Abstract
Macrophages (Mφ) may play an important role in the pathogenesis of invasive meningococcal infection. Previously, we have shown that the class A Mφ scavenger receptor (SR-A) is a major nonopsonic receptor for Neisseria meningitidis on Mφ. SR-A contributes to host defense by binding proinflammatory polyanionic ligands such as lipopolysaccharide (LPS) and by the uptake and killing of live organisms. SR-A-deficient mouse Mφ display a substantial reduction in the number of meningococci ingested compared to wild-type Mφ, and SR-A is required for meningococcal phagocytosis but not for the release of tumor necrosis factor alpha. Although soluble lipid A and lipidIVA are reported as ligands for SR-A, we demonstrated that LPS and LPS expression were not essential for the uptake of whole meningococci. In the present study, we set out to discover protein ligand(s) for SR-A in N. meningitidis lysates and outer membrane vesicles. Using various microbial mutant strains, we determined that molecules comprising the membrane capsule and pili, as well as the abundant surface Opa proteins were not essential for SR-A recognition. We developed a binding assay to detect SR-A ligands and identified three candidate proteins expressed on intact organisms, namely, NMB1220, NMB0278, and NMB0667. Soluble forms of these ligands were shown to block the binding of meningococci to CHO cells stably transfected with SR-A. Furthermore, NMB1220 was endocytosed by SR-A on Mφ and prevented internalization of soluble acetylated low-density lipoprotein. Thus, we have identified novel, unmodified protein ligands for SR-A that are able to inhibit meningococcal interactions with macrophages in vitro.
Neisseria meningitidis is a commensal organism of the nasopharynx which can occasionally cross this barrier to infect individuals, causing classical meningitis or potentially fatal septicemia. Meningococcal infections are particularly life-threatening in infants once protection from maternal antibodies diminishes (2, 32). Many aspects of the host response to meningococcal infection remain obscure, especially those governing the initial host-microbe interactions which result in infection, colonization, or clearance. Also, it is unclear what factors lead to the progression of fulminant septicemia in some individuals but not others.
The role of macrophages (Mφ) in this disease has received little attention; however, they are important during the initial stages of infection (27), dissemination (16), inflammation (3, 26) and possibly in the generation of acquired immunity. Mφ are found systemically, in the nasopharynx, and at the blood-brain interface; also, monocytes are readily recruited to sites of infection and local inflammation (7). Mφ express many pattern recognition receptors capable of distinguishing the presence of pathogens directly, including the Toll-like receptors (TLR), CD14, and the scavenger receptors (SR) (17, 21). Recently, we found that bone marrow culture-derived Mφ bind unopsonized N. meningitidis almost exclusively via the class A Mφ scavenger receptor (SR-A), a trimeric transmembrane glycoprotein expressed on mature Mφ but not on circulating monocytes (18). Three naturally occurring alternative splice variants—SR-AI, SR-AII, and SR-AIII—exist, although only SR-AI and SR-AII have been shown to be functional (8). Ligand-binding studies have shown a diverse range of polyanionic molecules are recognized by the receptor, including modified (acetylated and oxidized) but not native low-density lipoprotein (LDL), polyribonucleic acids [poly(I)] and polysaccharides, such as dextran sulfate (13). Proteins modified by maleylation are also bound by the receptor (1, 19); however, it is important to note that not all polyanionic molecules are recognized since there is an undetermined structural requirement.
Previously, we showed that bone marrow culture-derived Mφ (BMMφ) bind N. meningitidis predominantly via SR-A (18). Mφ lacking the receptor have a 90% reduction in ingestion of meningococci compared to wild-type controls. Our data demonstrated that, whereas SR-A was required for bacterial entry, it was not responsible for Mφ activation and the release of the proinflammatory cytokine tumor necrosis factor alpha (TNF-α), which required a functional TLR4 and bacterial lipopolysaccharide (LPS). Also, all of the Neisseria species tested, including N. gonorrhoeae and N. lactamica, were recognized similarly by SR-A. LPS was a candidate meningococcal ligand for SR-A since soluble lipid A had previously been shown to be recognized by the receptor (9). However, using various mutants, including one without any LPS expression, we showed that the requirement for SR-A was unchanged (18). These data suggested the presence of novel ligand(s) for SR-A on N. meningitidis.
We set out to identify the ligand(s) for SR-A on meningococci. The cell surface of Neisseria is heterogeneous, and the bacteria express a large number of surface molecules that undergo phase variation. In addition to LPS, potential candidate ligands for SR-A include capsular polysaccharide, pili, porins, and Opa proteins, and all of these are important during colonization and infection. For example, capsule polysaccharide is required for serum resistance (4, 14), and pili are required for initial attachment to epithelial cells via CD46 (11, 12, 33, 34). However, using various microbial surface mutants, we determined that these molecules were not recognized by SR-A. Therefore, we developed a binding assay to identify SR-A ligands and have isolated three candidate surface molecules: NMB1220, NMB0278, and NMB0667. Further study has revealed that these proteins are able to block the binding of meningococci and acetylated low-density lipoprotein (AcLDL) to SR-A to various degrees. However, only NMB1220 induced endocytosis by SR-A in Mφ. Thus, we have identified novel, unmodified protein ligands for the receptor that are able to inhibit meningococcal interaction with host macrophages.
MATERIALS AND METHODS
Chemicals and reagents.
Unless stated otherwise, all chemicals were from Sigma. DiI (1,1′-dioctadecyl-1-3,3,3′,3′-tetramethylindocarbocyanine perchlorate)-labeled AcLDL and AcLDL were obtained from Autogen-Bioclear (Wiltshire, United Kingdom). Rhodamine green X (RdGnX) was obtained from Molecular Probes (Eugene, OR); a 30% (wt/vol) solution of acrylamide-bisacrylamide (37.5:1) was from NBL Gene Science, Ltd. (Cramlington, United Kingdom). TMB substrate reagent set was purchased from BD Biosciences Pharmingen (San Diego, CA). Nonfat powder milk (Marvel Original) was purchased from local supermarkets; Hybond C+ membrane, ECL reagent, and Rainbow molecular weight protein markers were from Amersham Life Sciences, Ltd. (Buckinghamshire, United Kingdom).
Cell culture.
BMMφ were prepared as described previously (18). Mφ were routinely cultured in RPMI supplemented with 50 IU of penicillin G/ml, 50 μg of streptomycin/ml, and 2 mM l-glutamine (PSG); 10% fetal calf serum; and 15% L-cell conditioned medium. Before use in assays, the Mφ were plated in bacteriologic plastic (Greiner, Gloucester, United Kingdom) dishes at the appropriate density. CHO wild-type (WT) cells were cultured in Ham F-12 medium supplemented with 10% fetal calf serum and PSG. CHO cells stably expressing hSR-AI and hSR-AII have been described previously (6), and high levels of SR-A expression were achieved by culture in MAC medium: Ham F-12 medium supplemented with 3% LDL-deficient fetal calf serum, 250 μM mevalonate, 40 μM mevastatin, and 3 μg of AcLDL/ml.
Bacteria.
