterium species by growth characteristics (pigment, growth rate, colonial morphology) and biochemical tests are often difficult to interpret, even for some very common species. The tests are cumbersome and time-consuming, require expertise, lack sensitivity and reproducibility (21), and often use nonstandardized reagents. Rapid biochemical methods, such as high-performance liquid chromatography (HPLC) of mycolic acids and gas chromatography (GC) of fatty acids, are available only in specialized laboratories (2).
The rapid identification of mycobacteria to the species level is recommended in clinical laboratories to ensure accurate diagnosis and effective therapy. This has stimulated the development in recent years (1, 4-9, 11-20, 22, 25, 29, 30) of a number of rapid and more accurate identification tools based on molecular technology.
Several DNA sequences or genes have been targeted for the differentiation of Mycobacterium species, including the 16S rRNA gene (8, 9), the 23S rRNA gene (13, 15), the 16S-23S rRNA gene spacer region (15), the 32-kDa protein gene (22), dnaA (14), dnaJ (25), gyrB (4), recA (1), rpoB (6, 17), secA1 (29), and sod (30). There are at least two commercially available systems for the identification of Mycobacterium spp. based on line probe hybridization, namely, the INNO-LiPA Mycobacterium system (Immunogenetics, Ghent, Belgium), which targets the 16S-23S rRNA gene spacer region and which identifies 16 Mycobacterium species, and the GenoType Mycobacterium CM/AS system (Hain Life Science, Nehren, Germany), which targets the 23S rRNA gene and which identifies 13 species (14). None of these methods can differentiate all of the Mycobacterium species commonly isolated in the clinical laboratory; more discriminatory methods are still needed for clinical and epidemiological investigations.
In this study, we developed a PCR-reverse line blot hybridization (PCR-RLB) assay based on the 16S-23S rRNA gene internal transcribed spacers (ITSs) for the identification of a larger number of mycobacteria to the species or the subspecies level.
MATERIALS AND METHODS
Mycobacterial reference strains.
Reference strains of the following Mycobacterium species were included in this study: (i) the members of the M. tuberculosis complex M. africanum ATCC 25420, M. bovis ATCC 19210, M. bovis BCG (National Institute for the Control of Pharmaceutical and Biological Products, China), M. microti ATCC 19422, and M. tuberculosis H37Rv and (ii) the nontuberculous mycobacteria M. abscessus ATCC 19977, M. agri ATCC 27406, M. aichiense ATCC 27280, M. asiaticum ATCC 25276, M. aurum ATCC 23366, M. austroafricanum ATCC 33464, M. avium ATCC 25291, M. chelonae subsp. chelonae ATCC 35752, M. chitae ATCC 19627, M. chubuense ATCC 27278, M. diernhoferi ATCC 19340, M. duvalii ATCC 43910, M. fallax ATCC 35219, M. farcinogenes ATCC 35753, M. flavescens ATCC 14474, M. fortuitum subsp. fortuitum ATCC 6841, M. gadium ATCC 27726, M. gastri ATCC 15754, M. gilvum ATCC 43909, M. gordonae ATCC 14470, M. hemophilum ATCC 29548, M. intracellulare ATCC 13950, M. kansasii ATCC 12478, M. komossense ATCC 33014, M. malmoense ATCC 29571, M. marinum ATCC 927, M. neoaurum ATCC 25795, M. nonchromogenicum ATCC 19530, M. obuense ATCC 27023, M. parafortuitum ATCC 19686, M. phlei ATCC 11758, M. porcinum ATCC 33776, M. pulveris ATCC 35154, M. rhodesiae ATCC 27024, M. scrofulaceum ATCC 19981, M. senegalense ATCC 35796, M. shimoidei ATCC 27962, M. simiae ATCC 25275, smegmatis ATCC 19420, M. szulgai ATCC 35799, M. terrae ATCC 15755, M. thermoresistibile ATCC 19527, M. tokaiense ATCC 27282, M. triviale ATCC 23292, M. ulcerans ATCC 19423, M. vaccae ATCC 29678, and M. xenopi ATCC 19250).
Nonmycobacterial reference strains.
Reference strains of the following nonmycobacterial species were included: Corynebacterium paurometabolum ATCC 8368, Klebsiella pneumoniae subsp. pneumoniae ATCC 25304, Legionella pneumophila subsp. pneumophila ATCC 33153, Mycoplasma pneumoniae ATCC 29342, Nocardia asteroides ATCC 19247, Nocardia brasiliensis ATCC 19296, Nocardia farcinica ATCC 3308, Nocardia transvalensis ATCC 6856, Rhodococcus equi ATCC 6939, Rhodococcus erythropolis ATCC 25544, Rhodococcus rhodochrous ATCC 13808, Rhodococcus sputi ATCC 29627, Rhodococcus wratislaviensis ATCC 51786, and Streptococcus pneumoniae SSISP pn2L.
Clinical isolates.
