Abstract
The complete nucleotide sequence was determined for pMAR7, an enteropathogenic Escherichia coli (EPEC) adherence factor (EAF) plasmid that contains genes encoding a type IV attachment pilus (Bfp) and the global virulence regulator per. Prototypic EAF plasmid pMAR7 is self-transmissible, unlike the smaller EAF plasmid pB171, which has no genes encoding conjugative functions. The tra locus, a highly conserved 33-kb segment found in pMAR7, is similar to the tra (conjugation) region of the F plasmid. ISEc13 copies flanking the pMAR7 tra region could potentially mobilize or delete the tra genes. Hybridization of 134 EPEC strains showed that a complete tra region is present only in strains of the EPEC1 clonal group. This study confirms EPEC's potential for dissemination of virulence attributes by horizontal transfer of the EAF plasmid.
Enteropathogenic Escherichia coli (EPEC) are an important cause of acute and persistent diarrhea in infants in developing countries. EPEC strains produce a characteristic intestinal histopathology called the attaching-and-effacing (A/E) lesion, which is characterized by intimate bacterial adherence to the epithelial membrane, effacement of the microvilli, and striking cytoskeletal changes (18). The A/E phenotype is encoded by a 35-kb chromosomal pathogenicity island (PAI) known as the locus of enterocyte effacement (LEE) (17). The LEE encodes a type III secretion system that translocates virulence effector proteins into host epithelial cells and the surface protein intimin, mediating intimate adherence to host cells (reviewed in reference 12). The A/E histopathology and the presence of LEE are also characteristic of other diarrheagenic strains, including enterohemorrhagic E. coli (EHEC).
The large plasmid pMAR2, the first genetic element implicated in EPEC disease, is present in the prototypic EPEC strain E2348/69 (O127:H6) (2). Adherence of EPEC to HEp-2 epithelial cells and in vivo adherence to piglet intestinal epithelium was shown to be dependent on this self-transmissible plasmid. Transfer of this plasmid to a nonadherent E. coli K-12 strain conferred the ability to adhere to epithelial cells. Of 32 adherent strains with classical EPEC serotypes, 31 possessed similar large plasmids, subsequently named EAF for EPEC adherence factor plasmids. Adult volunteers developed diarrhea in nine of ten cases challenged with wild-type E2348/69 (13) but in only two of nine controls who ingested the plasmid-cured derivative. Antibodies to an outer membrane protein expressed by the parent strain were detected after treatment with wild type but not the plasmid-cured derivative. Expression of the protein, later identified as intimin (11), was also linked to the plasmid. A 1-kb region of pMAR2, called the EAF probe, was the first molecular diagnostic tool for EPEC (19).
The type IV pilus named the bundle-forming pilus (BFP) (8) encoded in a 14-gene locus, was the first virulence factor to be identified for the EAF plasmid (24, 25). The rope-like filaments of BFP connect EPEC bacteria into microcolonies in a pattern called localized adherence. A regulatory locus, per (plasmid-encoded regulator) (10), was shown to activate expression of the chromosomal eae gene encoding intimin. One of the three per open reading frames (ORFs), perA, encodes an AraC-family transcriptional activator (10). Tobe et al. reported that this locus (called bfpTWV in their study) also activates expression of the bfp genes (28). In studies with isogenic bfpA and perA mutants, experiments with human volunteers showed that PerA and BFP contribute to diarrhea and are therefore important in virulence (3).
Two divergent clonal groups exist among EPEC strains containing the EAF plasmid, designated EPEC1 and EPEC2 (22, 30, 31), distinguished by multilocus enzyme electrophoresis analysis of allelic differences among housekeeping genes. The EPEC1 group includes widespread serotypes such as O55:H6 and O119:H6, whereas EPEC 2 consists of serotypes with more limited occurrence such as O111:H2 and O114:H2. Strains that possess the LEE but lack the EAF plasmid are termed typical EPEC (20) and are found in developing countries, whereas EPEC that lack the EAF plasmid, called atypical EPEC, predominate in industrialized countries (29). Whittam and coworkers showed that horizontal transfer of the EAF plasmid, insertion of the LEE into different chromosomal sites, acquisition of Shiga toxin-encoding bacteriophage, and transfer of an EHEC virulence plasmid all contributed to the evolution of EPEC and EHEC strains (22).
Plasmid sequencing and analysis.