N. meningitidis MC58, surface mutants (MC58⊄3 to MC58⊄10), H44/76, and H44/76lpxA were obtained and treated as described previously (15, 23). All Neisseria strains were maintained in liquid nitrogen and used after a single subculture. Neisseria strains were grown on brain heart infusion media (Merck), supplemented with Levinthal's reagent (10% [vol/vol]) and agar (1% [wt/vol]; Bioconnections) at 37°C in 5% CO2 (vol/vol). For fluorescent labeling, N. meningitidis was fixed with 70% ethanol and labeled with RdGnX according to the manufacturer's protocol.
Purification of meningococcal proteins.
The purified N. meningitidis proteins used were expressed in Escherichia coli as previously described (22). In brief, for soluble proteins E. coli BL21(DE3) harboring the gene of interest was grown at 37°C to an optical density at 550 nm (OD550) of 0.6 to 0.8 in Luria broth medium containing 100 mg of ampicillin/ml. Expression of the recombinant protein was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and the culture shaken for 3 h at 37°C. After induction, the bacteria were pelleted by centrifugation at 8,000 × g for 15 min at 4°C and resuspended in 50 mM phosphate buffer (pH 8) containing 300 mM NaCl; 10 mM imidazole; and a complete, EDTA-free protease inhibitor (Roche, Mannheim, Germany). All subsequent procedures were performed at 4°C. Cells were disrupted by sonication before debris and membranes were pelleted by centrifugation at 16,000 × g for 30 min and discarded. The supernatant was loaded onto a metal-chelate affinity chromatography column and extensively washed with 10 mM imidazole, 20 mM imidazole, and 50 mM imidazole in the phosphate buffer used above for the pellet resuspension. Proteins bound to the column were purified in a single-step elution with 250 mM imidazole in phosphate buffer. For insoluble protein purification, fusion proteins were produced as insoluble inclusion bodies, solubilized with urea, and renatured after purification as described previously (5). The fusion proteins were purified by affinity chromatography on Ni2+-conjugated chelating fast flow Sepharose (Pharmacia). The purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of 10 μg samples of each protein, and the protein concentration was quantified by using Bradford reagent (Bio-Rad).
Uptake assays.
All cells were plated 24 to 48 h before use. Unless stated otherwise, BMMφ were plated in six-well bacteriologic plastic dishes at a density of 106 Mφ per well, and CHO cells were plated in 24-well dishes at 2 × 105 cells per well. During the assay all cells were cultured in OptiMEM culture medium. Before use, the Mφ and CHO cells were washed twice in phosphate-buffered saline (PBS) and then incubated in culture medium containing fluorescence-labeled bacteria as indicated. As required, cells were preincubated for 30 min with inhibitor, which was retained throughout the assay. Unless otherwise stated, poly(I) and its cognate nonligand, poly(C) (polycytidylic acid) were used at 50 μg/ml; the rat anti-murine SR-A antibody, 2F8, and its isotype-matched control, CAMPATH 1G, were used at 15 μg/ml, and fucoidan was used at 20 μg/ml. The endocytosis of 2.5 μg of DiI-labeled acetylated low density lipoprotein (DiIAcLDL)/ml was examined as a functional control for SR-A in assays. After incubation with bacteria, the culture medium was removed, and the cells were washed three times in PBS. Cells for flow cytometry were harvested with 0.1% (wt/vol) trypsin and 10 mM EDTA in PBS for CHO cells or PBS containing 10 mM EDTA and 4 mg of lidocaine-HCl/ml for Mφ and then fixed with 4% (vol/vol) formaldehyde in PBS. For the detection of His protein uptake by cells, the cells were permeabilized with PBS supplemented with 0.2% saponin, 1% bovine serum albumin (BSA), 5 mM EDTA, 5% normal goat serum, 5% normal rabbit serum, and 10 μg of 2.4G2 (a rat anti-mouse Fc receptor antibody)/ml after fixation. HIS was detected by using a goat anti-His fluorescein isothiocyanate-labeled antibody. Fluorescence was analyzed on a FACScan (Becton Dickinson, Mountain View, CA) using the FL-1 or FL-2 photomultiplier where appropriate, and the results were analyzed by using CellQuest software. The mean fluorescence of unloaded control cells was subtracted from the mean fluorescence of each assay condition, and the average was determined. The results are representative of at least three independent experiments. The statistical significance of results was determined by using a paired Student t test, and significance was tested at the 95% confidence level (P ≤ 0.05).
Cell lysates.
Cells were cultured in a 15-cm petri dish and, after five washes in ice-cold PBS, were lysed by using 1 ml of NP-40 protein lysis buffer (150 mM NaCl, 10 mM EDTA, 10 mM NaN3, 10 mM Tris [pH 8.0], 1 mM phenylmethylsulfonyl fluoride, 5 mM iodoacetamide, and 1% Nonidet P-40) per 107 cells. Lysates were then centrifuged for 15 min at 12,000 × g at 4°C to remove nuclei and cellular debris. Supernatant aliquots were stored at −80°C until required. The protein concentrations of the lysates were measured by using a bicinchoninic acid protein assay reagent Kit (Pierce Chemical Company, Chester, United Kingdom), and the absorbance at 550 nm was determined by using an Anthos HTII automated plate reader. Bacterial lysates were prepared by repeated freeze-thaw cycles at −20°C in the presence of protease inhibitors after washes with PBS.
Far-Western blots.
Approximately 10 to 20 μg of total cellular protein was analyzed per lane on an SDS-10% PAGE gel, and separated proteins were transferred electrophoretically to Hybond C+ nitrocellulose membranes. Blotted membranes were placed in blocking buffer (PBS plus 5% [wt/vol] powdered milk and 0.1% Tween 20) for at least 1 h. The blots were overlaid with postnuclear supernatant prepared from WT or SR-A−/− BMMφ (700 μg/ml as determined by titration experiments) for 2 h in the presence of 5 mM EDTA unless otherwise stated. Membranes were washed three times in blocking buffer and then incubated for 1 h in buffer containing 2F8 (10 μg/ml). After three further washes, the blots were incubated with horseradish peroxidase (HRP)-conjugated anti-rat antibody (1:1,000 in blocking buffer) for an hour. SR-A binding was visualized by chemiluminescence (ECL, Amersham Life Science, Ltd.). Bands detected on blots overlaid with WT, but not SR-A−/−, postnuclear supernatant were deemed to be candidate SR-A ligands.
SR-A-binding enzyme-linked immunosorbent assay (ELISA).