A total of 219 mycobacterial isolates were obtained from the following sources: Center for Infectious Disease and Microbiology (CIDM), Institute of Clinical Pathology and Medical Research, Westmead, Australia (n = 85); the Institute of Tuberculosis, Shenzhen Chronic Disease Hospital, China (n = 64); and the Clinical Laboratory, Guangzhou Thoracic Hospital, China (n = 70) (Table 1). They included isolates of the M. tuberculosis complex and the following nontuberculous Mycobacterium species: M. abscessus, M. avium, M. chelonae, M. fortuitum, M. gordonae, M. intracellulare, M. kansasii, M. lentiflavum, M. malmoense, M. nonchromogenicum, M. phlei, M. scrofulaceum, M. smegmatis, M. szulgai, M. triplex, M. ulcerans, and M. xenopi. All isolates were from different patients. Skin biopsy specimens from three patients with presumed leprosy which had been shown to contain acid-fast bacilli by microscopy were included in the study.
TABLE 1.
Identification of 34 Mycobacterium species, by PCR-RLB
| Species | Total no. of isolates | Identification methoda (no. of isolates) | PCR result | RLB result with probe MYCb | Species-specific probe used for RLBc (no. of isolates) | ITS sequencing result (no. of isolates) |
|---|---|---|---|---|---|---|
| M. abscessus | 12 | GC (8) | + | + | ABS (12) | NDd |
| HPLC (4) | ||||||
| M. chelonae complex | 28 | GC (28) | ND | |||
| M. abscessus | + | + | ABS (26) | ND | ||
| M. chelonae a/b | + | + | CHE1 (2) | ND | ||
| M. chelonae | 4 | HPLC (4) | + | + | CHE1 (3), CHE and FOR1 (1) | ND |
| M. chelonae a/b | Uninterpretablee (1) | |||||
| M. chelonae c | None found | ND | ||||
| MAC | 3 | GC (3) | + | + | INT (3) | ND |
| M. avium | 23 | HPLC (23) | + | + | AVI (11), INT (10), AVI and KAN1 (2) | M. intracellulare (1), uninterpretable (2) |
| M. fortuitum complex | 29 | GC (22) | + | + | FOR1 (12); FOR2 and FOR3 (1); FOR1, FOR2, and FOR3 (9) | Uninterpretable (2) |
| HPLC (7) | + | + | FOR1 (4); FOR2 and FOR3 (1); FOR1, FOR2, and FOR3 (2) | Uninterpretable (3) | ||
| M. gordonae | 28 | GC (20) | + | + | GOR (16); GOR and ABS (2); GOR and FOR1 (1); GOR and FOR1, FOR2, and FOR3 (1) | Uninterpretable (4) |
| HPLC (8) | + | + | GOR (8) | ND | ||
| M. intracellulare | 2 | HPLC (2) | + | + | AVI (1), INT (1) | M. avium (1), M. intracellulare (1) |
| M. kansasii | 3 | GC (1) | + | + | KAN1 (1) | ND |
| HPLC (2) | + | + | KAN1 (1), KAN2 (1) | ND | ||
| M. lentiflavum | 2 | HPLC (2) | + | + | LEN (2) | M. lentiflavum (2) |
| M. leprae | 3f | ND | + | + | LEP (2), MTBC and INT (1) | M. leprae (2), uninterpretable (1) |
| M. malmoense | 1 | HPLC (1) | + | + | MAL (1) | ND |
| M. nonchromogenicum | 11 | GC (11) | + | + | − (10),g FOR1 (1) | M. fortuitum a (1) |
| M. phlei a | 12 | GC (12) | + | + | PHL1 (12) | ND |
| M. phlei b | None found | |||||
| M. scrofulaceum | 15 | GC (8) | + | + | SCR (7), SCR and INT (1) | Uninterpretable (1) |
| HPLC (7) | + | + | SCR (7) | ND | ||
| M. smegmatis | 2 | HPLC (2) | + | + | SME (2) | ND |
| M. szulgai | 1 | GC (1) | + | + | SZU (1) | ND |
| M. triplex | 2 | HPLC (2) | + | + | TRI (2) | M. triplex (2) |
| M. tuberculosis complex | 39 | GC (20) | + | + | MTBC (39) | ND |
| HPLC (19) | ||||||
| M. ulcerans/M. marinum | 1 | HPLC (1) | + | + | ULC/MAR (1) | ND |
| M. xenopi | 1 | HPLC (1) | + | + | XEN (1) | ND |
| Total | 222h |
GC was performed with isolates obtained from Shenzhen Chronic Disease Hospital and Guangzhou Thoracic Hospital, China; HPLC was performed with isolates from the Mycobacterium Reference Laboratory, Centre for Infectious Diseases and Microbiology, ICPMR, Westmead, Australia.
MYC, Mycobacterium genus-specific probe; +, positive hybridization.
For the specificities of the probes on RLB, see Table 2.
ND, not done; sequencing was not performed if the HPLC or GC and PCR-RLB assay results were the same.
All cultures in which mixed species were present, as identified by the PCR-RLB assay, produced uninterpretable sequencing results.
Three clinical biopsy specimens with acid-fast bacilli on microscopy presumed to be M. leprae.
No probes specific for M. nonchromogenicum were included in the RLB.