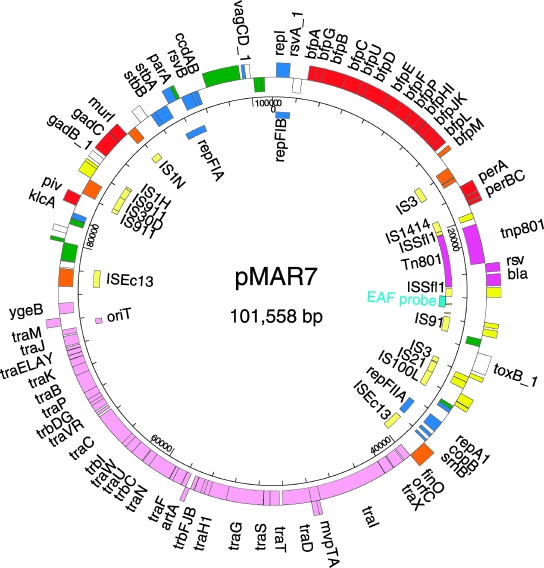
The DNA sequence of the EAF plasmid from an EPEC2 strain, B171 (O111:NM), was previously determined (27). Plasmid pB171 is 69 kb in length, which is considerably smaller than the ∼95-kb EPEC1 plasmid pMAR2 (2). No genes encoding DNA transfer functions (tra) were found in pB171, an observation at variance with the fact that EPEC1 EAF plasmids are self-transmissible. To compare the EPEC1 and EPEC2 plasmids, we determined and analyzed the complete nucleotide sequence of the EAF plasmid from E2348/69. The sequenced plasmid pMAR7 was derived from the native plasmid pMAR2 by transposition of Tn801 from plasmid pMR5 (2) to provide a selectable marker, ampicillin resistance. Annotations were assigned using ASAP (for “a systematic annotation package”) (9), and the annotated sequence was deposited in GenBank under accession number DQ388534. Protein coding sequences are in Tables S1 and S2 in the supplemental material, and the fully annotated sequences are accessible via the Enteropathogen Resource Integration Center-Bioinformatics Resource Center (ERIC-BRC/ASAP) database (http://www.ericbrc.org/portal/eric/). The circular map shown in Fig. 1 consists of 101,558 bp containing 115 protein-coding ORFs and one regulatory RNA gene, finP. Genes shared by pMAR7 and pB171 are colinear. For 90 ORFs (78%), putative functions could be assigned; 8 ORFs with experimental evidence of expression had no known function (uncharacterized proteins), and the remaining 17 ORFs were similar to conserved hypothetical proteins. Unique identifiers (MAR###) were assigned to each ORF.
FIG. 1.
Map of pMAR7. Outer circle, ORFs, and their orientations color coded by functional category: red, known or putative virulence-associated proteins; pink, conjugal DNA transfer; yellow, IS-related or transposase fragments; orange, intact IS or transposase; blue, plasmid replication, maintenance, or other DNA metabolic functions; green, conserved hypothetical; purple, Tn801 insertion associated; white, pseudogenes. The inner circle shows the scale in base pairs. Individual gene descriptions are available online (see Table S1 in the supplemental material) or in the ERIC-BRC ASAP database (https://ericbrc.org/asap). The three replication regions, oriT, and the EAF probe are indicated. The figure was generated using the program Genvision (DNASTAR, Madison, WI).
A total of 61 pMAR7 ORFs (55%) were previously identified in pB171. Proteins involved in plasmid replication and DNA maintenance (Fig. 1) are encoded by 16 ORFs (14% of pMAR7), constituting three putative replication regions characteristic of IncF plasmids, RepFIB (MAR001), RepFIIA (MAR039), and RepFIA (MAR103) as in pB171. A double-partition (par) locus has been described for pB171 (6), and this region is almost identical in pMAR7. Genes encoding the major virulence factors account for 14 (bfp) and 3 (per) ORFs in both plasmids (10, 24, 25, 27). These loci in the two plasmids share 99% sequence similarity, and most of the BFP proteins are 97 to 100% identical. MAR031 encodes a 465-amino-acid protein that is 97% identical to the C terminus of ToxB encoded on the large virulence plasmid pO157 of EHEC EDL933 (5). However, the pO157 ToxB protein is huge (362 kDa) and probably multifunctional (26). MAR031 represents only 15% of the 3,169-residue ToxB. The corresponding region in pB171 is further broken into two adjacent ORFs encoding 451- and 168-amino-acid ToxB-like proteins. Although single domain-specific functions for MAR031 or the pB171 protein fragments cannot be ruled out, it is unlikely that they are functional proteins.
Intact tra region.