We coated 96-well OPTI-EIA plates overnight with candidate ligands (at 10 μg/ml unless stated otherwise) diluted in PBS at 4°C in replicates of 10. The wells were blocked with 10% LPS-free BSA before incubation with postnuclear supernatant prepared as described above from WT or SR-A−/− BMMφ for 2 h in the presence of 5 mM EDTA. Five wells of each candidate were incubated with WT or SR-A−/−lysate. The wells were washed five times with PBS containing 0.1% Tween 20 and then incubated with 10 μg of 2F8/ml for 2 h at room temperature. Binding of the SR-A antibody was detected by incubating the wells with an HRP-conjugated anti-rat antibody (1:1,000 dilution) and visualized by using TMB reagent according to the manufacturer's instructions, and the absorbance was read at 450 nm. AcLDL or maleylated BSA (malBSA) were included in all assays as a positive control for SR-A. Candidate molecules that were positive when incubated with WT Mφ lysates but negative when the SR-A−/− preparation was used were considered to be potential ligands. The average of the five replicates for each condition was plotted, and the results are representative of at least three independent experiments. The statistical significance of results was determined by using the paired Student t test, and the significance was tested at the 95% confidence level (P ≤ 0.05). Note that, where appropriate, we verified that ligands had coated all of the wells equally by using direct ELISA detection of His-tagged proteins with an anti-His antibody (10 μg/ml) and a goat anti-mouse HRP-labeled (1:1,000 dilution) secondary antibody.
RESULTS
SR-A recognition of N. meningitidis is independent of LPS, capsule, pili, or Opa or Opc proteins.
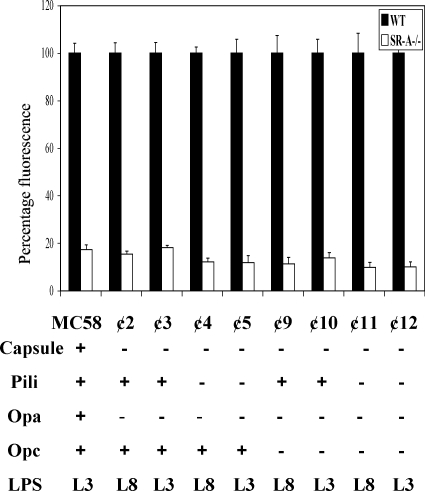
Previously, we have shown that SR-A recognition of unopsonized Neisseria was independent of LPS or LPS expression and present on all strains tested (18). Therefore, to identify the ligand(s) for SR-A on N. meningitidis, we tested potential cell surface ligands by comparing the ability of mutants to bind to WT and SR-A−/− BMMφ. Candidates included the polysaccharide capsule, pili, and outer membrane proteins. WT and SR-A−/− cells were incubated with ethanol-fixed fluorescence-labeled control N. meningitidis or a series of mutant strains (Fig. 1) in the absence of serum for 2 h at 37°C. We have found previously that SR-A recognition of meningococci is not affected by ethanol fixation (18). Association of bacteria with Mφ was measured by flow cytometry. To compare different strains of N. meningitidis, binding to WT Mφ was set at 100%, and the uptake by SR-A−/− cells is presented as a percentage of control Mφ. Compared to control N. meningitidis, mutants lacking pili and capsule had minor variations in overall association with Mφ; however, the difference in uptake between WT and SR-A−/− cells remained ca. 80 to 90%. Similarly, various Opa and Opc mutants had no major reductions in SR-A-mediated binding. Furthermore, several of the mutants included in the study are double and triple mutants with ⊄11 and ⊄12, for example, lacking the expression of capsule, pili, Opa and Opc; however, SR-A-mediated recognition was unchanged in these strains (Fig. 1). Thus, our data show that pili, capsule, and the outer membrane Opa and Opc proteins do not appear to be essential for SR-A-mediated detection of N. meningitidis.
FIG. 1.
The absence of capsule, pili, or Opa proteins does not alter N. meningitidis binding to SR-A. BMMφ from WT and SR-A−/− mice were seeded on six-well bacteriologic plastic plates at 106 cells/well and incubated with RdGnX-labeled ethanol-fixed MC58 or natural mutants lacking capsule, pili, Opa, or Opc for 2 h at 37°C in the absence of serum. The Mφ were washed to remove extracellular bacteria, detached by using lidocaine-EDTA, and fixed with 4% formaldehyde before analysis by flow cytometry. The average increase in fluorescence for triplicate wells of MC58 binding to WT BMMφ was set at 100%, and all other values are expressed as a percentage of the control value and are plotted. All results are from individual experiments that are representative of at least three similar assays.
Characterization of Neisserial ligands for SR-A using far-Western blots.
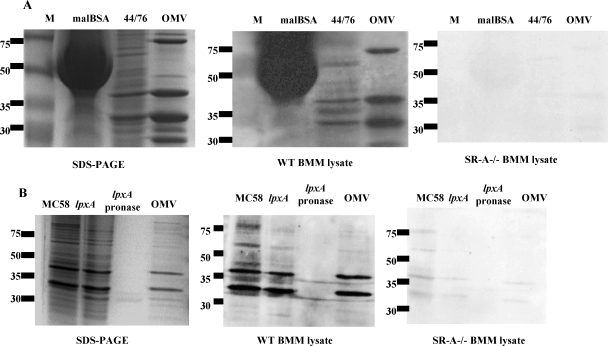
Previously, we observed that paraformaldehyde inactivated the ligand for SR-A on N. meningitidis. No difference in uptake was observed between WT and SR-A−/− cells when incubated with N. meningitidis fixed with paraformaldehyde (not shown). Also, limited trypsin digestion of N. meningitidis prior to use resulted in loss of SR-A recognition (not shown). These data suggest that the ligand(s) for SR-A may contain protein, so a far-Western blot system was set up to learn more about the nature of the ligand(s). N. meningitidis proteins derived from a WT bacterial lysate were separated on a SDS-PAGE gel. Each sample was run in triplicate, and one replicate was used for protein visualization, while the others were transferred to nitrocellulose, which was divided in two and overlaid with postnuclear lysate derived from WT or SR-A−/− Mφ. Binding of SR-A to the separated N. meningitidis proteins was detected by blotting with 2F8, a specific anti-SR-A monoclonal antibody, before detection with an HRP-coupled secondary antibody. Bands detected on the blots were compared to the SDS-PAGE gel for information on molecule size. N. meningitidis proteins that bound SR-A should only be detected when the blots were overlaid with WT, but not SR-A−/− postnuclear cell lysate. For example, malBSA, a known SR-A ligand, was detected strongly by the WT Mφ lysate but not when overlaid with the SR-A−/− cell preparation (Fig. 2A). Interestingly, several N. meningitidis proteins, falling mostly within the 75- to 30-kDa range, were recognized, showing that there were a number of candidates for SR-A. Detection by SR-A in this system was reduced when we titrated both the N. meningitidis preparation and the Mφ cell lysate (not shown). Since SR-A-mediated binding is resistant to EDTA, we added 10 mM EDTA to the Mφ lysate and showed that the inclusion of the chelator did not alter SR-A-mediated recognition of the N. meningitidis proteins (Fig. 2B). To confirm that the potential ligands were proteins, we analyzed the effect of protein digestion. Prior to separation by SDS-PAGE, N. meningitidis cell preparations were preincubated with pronase for 2 h at 37°C (Fig. 2B). Compared to a control N. meningitidis cell preparation, no binding to SR-A was observed when the protease digested sample was overlaid with WT lysate, confirming that potential ligands were proteins or protein associated.
FIG. 2.