The total includes 219 clinical isolates and three skin biopsy specimens.
Mycobacteria were grown on Löwenstein-Jensen (L-J) slants with weekly inspection until visible growth was detected or in 7H12 broth (MGIT liquid medium; Becton Dickinson) until a positive growth index was signaled by the automated system. Colonies were scraped from the L-J slants and suspended in 1 ml Tris-HCl buffer (10 mM; pH 7.5). The bacteria were killed by heating them to 100°C for 30 min and were then stored at 4°C. The clinical isolates used had been identified to the species level by standard biochemical procedures, and their identities were confirmed by HPLC (CIDM isolates) or GC (isolates from both Chinese laboratories) (2).
Probe and primer design.
Primers Sp1 (5′-ACC TCC TTT CTA AGG AGC ACC-3′) and Sp2 (5′-GAT GCT CGC AAC CAC TAT YCA-3′), which target the 16S-23S ITS region of the M. tuberculosis spacer sequence (GenBank accession number L15623), were modified from a previous publication (19); 35 species-specific probes based on the published 16S-23S ITS sequences of various Mycobacterium species were designed (Table 2). All probes and primers were checked for specificity against all sequences in GenBank by using QueryBD, WebANGIS GCG, SeqSearch, and Browse code in the Australian National Genomic Information Services (ANGIS) programs (http://www1.angis.org.au/WebANGIS/WebFM). The probes were designed to have similar melting temperatures above 62°C, and their lengths varied from 20 bp to 27 bp. All oligonucleotide probes and primers were synthesized with a 5′-terminal amino group and biotin labels (Sigma-Aldrich, St. Louis, Mo.), respectively.
TABLE 2.
Specificities and sequences of probes used for Mycobacterium PCR-RLB assay
| Probe | Strain | Melting temp (°C) | GenBank accession no. | Sequence |
|---|---|---|---|---|
| MYC | Mycobacterium genus | 68.59 | L15623 | 301-GTGGTGGGGTGTGGTGTTTG-320 |
| ABS | M. abcessus | 65.37 | AJ314869 | 32-GGG AAC ATA AAG TAG GCA TCT GTA GTG-58 |
| ASI | M. asiaticum | 72.03 | AF191087 | 35-TGC AGG CCG TGT GGA GTT CTC-55 |
| AVI | M. avium | 69.12 | AJ314863 | 126-AAC ACT CGG TCC GTC CGT GT-145 |
| CHE1 | M. chelonae a/ba | 62.81 | AJ314873 | 33-GGA ACA TAA AGC GAG TTT CTG TAG-56 |
| CHE2 | M. chelonae c | 67.75 | AJ291584 | 55-AGT GGA TGC ATG CTT GGT GAA-75 |
| DIE | M. diernhoferi | 66.16 | AF186463 | 221-GTT CGG ACG GTG TCT GTT GTT-241 |
| FOR1 | M. fortuitum a/c | 66.11 | AJ291587/89 | 89-ACA AAC TTT TTT GAC TGC CAG ACA C-113 |
| FOR2 | M. fortuitum b | 72.55 | AJ291588 | 112-TTT GCG GTG ATG GGA CTG CC-131 |
| FOR3 | M. fortuitum e | 68.51 | AJ291591 | 107-GAT TTT GCG GTG ATG GGA CC-126 |
| FLA | M. flavescens | 65.20 | AJ291586 | 121-GGA ACA CTG CTT TGA GGA ATC AT-143 |
| GAS | M. gastri | 71.46 | X97633 | 79-GTG CGC AAC AGC AAG CAA GC-98 |
| GIL | M. gilvum | 71.85 | AJ314876 | 138-CGG TTT TCC GGG GTT GTG GT-157 |
| GOR | M. gordonae | 67.27 | AJ315574 | 43-CGT GAG GGG TCA TCG TCT GTA-63 |
| HAE | M. hemophilum | 68.86 | AY579400 | 130-CAC TCA GGC TTG TCC CAT GTT G-151 |
| INT | M. intracellulare | 69.21 | AJ306711 | 133-GGT CGA TCC GTG TGG AGT CC-152 |
| KAN1 | M. kansasii a/b | 66.72 | L42263 | 213-GGG TGC GCA ACT GTA AAT GAA T-234 |
| KAN2 | M. kansasii c | 67.74 | L42264 | 150-AAA AGT GCC CCA ATT GGT GG-169 |
| LEN | M. lentiflavum | 69.65 | AF318174 | 266-CCG ACT TTG GTC GAC GTG GT-285 |
| LEP | M. leprae | 65.48 | X56657 | 172-TTG TCC CTC ATC TTT GGT GGT-192 |
| MAL | M. malmoense | 73.38 | Y14184 | 126-AAC ACT CGG CCA GTC CGC GT-145 |
| PHL1 | M. phlei a | 70.80 | AJ291596 | 128-AAC ACC GTG TCG AGG ACT GCC-148 |
| PHL2 | M. phlei b | 66.89 | AJ291597 | 107-AAA CGT TTG TCG TTC GGT GG-126 |
| SCR | M. scrofulaceum | 67.40 | AJ314884 | 129-ACT CGG CTC GTT CTG AGT GGT-149 |
| SEN | M. senegalense | 66.28 | Y10385 | 55-GTG AGG AGT CTG TGC GCT GTA G-72 |
| SHI | M. shimoidei | 66.34 | X99219 | 84-TGC ACA ACA ACA AGC GAG AAG-104 |
| SIM | M. simiae | 65.16 | Y14186 | 138-ACT TCG GTT GAA GTG GTG TCC-158 |