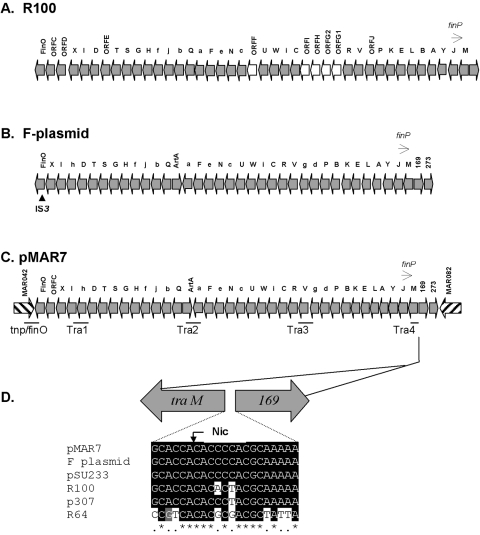
A segment of 50 ORFs in pMAR7 is absent from pB171 (Table S1 in the supplemental material). Thirty-eight of these ORFs are very similar in genetic organization and sequence to the tra regions responsible for conjugal transfer in plasmids F and R100 (1). The transfer regions of F, R100, and pMAR7 are compared in Fig. 2. All 36 genes of the 33.3-kb F-transfer system (7) are present in pMAR7 and are apparently complete and functional. The 3′ end of the pMAR7 operon, however, has unique sequence similarity to plasmid R100. In nearly all other differences between the R100 and F tra regions, pMAR7 more closely resembles the F plasmid, the pMAR7 proteins being 70 to 100% identical to F proteins except TrbJ (66%), TraQ (58%), and TraS (25% identity with F but 83% with R100).
FIG. 2.
Transfer operons of the pMAR7, R100, and F plasmids. The transfer operons of plasmid R100 (A) plasmid F (B), and pMAR7 (C) are compared. Orientation of the ORFs is that in pMAR7. Transcription proceeds from right to left; tra genes are in uppercase, trb genes are in lowercase, and antisense RNA finP is indicated above the lines. ORFs are not to scale. (D) Comparison of putative oriT region of conjugative plasmids aligned at nic site (arrow) according to Frost et al. (7). Sequences were aligned by CLUSTAL W and highlighted by Boxshade. Source or reference for sequences: pMAR7 (the present study); R100 (NC_002134), F (NC_002483), R64 (AB027308), p307 (X06534), pSU233 (X55896).
oriT (Fig. 1) is the origin of conjugative transfer in the F plasmid, where the DNA is nicked and a single strand of plasmid DNA begins to transfer from the donor to the recipient. oriT is a 463-nucleotide segment immediately upstream of traM (7). It is highly conserved in pMAR7, with perfect identity to the nicking-site region of F (Fig. 2D). Upstream of oriT, the pMAR7 ORFs are orthologs of the corresponding F plasmid ORFs. MAR083 to MAR087 are similar to leading-region ORFs of pO157 (5, 15).
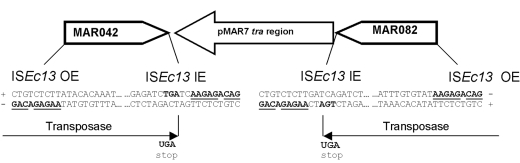
An unusual feature of the pMAR7 tra region is that it is flanked by two previously unreported IS4 family elements (14) designated ISEc13 that are nearly identical and in inverse orientations (Fig. 3). Their inverted repeats of 23 and 21 nucleotides are very similar to ISPlu9 and IS50. No direct repeats were observed. These elements appear to form an unusual Tn5-like composite transposon. Although there has never been a report of a Tn5 containing a sequence as large as the tra region (W. Reznikoff, unpublished data), the possibility remains that the IS-bracketed region could be transposed as an intact unit or island.
FIG. 3.
ISEc13 flanking tra region of pMAR7. The outer end (OE) 19-bp terminal inverted repeat begins 165 bp upstream of the ORF (MAR042 and MAR082). The transposase TGA stop codon is within the 19-bp inner end (IE).
Distribution of tra genes in EPECs.
We analyzed 134 EPEC strains representing the two major clonal groups of typical EPEC and strains representing atypical EPEC. Four DNA probes (TRA1 to TRA4) were prepared that spanned the pMAR7 transfer region (Fig. 2C). Probe TRA4 encompassed the 5′ origin of transfer, and a fifth probe, tnp/finO, covered the opposite (3′) end of the region. Colony hybridization was used to detect sequences homologous to these probes plus the EAF probe (19).