Overlay blots reveal several ligands for SR-A ranging between 30 and 75 kDa that are EDTA resistant, protease sensitive, and present in multiple strains. (A) Preparations of control N. meningitidis (H44/76) lysates and malBSA were separated on a SDS-10% PAGE gel (left panel) and transferred to nitrocellulose (middle and right panels). The middle panel shows nitrocellulose overlaid with WT Mφ postnuclear cell lysate, and the right panel shows nitrocellulose overlaid with SR-A−/− cell lysate. The blots were washed, and SR-A binding was detected by incubating the membranes with 2F8 (10 μg/ml) and an anti-rat immunoglobulin G-HRP and visualized by chemiluminescence. (B) Bacterial lysates from H44/76, lpxA, or OMV were analyzed as described above except that the lpxA lysate was divided and half incubated with pronase for 2 h at 37°C before use. The left panel is the corresponding SDS-PAGE gel for the middle and right panels. The middle panel shows one-half overlaid with WT Mφ cell lysate, and right panel shows one-half overlaid with SR-A−/− lysate, both in the presence of 5 mM EDTA. All results are from individual experiments that are representative of at least three similar assays.
The ligand for SR-A should be expressed on the bacterial cell surface. The bacterial lysates used above were derived from whole bacteria. To identify ligands present on the outer surface, we compared preparations derived from whole organisms to those from outer membrane vesicles (OMV) (Fig. 2B). As with whole bacteria, several major bands between 75 and 30 kDa were observed when OMV were overlaid with WT but not SR-A−/− Mφ lysates. A few bands appeared to be enriched in the OMV fraction compared to the whole bacterial cell extract, whereas others were only present in the whole N. meningitidis preparation. We compared N. meningitidis proteins derived from the LPS mutant of N. meningitidis (lpxA) to determine whether SR-A bound proteins similar to those detected using control H44/76 (WT) organisms. Figure 2B shows that the same bands are recognized by SR-A in both the WT and the LPS-free bacteria, confirming our previous data (18). We also observed similar potential ligands derived from N. gonorrhoeae and two strains of N. lactamica, confirming previous data on binding of these strains to SR-A (not shown). However, minor variations in the sizes of the bands were observed.
Thus, our data show that SR-A binds to a number of candidate N. meningitidis molecules. Therefore, to identify the ligands detected by far-Western blot, we pursued a candidate ligand approach based on the molecular weight information generated above and sequence information gathered from the N. meningitidis genome.
SR-A binding ELISA.
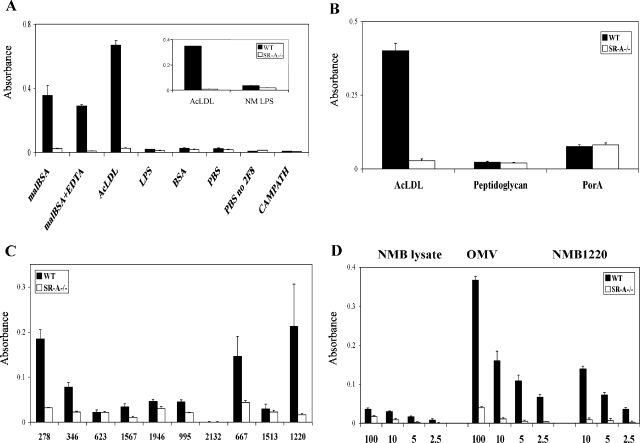
To narrow the search for the SR-A ligand(s), we analyzed the N. meningitidis genome for predicted outer membrane molecules between 30 and 70 kDa and determined their ability to bind SR-A in a novel binding ELISA-type assay (see Table 1 for a list of candidates analyzed with predicted size and annotation). The genes of the outer membrane proteins were cloned and fused with GST or HIS and purified as described previously (22). The candidate ligands were selected for their size and because genome sequence analysis predicted them to be outer membrane proteins, which was confirmed by flow cytometric analysis for surface expression (22, 31). For the SR-A binding ELISA, purified candidate molecules were used to coat wells of a 96-well plate and then incubated with postnuclear cell lysate from WT or SR-A−/− Mφ. Specific SR-A binding to the coated ligands was detected by using 2F8 to identify bound receptor in a colorimetric reaction. Coated wells incubated with SR-A−/− cell lysate were used as a negative control. Using this method we could show specific receptor binding to malBSA and AcLDL, but not native BSA, when coated wells were incubated with WT Mφ lysate but not with SR-A−/− preparations (Fig. 3A). Since soluble LPS is a reported ligand for SR-A, we preincubated wells with phosphate-buffered saline (PBS) or purified LPS from E. coli or N. meningitidis and could not detect an SR-A-dependent signal when incubated with WT Mφ lysates, demonstrating that samples contaminated with LPS would not result in false positives. SR-A detection of positive control ligands was unaltered in the presence of EDTA and could be titrated by using decreasing concentrations of candidate ligand and Mφ lysate. Furthermore, preincubation of the WT lysate with AcLDL blocked subsequent detection of malBSA-coated wells (data not shown). Substitution of CAMPATH, an isotype-matched control antibody for 2F8, resulted in loss of detection of the modified BSA by the WT Mφ lysate.
TABLE 1.
N. meningitidis proteins in this study
| Protein | Molecular mass (kDa) | Annotation |
|---|---|---|
| NMB0278 | 24 | Thiol:disulfide interchange protein DsbA |
| NMB0346 | 28 | Hypothetical protein |
| NMB0623 | 40 | Spermidine/putrescine ABC transporter |
| NMB0667 | 47 | Hypothetical protein |
| NMB0995 | 20 | Mφ infectivity potentiator-related protein |
| NMB1220 | 33 | Stomatin/Mec-2 family protein |
| NMB1513 | 23 | Conserved hypothetical protein |
| NMB1567 | 28 | Mφ infectivity potentiator |
| NMB1946 | 30 | Lipoprotein/similar to HlpA of Haemophilus influenzae; belongs to the NlpA family of lipoproteins |
| NMB2132 | 42 | Lipoprotein; low similarity to transferrin-binding proteins |
FIG. 3.
A newly developed SR-A binding assay identified protein ligands for SR-A. (A) Quintuplicate wells on a 96-well plate were coated overnight with 10 μg of known ligands for SR-A (malBSA, AcLDL) and compounds that do not bind the receptor (BSA)/ml. Uncoated wells incubated with PBS alone served as a control for nonspecific binding. Next, the wells were incubated with WT or SR-A−/− BMMφ postnuclear lysate, and binding to the receptor was detected with 2F8. CAMPATH 1G is an isotype-matched control for 2F8, and wells coated with malBSA, overlaid with WT Mφ lysate, and detected with CAMPATH showed no binding. Note that the wells coated with LPS were not detected by SR-A. The average absorbance for each compound is shown. Assays are from a single experiment and are representative of at least three similar assays. (B) Purified AcLDL, B. subtilis peptidoglycan, or PorA were used to coat the wells of a 96-well plate, and SR-A binding was tested as described above. (C) Purified N. meningitidis proteins were tested in the SR-A ELISA for potential ligands. Wells of a 96-well plate were coated with 10 μg of various purified His- or glutathione S-transferase-tagged N. meningitidis surface proteins/ml. SR-A binding was detected as described above using Mφ lysates from WT and SR-A−/− BMMφ. Some proteins were not ligands for SR-A (NMB0623, NMB1513, and NMB2132), whereas others showed a definite interaction with SR-A (NMB0278, NMB0667, and NMB1220). (D) Various concentrations of whole N. meningitidis lysates from MC58, OMV preparations, or purified NMB1220 were used to coat quintuplicate wells of a 96-well plate. SR-A-mediated binding was assayed as described above. The results are from a single assay representative of at least three similar experiments.