| SME | M. smegmatis | 68.82 | AJ291599 | 91-AAC GTT GAG ATG CGG TGT GGT-110 |
| SZU | M. szulai | 69.36 | X99220 | 131-AGG CTT GGC CAG AGC TGT TGT-1513 |
| TER | M. terrae | 65.90 | AJ314868 | 93-GTG CAG AGG AAT TAC GAA CAA CAA-116 |
| TRI | M. triplex | 69.21 | AF214587 | 84-CAA CAG CAG ACA ATC GCC AGA C-105 |
| MTBC | M. tuberculosis complex | 68.04 | AJ315568 | 79-TGC ATG ACA ACA AAG TTG GCC-99 |
| ULC/MAR | M. ulcerans/M. marinum | 61.52 | X99217 | 124-AAC ATC TCT GTT GGT TTC GG-143 |
| VAC | M. vaccae | 72.60 | AJ291600 | 134-AAT GCC GGC GAG GGA AAT CA-153 |
| XEN | M. xenopi | 69.39 | AJ314866 | 66-TGT TGG GCA GCA GGC AGT AAC-86 |
Identification of subtypes a and b of M. chelonae, M. fortuitum, M. kansasii, and M. phlei was based on sequence variations in their ITS regions.
DNA extraction and PCR amplification.
The extraction of DNA was carried out with a commercially available kit (InstaGene Matrix; Bio-Rad Laboratories), according to the manufacturer's specifications. Amplification of an ITS segment was performed with primers Sp1 and Sp2 in a 25-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, 200 μM (each) deoxynucleoside triphosphate (dATP, dGTP, dCTP, and dUTP), 25 pmol of each primer, 0.75 U of Taq DNA polymerase (all reagents were from QIAGEN, Pty. Ltd., Australia), and 5 μl of DNA. The thermal profile involved initial denaturation for 5 min at 96°C and 35 cycles with the following steps: 30 s of denaturation at 94°C, 30 s of annealing at 60°C, and 1 min of extension at 72°C. The final extension was for 10 min at 72°C. The PCR products were analyzed with SRBR Safe DNA gel stain (Molecular Probes Europe BV, The Netherlands).
Nucleotide sequencing.
The purified PCR products were directly sequenced with forward primers Sp1 and Sp2 by using an Applied Biosystems model 373A DNA sequencer (Perkin-Elmer Applied Biosystems) and a BigDye Terminator cycle sequencing kit. Sequences were identified by using the FastA program group accessed through WebANGIS.
RLB assay.
The RLB assay was performed by using a system which has previously been described in detail (28, 31). Briefly, the slots in a Miniblotter 45 apparatus (Immunetics) were filled with 150 μl of optimized concentrations of probe solution. Each PCR product was denatured and immediately chilled on ice. Hybridization was performed at 60°C for 40 min. The washed membrane was incubated in peroxidase-labeled streptavidin conjugate (Roche, Mannheim, Germany) at 42°C for 40 min. The washed membrane was then incubated in chemiluminescence blotting substrate (ECL Direct system; Roche) for 1 min, covered with Hyperfilm X-ray film (Amersham) for detection of chemiluminescence, and exposed to the X-ray film for 7 min.
Sensitivities of species-specific PCR and ITS PCR-RLB assay.
M. tuberculosis H37Rv and M. abscessus stock suspensions were prepared from cultures after 2 and 3 weeks of growth, respectively, on Löwenstein-Jensen slants and adjusted to a 0.5 McFarland standard. DNA was extracted as described above. The detection limits of the individual species-specific PCR and the PCR-RLB assay were determined by amplification and hybridization of the DNA of each species serially diluted from 5 ng/μl to 500 fg/μl with molecular biology-grade water and tested in parallel.
RESULTS
Sensitivities of PCR-RLB assay.
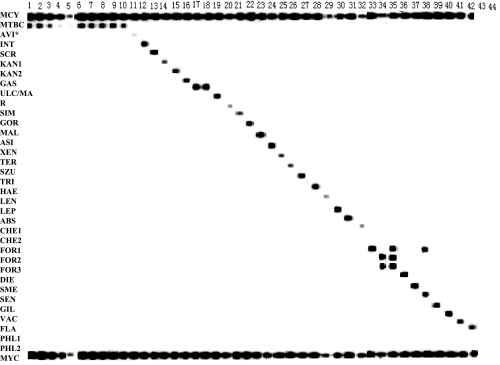
The detection limits of the species-specific PCRs for both M. tuberculosis H37Rv and M. abscessus DNA were 5 pg/μl (data not shown). The PCR-RLB assay was 10 times more sensitive than the species-specific PCR assays for both M. tuberculosis H37Rv (Fig. 1) and M. abscessus DNA (data not shown); i.e., it was able to detect 500 fg/μl. The MYC (genus-specific) probe was more sensitive (50 fg/μl) than the MTBC (species-specific) probe for the detection of M. tuberculosis H37Rv (Fig. 1) but was less sensitive (500 fg/μl) than the ABS probe (50 fg/μl) for the detection of M. abscessus (data not shown).
FIG. 1.
Hybridization results with Mycobacterium species and other bacteria by RLB assay. Lanes 1 to 5, dilutions of M. tuberculosis H37Rv (500 pg/μl, 50 pg/μl, 5 pg/μl, 500 fg/μl, and 50 fg/μl, respectively); lane 6, M. tuberculosis H37Rv; lane 7, M. bovis; lane 8, M. microti; lane 9, M. africanum; lane 10, BCG; lane 11, M. avium; lane 12, M. intracellulare; lane 13, M. scrofulaceum; lane 14, M. kansasii a/b; lane 15, M. kansasii c; lane 16, M. gastri; lane 17, M. ulcerans; lane 18, M. marinum; lane 19, M. simiae; lane 20, M. gordonae; lane 21, M. triplex; lane 22, M. malmoense; lane 23, M. asiaticum; lane 24, M. shimoidei; lane 25, M. xenopi; lane 26, M. terrae; lane 27, M. szulai; lane 28, M. lentiflavum; lane 29, M. hemophilum; lane 30, M. lepre; lane 31, M. abcessus; lane 32, M. chelonae a/b; lane 33, M. fortuitum a/c; lane 34, M. fortuitum b; lane 35, M. fortuitum e; lane 36, M. diernhoferi; lane 37, M. smegmatis; lane 38, M. senegalense; lane 39, M. gilvum; lane 40, M. vacca; lane 41, M. flavescens; lane 42, M. phlei a; lane 43, R. equi; lane 44, N. farcinica. *, a weakly hybridizing band (lane 11, M. avium) was due to a low input DNA concentration.
Specificity of species-specific mycobacterial PCR-RLB assay.
PCR amplification with primers Sp1 and Sp2 of DNA from all 78 mycobacterium reference strains produced single bands ranging in length from 200 to 320 bp. The amplicons from the slowly growing Mycobacterium species were smaller (200 to 250 bp) than those from the rapidly growing species (250 to 320 bp). M. xenopi produced the smallest product (∼200 bp), and M. gilvum produced the largest product (∼320 bp).
There were cross-reactions between the Mycobacterium genus-specific primers (Sp1 and Sp2) and DNA extracts from Nocardia spp. and Rhodococcus spp., which produced two to three copies of the amplicon (Fig. 2). However the genus-specific probe (MYC) hybridized only with Mycobacterium spp., and with a few exceptions, all species-specific probes hybridized only with the corresponding species. The exceptions were as follows: the probe for M. marinum cross-reacted with M. ulcerans because the ITS regions of these two species are identical, one M. senegalense reference strain (ATCC 35796) reacted with the FOR1 probe (specific for M. fortuitum a/c and M. senegalense), and one M. fortuitum subsp. fortuitum (ATCC 6841) reference strain did not react with the SEN probe (specific for M. senegalese) (Fig. 1). The PCR-RLB assay allowed differentiation of M. chelonae from M. abscessus, M. gastri from M. kansasii, and M. avium from M. intracellulare. M. tuberculosis complex subspecies could not be distinguished from each other.
FIG. 2.
PCR results for Mycobacterium species and other bacteria obtained by using primers Sp1 and Sp2 (based on the 16S-23S rRNA gene ITS). Lanes: M, DNA size standards (Hinf DNA; 24, 40, 66, 82, 118, 140, 151, 200, 249, 311, 413, 417, 427, 500, 553, 713, and 726 bp); 1, M. smegmatis; 2, M. triplex; 3, M. fortuitum; 4, M. scrofulaceum; 5, M. flavescens; 6, M. lepre; 7, M. tuberculosis; 8, M. simiae; 9, M. gilvum; 10, M. ulcerans; 11, M. abcessus; 12, M. szulai; 13, M. avium; 14, N. farcinica; 15, R. equi; 16, K. pneumoniae. Note the cross-reactions with Nocardia and Rhodococcus species, which are resolved by RLB assay.
PCR-RLB assay with clinical isolates of mycobacteria.