PorA is a major outer membrane protein and falls within the predicted size range of our candidate ligands (24). Also, peptidoglycan is a major structural component of gram-positive bacterial cell walls and, to a lesser extent, gram-negative bacterial cell envelopes and is a candidate for binding to SR-A. Therefore, to test whether PorA and peptidoglycan were responsible for the SR-A binding observed in the overlay blots, we tested them in the SR-A ligand ELISA. Figure 3B shows that neither PorA nor peptidoglycan (derived from B. subtilis) were recognized by SR-A in this system. Next, we tested a range of 10 potential candidate outer membrane proteins from N. meningitidis (Table 1). ELISA plates were coated with 10 μg of purified protein/ml overnight before SR-A binding was detected by using WT Mφ postnuclear cell lysate. Figure 3C shows a series of potential candidate proteins tested, and a range of SR-A-mediated binding of these molecules was observed. For example, NMB2132 showed no recognition by SR-A, NMB0995 and NMB0346 showed intermediate detection by the receptor, whereas NMB1220, NMB0667, and NMB0278 were taken to be positive. To help confirm the specificity, we showed that the ligands tested above behaved similarly to that shown in Fig. 3C when we assessed SR-A binding by using far-Western blots (see Fig. S1 in the supplemental material). Furthermore, we showed that SR-A-mediated binding to the positive ligands could be titrated and, by using decreasing concentrations of whole bacterial lysates, OMV preparations, and purified proteins, we could show an enrichment of the SR-A ligand from the whole bacterial lysate to outer membrane vesicles to purified protein (Fig. 3D). However, we wanted to show that in whole Mφ these ligands could bind SR-A.
NMB1220, but not NMB0278, is endocytosed by SR-A.
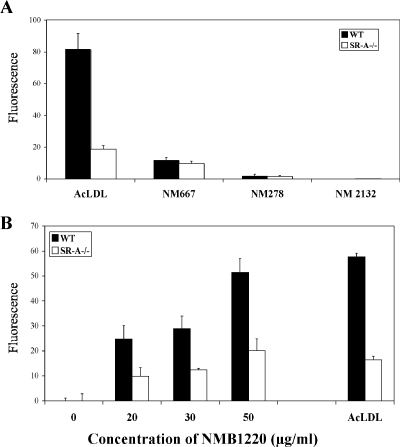
To establish whether SR-A in whole Mφ could specifically bind and internalize NMB1220, NMB0667, and NMB0278, we compared the endocytosis of the purified proteins by WT and SR-A−/− Mφ. BMMφ were incubated with His-tagged NMB1220, NMB0667, and NMB0278 for 45 min before the cells were washed, and the N. meningitidis proteins were detected by using an anti-His tag antibody (Fig. 4A). Flow cytometry showed that although SR-A−/− Mφ could internalize some NMB1220, it was reduced by ca. 50% compared to WT Mφ over a range of protein concentrations (Fig. 4B). Interestingly, no difference in NMB0667 uptake was observed, and NMB0278 was not endocytosed by WT or SR-A−/− Mφ, which may be due to the soluble protein binding SR-A and not triggering internalization or because the protein did not bind the receptor in the form that it has been presented. Therefore, to begin to address these issues, we examined the ability of the purified proteins to block binding and endocytosis of previously described ligands for SR-A. Furthermore, we wanted to confirm that human SR-A was able to bind the ligands identified above. Thus, we used a well-established transfected cell system to examine the uptake of the ligands described above by human SR-A expressed by CHO cells.
FIG. 4.
NMB1220, but not NMB0278, NMB0667, or NMB2132, is endocytosed via SR-A expressed by BMMφ. (A) BMMφ from WT and SR-A−/− animals were seeded in six-well plates as described in Fig. 1 before incubation with 10 μg/ml of purified His-tagged NMB0278, NMB0667, or NMB2132 or DiIAcLDL for 45 min at 37°C. The cells were washed with PBS, detached by using lidocaine-EDTA, and fixed with 4% paraformaldehyde. The uptake of N. meningitidis proteins was measured by using a mouse anti-His-tagged monoclonal antibody, followed by a goat anti-mouse immunoglobulin G-phycoerythrin. An isotype-matched control antibody showed no binding to the Mφ (not shown). Cells were analyzed by flow cytometry, and the average fluorescence of triplicate conditions was plotted. (B) BMMφ were treated as described above except that the cells were incubated with increasing concentrations of His-tagged NMB1220 for 45 min at 37°C before detection of endocytosis using the anti-His antibody system described above. The results are from a single experiment but represent at least three similar assays.
N. meningitidis proteins block the endocytosis of AcLDL and binding of MC58 bacteria to CHO cells stably transfected with human SR-A.
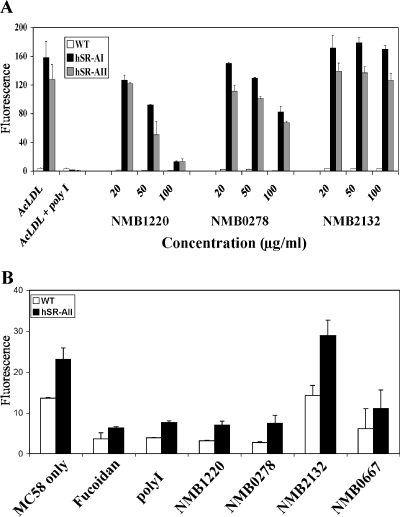
We used transfected cells to study SR-A in isolation and to determine whether the candidates mentioned above bound human SR-A, as well as the murine receptor. We tested the ability of the candidate N. meningitidis ligands and NMB2132, a negative control as determined by the SR-A binding ELISA, to block the endocytosis of known ligands by CHO cells stably transfected with human SR-AI or SR-AII. We have previously used these cells to study the interaction of SR-A with AcLDL, E. coli, and whole N. meningitidis. Endocytosis by SR-A in this system is well established and characterized (8, 18). WT, CHO-hSR-AI, and CHO-hSR-AII cells were preincubated with increasing concentrations of NMB1220, NMB0278, and NMB2132 for 30 min before the addition of DiI-labeled AcLDL (Fig. 5A). Poly(I) (polyinosinic acid) was used as a positive control to block AcLDL binding to the receptor and completely inhibited the uptake of the modified lipid by the transfected cells but not by WT CHO controls. Similarly, with increasing concentrations of NMB1220 and NMB0278, the endocytosis of AcLDL was inhibited. At 100 μg/ml NMB1220 was able to block >90% of AcLDL internalization. NMB0278 showed 50% blocking at 100 μg/ml, suggesting that it bound the receptor with less affinity than poly(I) and NMB1220 or that it had a binding site different from that of NMB1220 or AcLDL. NMB2132 had no effect on the endocytosis of AcLDL, confirming that it did not bind SR-A.