A total of 219 clinical isolates were tested “blind” (i.e., without knowledge of the species identity) by the PCR-RLB assay. The PCR-RLB assay was repeated with the few isolates for which there were discrepancies between the results of the PCR-RLB assay and those of GC or HPLC (Table 2). The PCR-RLB assay (including duplicate tests) and sequencing results were identical for all isolates. Three isolates that were identified as members of the M. avium-M. intracellulare complex by GC were identified as M. intracellulare by the PCR-RLB assay. Of 23 isolates identified as M. avium by HPLC, 11 were identified as M. avium, 10 were identified as M. intracellulare, and 2 were identified as M. avium mixed with M. kansasii a/b by the PCR-RLB assay. Of 28 isolates identified as M. chelonae-M. abscessus complex by GC, 26 were identified as M. abscessus, 1 was identified as M. chelonae a/b, and 1 was identified as M. chelonae c by the PCR-RLB assay. Of four isolates identified as M. chelonae by HPLC, three were identified as M. chelonae a/b and one was identified as M. chelonae c by the PCR-RLB assay. Two clinical isolates that had been reported as resembling M. fortuitum by HPLC were shown by the PCR-RLB assay and sequencing to be M. triplex, and the identities of two clinical isolates presumptively identified as M. lentiflavum by HPLC were confirmed to be M. lentiflavum by PCR-RLB and sequencing.
In all, the PCR-RLB assay identified 22 mixed cultures (Table 1). In addition to two with a mixture of M. avium and M. kansasii, there were three in which two potentially pathogenic species were identified: M. chelonae and M. fortuitum, M. tuberculosis complex and M. intracelluare, and M. scrofulaceum and M. intracelluare. Four cultures initially identified as M. gordonae (usually a contaminant) were mixed with another species, and 13 cultures apparently contained mixtures of M. fortuitum subtypes.
DNA from two of the three biopsy specimens from patients with clinical leprosy hybridized with the LEP probe and were confirmed by sequencing to contain M. leprae. The other hybridized with both the MTB and the INT probes, but the sequencing result was uninterpretable, suggesting that both M. tuberculosis and M. intracellulare were present. In addition to this clinical specimen, the PCR-RLB assay identified mixed species in 14 separate clinical isolates and the sequencing results were uninterpretable.
DISCUSSION
In comparison to conventional biochemical methods, GC and HPLC provide rapid results. However, both require expensive instrumentation, reproducible patterns depend on standardized conditions of growth, and interpretation of the results is not always easy. The PCR-RLB assay does not require standardized growth conditions, and it is more accurate and objective than either GC or HPLC. In our study, two clinical isolates which had presumptively been identified as M. fortuitum by HPLC were identified by the PCR-RLB assay as M. triplex, and this result was confirmed by sequencing. The PCR-RLB assay identified mixed mycobacterial cultures which had not been recognized by HPLC or GC and which gave uninterpretable sequencing results. Some previously unidentified species were potentially clinically significant. The finding of four mixed cultures initially identified as containing only M. gordonae indicates that this contaminant Mycobacterium species can mask potential pathogens. It is not clear whether the high proportion of mixed subtypes of M. fortuitum indicates that cross-reactions between probes occur or that mixtures of subtypes commonly occur in nature.
In addition, the PCR-RLB assay was able to discriminate between M. chelonae and M. abscessus, which could not be distinguished by GC, and between M. avium and M. intracellulare, which could not be distinguished by HPLC (Table 1).
Sequencing of the 16S rRNA gene is often regarded as the “gold standard” for the identification of mycobacteria (5), but it has a demonstrated lower variability with several examples of species difficult to separate from the genus Mycobacterium (10, 25). Some Mycobacterium species share the same 16S rRNA gene sequence (M. kansasii and M. gastri, M. senegalense and M. farcinogenes) or have very similar 16S rRNA gene sequences (e.g., M. malmoense and M. szulgai) (24).
The 16S-23S rRNA gene ITS region has been identified as a potentially suitable target for probes that can differentiate closely related species. It contains both conserved and highly variable signatures and is rather small. The 16S-23S rRNA gene ITS-based PCR produces a relatively small PCR product (200 to 350 bp) (Fig. 2). Roth et al. reported that PCR based on the 16S-23S rRNA gene ITS and restriction fragment length polymorphism (RFLP) analysis is a promising method with advantages over the previously used hsp65 gene-based method for the reliable and easy identification of mycobacteria (19, 20). However, it has been impeded by difficulties, such as minor differences in band sizes between some species and the occurrence of new patterns not previously reported (17, 27). Moreover, RFLP analysis cannot reliably detect mixtures of species within samples. Others have also found that the presence of more than one species in a culture or a clinical sample makes it difficult or impossible to identify the species present by sequencing or RFLP analysis of 16S-23S rRNA gene ITS PCR products (10).
In our study, from the analysis of 78 reference strains, 219 clinical isolates, and three skin biopsy specimens, the specificity of our PCR-RLB assay, based on the 16S-23S rRNA gene ITS region, was close to 100% (except that M. senegalense reference strain ATCC 35796 reacted with the FOR1 probe). Several other systems that are similar to our PCR-RLB assay system are available: the INNO-LiPA strip (version 2; Immunogenetics) (15, 23, 26), the GenoType Mycobacterium system (Hain Life Science) (13), and an oligonucleotide array (16) can identify 16, 13, and 19 Mycobacterium species, respectively. Our PCR-RLB assay based on the 16S-23S rRNA gene ITS can differentiate 34 mycobacterial species and subspecies by the use of 35 probes and has a high degree of specificity.
Our assay is as simple as other reverse hybridization methods, but it is more sensitive because it uses a nonprecipitating enhanced chemiluminescent substrate and Hyperfilm X-ray film. It successfully identified mycobacteria directly in skin biopsy specimens and has the potential to be used for the direct identification of Mycobacterium species from other types of clinical specimens. In addition, it has the advantage that the nylon filters can be stripped and reused at least 30 times and therefore is relatively inexpensive.
In conclusion, the PCR-RLB assay method described here is faster than conventional biochemical tests, is more accurate than HPLC or GC, is less expensive than sequencing, and does not require specialized instrumentation. It is more discriminatory than methods based on 16S rRNA gene-specific PCR or PCR-RFLP analysis of other genes (e.g., rpoB) and identifies more species than other PCR-RLB assays based on the 16S-23S rRNA gene ITS. It has the potential to be used to identify Mycobacterium species directly in clinical specimens.
Acknowledgments
We thank Zhihuei Liu (Guangzhou Thoracic Hospital, China) for GC; William Chew (Mycobacterium Reference Laboratory, Centre for Infectious Diseases and Microbiology, Sydney, Australia) for HPLC; and Guohua Mei, Feng Wang, and Junxin Huang (Shenzhen Chronic Disease Hospital, China), Guozhi Wang, Baowen Chen, and Miao Xv (National Institute for the Control of Pharmaceutical and Biological Products, China), and William Chew for providing isolates and reference strains. We thank Peter Jelfs for support and Mark Wheeler and Ilya Henner for help with DNA sequencing.
REFERENCES
- 1.Blackwood, K. S., C. He, J. Gunton, C. Y. Turenne, J. Wolfe, and A. M. Kabani. 2000. Evaluation of recA sequences for identification of Mycobacterium species. J. Clin. Microbiol. 38:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler, W. R., K. C. Jost, Jr., and J. O. Kilburn. 1991. Identification of mycobacteria by high-performance liquid chromatography. J. Clin. Microbiol. 29:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fletcher, H. A., H. D. Donoghue, J. Holton, I. Pap, and M. Spigelman. 2003. Widespread occurrence of Mycobacterium tuberculosis DNA from 18th-19th century Hungarians. Am. J. Phys. Anthropol. 120:144-152. [DOI] [PubMed] [Google Scholar]
- 4.Kasai, H., T. Ezaki, and S. Harayama. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim, B. J., S. K. Hong, K. H. Lee, Y. J. Yun, E. C. Kim, Y. G. Park, G. H. Bai, and Y. H. Kook. 2004. Differential identification of Mycobacterium tuberculosis complex and nontuberculous mycobacteria by duplex PCR assay using the RNA polymerase gene (rpoB). J. Clin. Microbiol. 42:1308-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim, H., S. H. Kim, T. S. Shim, M. N. Kim, G. H. Bai, Y. G. Park, S. H. Lee, C. Y. Cha, Y. H. Kook, and B. J. Kim. 2005. PCR restriction fragment length polymorphism analysis (PRA)-algorithm targeting 644 bp heat shock protein 65 (hsp65) gene for differentiation of Mycobacterium spp. J. Microbiol. Methods 62:199-209. [DOI] [PubMed] [Google Scholar]
- 7.Kim, S. Y., Y. J. Park, E. Song, H. Jang, C. Kim, J. Yoo, and S. J. Kang. 2006. Evaluation of the CombiChip MycobacteriaTM drug-resistance detection DNA chip for identifying mutations associated with resistance to isoniazid and rifampin in Mycobacterium tuberculosis. Diagn. Microbiol. Infect. Dis. 54:203-210. [DOI] [PubMed] [Google Scholar]
- 8.Kirschner, P., and E. C. Bottger. 1998. Species identification of mycobacteria using rDNA sequencing. Methods Mol. Biol. 101:349-361. [DOI] [PubMed] [Google Scholar]
- 9.Kox, L. F., H. M. Jansen, S. Kuijper, and A. H. Kolk. 1997. Multiplex PCR assay for immediate identification of the infecting species in patients with mycobacterial disease. J. Clin. Microbiol. 35:1492-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kox, L. F., J. van Leeuwen, S. Knijper, H. M. Jansen, and A. H. Kolk. 1995. PCR assay based on DNA coding for 16S rRNA for detection and identification of mycobacteria in clinical samples. J. Clin. Microbiol. 33:3225-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lappayawichit, P., S. Rienthong, D. Rienthong, C. Chuchottaworn, A. Chaiprasert, W. Panbangred, H. Saringcarinkul, and P. Palittapongarnpim. 1996. Differentiation of Mycobacterium species by restriction enzyme analysis of amplified 16S-23S ribosomal DNA r sequences. Tuber. Lung Dis. 77:257-263. [DOI] [PubMed] [Google Scholar]
- 12.Lebrun, L., F. X. Weill, L. Lafendi, F. Houriez, F. Casanova, M. C. Gutierrez, D. Ingrand, P. Lagrange, V. Vincent, and J. L. Herrmann. 2005. Use of the INNO-LiPA-MYCOBACTERIA assay (version 2) for identification of Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum complex isolates. J. Clin. Microbiol. 43:2567-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makinen, J., M. Marjamaki, H. Marttila, and H. Soini. 2006. Evaluation of a novel strip test Genotype Mycobacteriun CM/AS, for species identification of mycobacterial cultures. Clin. Microbiol. Infect. 12:481-483. [DOI] [PubMed] [Google Scholar]