FIG. 5.
NMB1220 and NMB0278 are able to block endocytosis and binding of known ligands to CHO cells transfected with human SR-A. (A) CHO WT, CHO hSR-AI, and CHO hSR-AII were preincubated with poly(I) or purified N. meningitidis proteins for 30 min before the addition of DiIAcLDL (5 μg/ml) and were maintained in their presence throughout the assay. The cells were washed and analyzed by flow cytometry. The graph shows the average increase in fluorescence from duplicate wells. (B) WT and hSR-AII cells were plated as described above and preincubated with Fucoidan, poly(I), or purified N. meningitidis proteins (100 μg/ml) for 30 min. Next, RdGnX-labeled, ethanol-fixed N. meningitidis was added (200 bacteria per cell for 1 h) while maintaining the inhibitor. The increase in fluorescence was measured by flow cytometry, and the average increase in fluorescence was plotted. The results are from a single assay that is representative of at least three similar experiments.
Next, we tested the ability of the candidate proteins to block the binding of whole N. meningitidis to SR-A (Fig. 5B). WT, hSR-AI (not shown), and CHO hSR-AII cells were preincubated with 100 μg/ml of poly(I), NMB1220, NMB0278, NMB0667, or NMB2132. Some background binding of N. meningitidis to WT CHO cells was observed in all cases; however, more association was observed with the SR-AII-transfected cells. Preincubation of the cells with poly(I) and all of the selected N. meningitidis candidates except NMB2132 blocked N. meningitidis binding to the cells. NMB0667, NMB1220, and NMB0278, but not NMB2132, also blocked meningococcal association with CHO cells stably expressing hSR-AI; however, as observed previously, the overall bacterial association with hSR-AI cells was decreased compared to hSR-AII-transfected CHO cells (data not shown) (20). Some bacterial association with WT CHO cells was also blocked in the presence of the control scavenger receptor inhibitors [poly(I) and fucoidan], as well as when the candidate ligands were used in the assay. This blocking could be due to the presence of unknown scavenger receptors expressed by the WT CHO cells, which also recognize the bacteria, or due to nonspecific inhibition of the bacteria binding to other nonscavenger receptors. Although not endocytosed and not an efficient inhibitor of soluble ligands, NMB0278 was able to block whole bacterial binding to the receptor with an efficiency similar to that of NMB1220 and poly(I).
Thus, our data show that, using a candidate ligand approach, whole cells, and a novel binding assay, we have identified at least one naturally occurring, unmodified protein that binds to and is endocytosed by SR-A. We have also identified two other molecules that were able to compete for bacterial binding and, to a lesser extent, for endocytic ligand uptake by SR-A.
DISCUSSION
We have previously shown SR-A to be a major nonopsonic receptor for meningococci on Mφ. We have now extended those studies to identify novel bacterial ligands for SR-A, namely, NMB1220, NMB0667, and NMB0278. Using a series of natural mutants of N. meningitidis, we showed that abundant, but highly variable capsule polysaccharide, pili, PorA, Opa, and Opc were not recognized by SR-A. Characterization by far-Western blot and an ELISA-based assay identified a number of surface-expressed candidates all recognized by the receptor to various extents. Further analysis showed that only one of the three candidates identified, NMB1220, was endocytosed when in soluble form, but all could block internalization of AcLDL by SR-A. Furthermore, NMB1220, NMB0667, and NMB0278 were able to inhibit the binding of N. meningitidis to CHO cells stably expressing human SR-A, whereas NMB2132, identified in our assays not to be a ligand for the receptor, could not block the interaction. Thus, we have identified novel molecules that are able to block meningococcal interactions with host cells via SR-A.
The surface structure of Neisseria is highly variable and many surface moieties are subject to phase variation (31). This sequence variation of surface-exposed proteins has hampered the identification of new vaccine candidates (28) and made searching for ligands more challenging. Previously, we determined that the ligand for SR-A on meningococci was conserved throughout the species since SR-A−/− Mφ were unable to ingest N. gonorrhoeae, N. lactamica, and N. sicca efficiently compared to WT control cells. Therefore, it was not surprising that we were able to show, using natural mutants and our binding ELISA, that capsule, pili, PorA, Opc, and Opa proteins were not essential for recognition by SR-A since they are all highly variable. However, our data do not exclude the possibility that these molecules contribute to the overall affinity of the organism for SR-A by steric interference or, conversely, increasing the overall avidity of binding.
The far-Western blot was used to characterize meningococcal ligands for SR-A. Protease digestion showed that these candidates were at least in part proteinaceous in nature. Analysis of the LPS-free mutant of N. meningitidis revealed no difference in the distribution of the potential candidate ligands compared to the control bacterial strain, confirming that the ligand was not LPS or related to LPS expression. The far-Western blot used total lysates of meningococci, making it possible that some of the candidates could be expressed intracellularly. To assess this, we separated the proteins from OMV and found that a number of the bands positive for SR-A binding were enriched in these vesicles. These data confirmed that some candidates were appropriately expressed on the surface of the bacteria and thus potentially available for SR-A binding in the whole bacteria. Furthermore, during infection meningococci are known to release large numbers of OMVs into the circulation (24). SR-A-mediated recognition of the OMVs by Mφ in the spleen and Kupffer cells in the liver could contribute to the resolution of inflammation by removing the inflammatory vesicles without proinflammatory cytokine release if they are bound by SR-A similarly to the whole bacteria.
Interestingly, the far-Western blot revealed that there was more than one potential SR-A ligand on meningococci, which was confirmed using our SR-A binding ELISA. In fact, we do not expect that we have found all of the molecules that bind SR-A on N. meningitidis. For example, Jeannin et al. (10) showed recently that OmpA from E. coli and Klebsiella pneumoniae bound to two other members of the scavenger receptor family, LOX-1 and SREC. Since meningococcal class 4 OMP shares structural homology with E. coli OmpA, it may also be a ligand for SR-A and remains to be tested. The multiplicity of ligands for SR-A on N. meningitidis was interesting, especially since those identified bound with lower apparent affinity compared to AcLDL. These data suggest that the selectivity of meningococci for SR-A could result from multiple low-affinity ligands that increase the overall avidity of the bacteria for the receptor.