- 14.Mukai, T., Y. Miyamoto, T. Yamazaki, and M. Makino. 2006. Identification of Mycobacterium species by comparative analysis of the dnaA gene. FEMS Microbiol. Lett. 254:232-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padilla, E., V. Gonzalez, J. M. Manterola, A. Perez, M. D. Quesada, S. Gordillo, C. Vilaplana, M. A. Pallares, S. Molinos, M. D. Sanchez, and V. Ausina. 2004. Comparative evaluation of the new version of the INNO-LiPA Mycobacteria and GenoType Mycobacterium assays for identification of Mycobacterium species from MB/BacT liquid cultures artificially inoculated with mycobacterial strains. J. Clin. Microbiol. 42:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park, H., H. Jang, E. Song, C. L. Chang, M. Lee, S. Jeong, J. Park, B. Kang, and C. Kim. 2005. Detection and genotyping of Mycobacterium species from clinical isolates and specimens by oligonucleotide array. J. Clin. Microbiol. 43:1782-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richter, E., S. Niemann, S. Rusch-Gerdes, and S. Hoffner. 1999. Identification of Mycobacterium kansasii by using a DNA probe (AccuProbe) and molecular techniques. J. Clin. Microbiol. 37:964-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richter, E., M. Weizenegger, A. M. Fahr, and S. Rusch-Gerdes. 2004. Usefulness of the GenoType MTBC assay for differentiating species of the Mycobacterium tuberculosis complex in cultures obtained from clinical specimens. J. Clin. Microbiol. 42:4303-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roth, A., M. Fischer, M. E. Hamid, S. Michalke, W. Ludwig, and H. Mauch. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth, A., U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer, and H. Mauch. 2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol. 38:1094-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanguinetti, M., B. Posteraro, F. Ardito, S. Zanetti, A. Cingolani, L. Sechi, A. De Luca, L. Ortona, and G. Fadda. 1998. Routine use of PCR-reverse cross-blot hybridization assay for rapid identification of Mycobacterium species growing in liquid media. J. Clin. Microbiol. 36:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soini, H., and M. K. Viljanen. 1997. Diversity of the 32-kilodalton protein gene may form a basis for species determination of potentially pathogenic mycobacterial species. J. Clin. Microbiol. 35:769-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Bottger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suffys, P. N., R. A. da Silva, M. de Oliveira, C. E. Campos, A. M. Barreto, F. Portaels, L. Rigouts, G. Wouters, G. Jannes, G. van Reybroeck, W. Mijs, and B. Vanderborght. 2001. Rapid identification of mycobacteria to the species level using INNO-LiPA Mycobacteria, a reverse hybridization assay. J. Clin. Microbiol. 39:4477-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takewaki, S., K. Okuzumi, I. Manabe, M. Tanimura, K. Miyamura, K. Nakahara, Y. Yazaki, A. Ohkubo, and R. Nagai. 1994. Nucleotide sequence comparison of the mycobacterial dnaJ gene and PCR-restriction fragment length polymorphism analysis for identification of mycobacterial species. Int. J. Syst. Bacteriol. 44:159-166. [DOI] [PubMed] [Google Scholar]
- 26.Tortoli, E., A. Mariottini, and G. Mazzarelli. 2003. Evaluation of INNO-LiPA MYCOBACTERIA v2: improved reverse hybridization multiple DNA probe assay for mycobacterial identification. J. Clin. Microbiol. 41:4418-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson, R. W., V. A. Steingrube, E. C. Bottger, B. Springer, B. A. Brown-Elliott, V. Vincent, K. C. Jost, Jr., Y. Zhang, M. J. Garcia, S. H. Chiu, G. O. Onyi, H. Rossmoore, D. R. Nash, and R. J. Wallace, Jr. 2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int. J. Syst. Evol. Microbiol. 51:1751-1764. [DOI] [PubMed] [Google Scholar]
- 28.Xiong, L., F. Kong, H. Zhou, and G. L. Gilbert. 2006. Use of PCR and reverse line blot hybridization assay for rapid simultaneous detection and serovar identification of Chlamydia trachomatis. J. Clin. Microbiol. 44:1413-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zelazny, A. M., L. B. Calhoun, L. Li, Y. R. Shea, and S. H. Fischer. 2005. Identification of Mycobacterium species by secA1 sequences. J. Clin. Microbiol. 43:1051-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zolg, J. W., and S. Philippi-Schulz. 1994. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J. Clin. Microbiol. 32:2801-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zwart, G., E. J. van Hannen, M. P. Kamst-van Agterveld, K. Van der Gucht, E. S. Lindstrom, J. Van Wichelen, T. Lauridsen, B. C. Crump, S. K. Han, and S. Declerck. 2003. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl. Environ. Microbiol. 69:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]