Although the far-Western blots helped characterize the ligands, to identify the exact molecules involved in SR-A binding, we developed a variation of this assay in an ELISA format. This assay provided a powerful new method that allowed quick and rapid screening of a large number of potential ligands and utilized available tools for SR-A. This assay can be used for the identification of nonbacterial scavenger receptor ligands and adapted for use with other surface receptors expressed by Mφ and other cell types. Previously, the N. meningitidis serogroup B genome sequence was mined for potential vaccine candidates by in silico identification of cell surface molecules that were then cloned, tagged, and purified (22). Utilizing this resource, we selected candidates based on known cell surface expression and their predicted size (22, 31). The assay provided us with a number of candidates for SR-A binding, two of which fell into the predicted size range (30 to 75 kDa) obtained from our far-Western blots. We observed a wide range of SR-A-dependent recognition with respect to their affinity of binding. Molecules such as NMB1220 were strongly positive, while some, such as NMB2132, were negative; others ranged in between. We were able to show an enrichment of SR-A binding from the whole bacteria cell lysates, OMVs, to the purified candidate ligands. Also, we were able to titrate SR-A binding and show that it was able to recognize the molecules in the presence of EDTA.
Due to the number of SR-A ligands on the meningococcal cell surface, it was unlikely that deletion mutants of single or even double ligands would show much reduction in binding to Mφ, especially since we do not understand the redundancies in this system. However, future work will entail evaluation of the ability of various mutants selectively deficient for the various candidate ligands to interact with SR-A in vitro and in vivo. Thus, to prove that these molecules were authentic ligands for the receptor, we utilized the purified proteins in whole-cell assays. All of the identified molecules were able to block the binding of whole bacteria to transfected cells, whereas NMB2132, a non-SR-A ligand used in our assays, could not. Also, NMB1220, NMB0667, and NMB0278 were able to inhibit the binding of these particles to SR-A more effectively than soluble ligand. Furthermore, by using cells transfected with human SR-A, we confirmed that the candidate ligands interact with the human, as well as the murine receptor. NMB1220 was the best inhibitor of the three candidates; it was able to block meningococcal-Mφ interaction at lower concentrations and was the only candidate that triggered endocytosis. Ingestion of NMB1220 by SR-A−/− Mφ was reduced by 50% compared to WT cells. Interestingly, AcLDL internalization is also reduced by 50% in SR-A−/− cells. Using soluble NMB1220, we did detect some endocytosis by SR-A−/− Mφ, suggesting the presence of alternate scavenger-type receptors on the Mφ. We found very little receptor-mediated endocytosis of NMB0278 and NMB0667. This could imply that these two molecules do not bind the receptor. Conversely, they could still bind but not trigger endocytosis. The ability of these ligands to block uptake, at least in part, suggests that this may be the case. Also, they may bind the receptor at a different site from the collagenous domain.
None of three candidate SR-A ligands have been reported to be phase variable (29, 30). Sequence analysis suggests that NMB0278 has homology to E. coli DsbA, which functions in disulfide bond formation and is the only one of the three SR-A ligands described here that is found on membrane blebs (25). NMB1220 has homology to stomatin found on red blood cells, whereas NMB0667 is a hypothetical protein of unknown function. Modeling based on related proteins revealed that NMB0667 and NMB0278 both had conformations containing negatively charged faces. SR-A is able to bind polyanions, but there is an undetermined structural requirement since not all polyanions are ligands for the receptor. Mutagenesis of targeted residues within this negatively charged face could provide valuable insight into this elusive requirement. We may then also be able to promote or reduce the attachment of meningococci to Mφ as required. NMB1220 still remains to be modeled to determine whether it contains a similar negatively charged face.
Further studies are required to determine whether NMB1220 is able to block N. meningitidis binding to other cell types in vitro and in vivo. Previously, it has been shown that NMB1220 antiserum displayed some bactericidal activity (22), but it is unclear whether this would be sufficient in vivo to inhibit interactions within the host. Our preliminary data in the whole animal suggested that N. meningitidis targeting of Mφ provided some selective advantage for the organism since SR-A−/− animals were more resistant to meningococcal infection (L. Peiser and S. Gordon, unpublished data). Therefore, it would be intriguing to determine whether blocking of SR-A with NMB1220 alone or in combination with NMB0667 and NMB0278 would increase animal survival in WT animals. Conversely, we found previously that SR-A was not required for macrophage secretory responses to meningococci (18), and SR-A-mediated recognition of NMB1220, NMB0667, and NMB0278 on whole bacteria and OMVs may help resolve inflammatory responses. However, further study is required to show clearance of OMVs by SR-A in vitro and in vivo and the possible consequences for inflammation. Nevertheless, we have identified possible new targets for vaccine development and have new aids for inhibiting the interaction of Mφ with N. meningitidis.
Supplementary Material
Acknowledgments
Work in the laboratory of S.G. was supported by the Medical Research Council, United Kingdom. L.P. was supported by the E. P. Abraham Junior Research Fellowship at St. Cross College. K.M., J.C.W., and E.R.M. were supported by the Medical Research Council, United Kingdom.
Editor: J. L. Flynn
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abraham, R., N. Singh, A. Mukhopadhyay, S. K. Basu, V. Bal, and S. Rath. 1995. Modulation of immunogenicity and antigenicity of proteins by maleylation to target scavenger receptors on macrophages. J. Immunol. 154:1-8. [PubMed] [Google Scholar]
- 2.Brandtzaeg, P. 1995. Pathogenesis of meningococcal infections, p. 71-114. In K. Cartwright (ed.), Meningococcal disease. John Wiley, Ltd., Chichester, United Kingdom.
- 3.Brandtzaeg, P., L. Osnes, R. Ovstebo, G. B. Joo, A. B. Westvik, and P. Kierulf. 1996. Net inflammatory capacity of human septic shock plasma evaluated by a monocyte-based target cell assay: identification of interleukin-10 as a major functional deactivator of human monocytes. J. Exp. Med. 184:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartwright, K. 1995. Meningococcal carriage and disease, p. 159-177. In K. Cartwright (ed.), Meningococcal disease. John Wiley, Ltd., Chichester, United Kingdom.
- 5.Coligan, J. E., B. M. Dunn, H. L. Ploegh, D. W. Speiches, and P. T. Wingfield (ed.). 1997. Current protocols in protein science, p. 6.0.1-6.7.14 and 9.14.11-9.14.16. Wiley, New York, N.Y.
- 6.Freeman, M., Y. Ekkel, L. Rohrer, M. Penman, N. J. Freedman, G. M. Chisolm, and M. Krieger. 1991. Expression of type I and type II bovine scavenger receptors in Chinese hamster ovary cells: lipid droplet accumulation and nonreciprocal cross competition by acetylated and oxidized low density lipoprotein. Proc. Natl. Acad. Sci. USA 88:4931-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon, S., L. Lawson, S. Rabinowitz, P. R. Crocker, L. Morris, and V. H. Perry. 1992. Antigen markers of macrophage differentiation in murine tissues, p. 1-37. In S. Russell and S. Gordon (ed.), Macrophage biology and activation, vol. 181. Springer-Verlag, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 8.Gough, P. J., D. R. Greaves, and S. Gordon. 1998. A naturally occurring isoform of the human macrophage scavenger receptor (SR-A) gene generated by alternative splicing blocks modified LDL uptake. J. Lipid Res. 39:531-543. [PubMed] [Google Scholar]
- 9.Hampton, R. Y., D. T. Golenbock, M. Penman, M. Krieger, and C. R. Raetz. 1991. Recognition and plasma clearance of endotoxin by scavenger receptors. Nature 352:342-344. [DOI] [PubMed] [Google Scholar]
- 10.Jeannin, P., B. Bottazzi, M. Sironi, A. Doni, M. Rusnati, M. Presta, V. Maina, G. Magistrelli, J. F. Haeuw, G. Hoeffel, N. Thieblemont, N. Corvaia, C. Garlanda, Y. Delneste, and A. Mantovani. 2005. Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity 22:551-560. [DOI] [PubMed] [Google Scholar]
- 11.Kallstrom, H., M. S. Islam, P. O. Berggren, and A. B. Jonsson. 1998. Cell signalling by the type IV pili of pathogenic Neisseria. J. Biol. Chem. 273:21777-21782 [DOI] [PubMed] [Google Scholar]
- 12.Kallstrom, H., M. K. Liszewski, J. P. Atkinson, and A. B. Jonsson. 1997. Membrane cofactor protein (MCP or CD46) is a cellular pilus receptor for pathogenic Neisseria. Mol. Microbiol. 25:639-647. [DOI] [PubMed] [Google Scholar]
- 13.Krieger, M., and J. Herz. 1994. Structures and functions of multiligand lipoprotein receptors: macrophage scavenger receptors and LDL receptor-related protein (LRP). Annu. Rev. Biochem. 63:601-637. [DOI] [PubMed] [Google Scholar]
- 14.Mackinnon, F. G., R. Borrow, A. R. Gorringe, A. J. Fox, and D. M. Jones. 1993. Demonstration of lipooligosaccharide immunotype and capsule as virulence factors for Neisseria meningitidis using an infant mouse intranasal infection model. Microb. Pathog. 15:359-366. [DOI] [PubMed] [Google Scholar]
- 15.McNeil, G., and M. Virji. 1997. Phenotypic variants of meningococci and their potential in phagocytic interactions: the influence of opacity proteins, pili, PilC, and surface sialic acids. Microb. Pathog. 22:295-304. [DOI] [PubMed] [Google Scholar]
- 16.McNeil, G., M. Virji, and E. R. Moxon. 1994. Interactions of Neisseria meningitidis with human monocytes. Microb. Pathog. 16:153-163. [DOI] [PubMed] [Google Scholar]
- 17.Mukhopadhyay, S., L. Peiser, and S. Gordon. 2004. Activation of murine macrophages by Neisseria meningitidis and IFN-γ in vitro: distinct roles of class A scavenger and Toll-like pattern recognition receptors in selective modulation of surface phenotype. J. Leukoc. Biol. 76:577-584. [DOI] [PubMed] [Google Scholar]
- 18.Peiser, L., M. P. De Winther, K. Makepeace, M. Hollinshead, P. Coull, J. Plested, T. Kodama, E. R. Moxon, and S. Gordon. 2002. The class A macrophage scavenger receptor is a major pattern recognition receptor for Neisseria meningitidis which is independent of lipopolysaccharide and not required for secretory responses. Infect. Immun. 70:5346-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peiser, L., and S. Gordon. 2001. The function of scavenger receptors expressed by macrophages and their role in the regulation of inflammation. Microbes Infect. 3:149-159. [DOI] [PubMed] [Google Scholar]
- 20.Peiser, L., P. J. Gough, T. Kodama, and S. Gordon. 2000. Macrophage class A scavenger receptor-mediated phagocytosis of Escherichia coli: role of cell heterogeneity, microbial strain, and culture conditions in vitro. Infect. Immun. 68:1953-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peiser, L., S. Mukhopadhyay, and S. Gordon. 2002. Scavenger receptors in innate immunity. Curr. Opin. Immunol. 14:123-128. [DOI] [PubMed] [Google Scholar]
- 22.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Arico, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 23.Plested, J. S., B. L. Ferry, P. A. Coull, K. Makepeace, A. K. Lehmann, F. G. MacKinnon, H. G. Griffiths, M. A. Herbert, J. C. Richards, and E. R. Moxon. 2001. Functional opsonic activity of human serum antibodies to inner core lipopolysaccharide (galE) of serogroup B meningococci measured by flow cytometry. Infect. Immun. 69:3203-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poolman, J. T., P. A. van der Ley, and J. Tommassen. 1995. Surface structures and secreted products of meningococci, p. 21-34. In K. Cartwright (ed.), Meningococcal disease. John Wiley, Ltd., Chichester, United Kingdom.
- 25.Post, D. M., D. Zhang, J. S. Eastvold, A. Teghanemt, B. W. Gibson, and J. P. Weiss. 2005. Biochemical and functional characterization of membrane blebs purified from Neisseria meningitidis serogroup B. J. Biol. Chem. 280:38383-38394. [DOI] [PubMed] [Google Scholar]
- 26.Pridmore, A. C., D. H. Wyllie, F. Abdillahi, L. Steeghs, P. van der Ley, S. K. Dower, and R. C. Read. 2001. A lipopolysaccharide-deficient mutant of Neisseria meningitidis elicits attenuated cytokine release by human macrophages and signals via Toll-like receptor (TLR) 2 but not via TLR4/MD2. J. Infect. Dis. 183:89-96. [DOI] [PubMed] [Google Scholar]
- 27.Read, R. C., A. Fox, K. Miller, T. Gray, N. Jones, R. Borrows, D. M. Jones, and R. G. Finch. 1995. Experimental infection of human nasal mucosal explants with Neisseria meningitidis. J. Med. Microbiol. 42:353-361. [DOI] [PubMed] [Google Scholar]
- 28.Ruggeberg, J. U., and A. J. Pollard. 2004. Meningococcal vaccines. Paediatr. Drugs 6:251-266. [DOI] [PubMed] [Google Scholar]
- 29.Saunders, N. J., A. C. Jeffries, J. F. Peden, D. W. Hood, H. Tettelin, R. Rappuoli, and E. R. Moxon. 2000. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol. 37:207-215. [DOI] [PubMed] [Google Scholar]
- 30.Snyder, L. A., S. A. Butcher, and N. J. Saunders. 2001. Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology 147:2321-2332. [DOI] [PubMed] [Google Scholar]
- 31.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 32.van Deuren, M., P. Brandtzaeg, and J. W. van der Meer. 2000. Update on meningococcal disease with emphasis on pathogenesis and clinical management. Clin. Microbiol. Rev. 13:144-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virji, M., H. Kayhty, D. J. Ferguson, C. Alexandrescu, J. E. Heckels, and E. R. Moxon. 1991. The role of pili in the interactions of pathogenic Neisseria with cultured human endothelial cells. Mol. Microbiol. 5:1831-1841. [DOI] [PubMed] [Google Scholar]
- 34.Virji, M., J. R. Saunders, G. Sims, K. Makepeace, D. Maskell, and D. J. Ferguson. 1993. Pilus-facilitated adherence of Neisseria meningitidis to human epithelial and endothelial cells: modulation of adherence phenotype occurs concurrently with changes in primary amino acid sequence and the glycosylation status of pilin. Mol. Microbiol. 10:1013-1028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.