The hybridization results enabled strains to be assigned to different classes (Table 1) . For class I strains, all probes were positive, indicating that they were EAF+, TRA1-4+, and finO/tnp+, suggesting that these strains possessed pMAR7-like plasmids. This class included EPEC1 serotypes O127:H6, O55:H6, and O142:H34 (47 strains). Class II strains, also from EPEC1, were EAF− and TRA2-4+ but were negative for the TRA1 and finO/tnp probes. The serotypes belonging to class II were O142:H6 and O119:H6, which also contained bpfA sequences that were more divergent than those found in other EPEC1 strains (4). Class III represented EAF+ TRA4− strains lacking oriT. The majority of strains in this group (23 of 27 tested) were negative for TRA probes, but 4 were positive for TRA2 and TRA3. O111:H2 and O111:H− serotypes were found in both groups of class III. Class IV strains were EAF- and finO/tnp-negative but TRA3/4 positive and either positive or negative for TRA1/2. These are atypical EPEC strains (lacking EAF sequences) and presumably were nonconjugative despite the presence of oriT; the colony hybridization technique would not distinguish chromosomal versus plasmid-borne TRA3/4 sequences. Class V strains were negative for all probes. These strains were atypical EPEC whose serotypes overlapped with class IV. The presence or absence of the intact tra region is consistent with reports (2, 16) that O127:H7 (class I) strains contained self-transmissible EAF plasmids, whereas EAF plasmids in O142:H6 (class II) and O111:H2 (class III) strains were not self-transmissible. We also examined a number of EHEC strains and found only partial, if any, tra regions. O157:H7 strains tested were negative for all TRA probes, while some (but not all) O26:H11, O111:H8, and O111:H− strains reacted with TRA1 to TRA4 but not tnp-finO.
TABLE 1.
Summary of tra region hybridization analysis of EPEC strains
| Class | Description | Serotype | Clonal group | No. of strains analyzed | Presence (+) or absence (−) of:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| EAF | tnp/finO | TRA1 | TRA2 | TRA3 | TRA4 | |||||
| I | EAF+ TRA1-4+ | O127:H6 | EPEC1 | 12 | + | + | + | + | + | + |
| O55:H6 | EPEC1 | 26 | + | + | + | + | + | + | ||
| O142:H34 | EPEC1 | 5 | + | + | + | + | + | + | ||
| O86:H34 | EPEC1 | 4 | + | + | + | + | + | + | ||
| II | EAF+ TRA4+ | O142:H6 | EPEC1 | 8 | + | − | − | + | + | + |
| O119:H6 | EPEC1 | 8 | + | − | − | + | + | + | ||
| III | EAF+ TRA4− | O111:H2, H− | EPEC2 | 4 | + | − | − | + | + | − |
| O111:H2, H− | EPEC2 | 23 | + | − | − | − | − | − | ||
| IV | EAF− TRA4+ | O128:H2 | Atypical | 12 | − | − | + | + | + | + |
| O55:H7 | Atypical | 4 | − | − | + | + | + | + | ||
| O119:H2 | Atypical | 6 | − | − | − | − | + | + | ||
| V | EAF− TRA1-4− | O55:H7 | Atypical | 7 | − | − | − | − | − | − |
| O125:H6 | Atypical | 8 | − | − | − | − | − | − | ||
| O128:H2 | Atypical | 2 | − | − | − | − | − | − | ||
| O111:H9 | Atypical | 5 | − | − | − | − | − | − | ||
Other differences between pMAR7 and pB171.
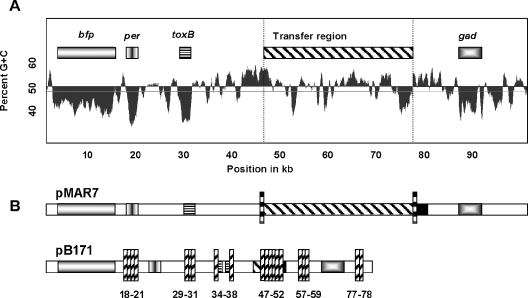
Apart from the tra region, several other ORFs were present in pB171 but not in pMAR7 (Fig. 4B). One cluster, located between the bfp and per operons, is 3.6 kb larger in pB171 and contains four genes not in pMAR7 encoding proteins similar to a stability locus of R100, a putative resolvase of Yersinia pestis pMT1, a serine acetyltransferase, and the putative chaperone trcP (27). Two pB171-specific regions designated IS629 by Tobe et al. contain the same three ORFs (ORF29pB171 to ORF31pB171 and ORF49pB171 to ORF51pB171). These are now identified as ORFs of ISEc8 (23). IS-related sequences accounted for 29.5% of the pB171 sequence and 18.6% of the pMAR7 sequence.
FIG. 4.
(A) G+C content of pMAR7. The G+C percentage (sliding window) is plotted against the sequence position. Regions with low G+C content, bfp, per, toxB, gad, and the transfer region are indicated. The scale is in kilobases. (B) Comparison of pMAR7 and pB171. Marked pMAR7 segments are aligned with the G+C content diagram, and homologous pB171 regions are indicated with similar patterns. IS elements flanking the tra genes are indicated with vertical checked bars. pB171-specific ORFs are indicated by vertical hash bars with ORF numbers below the bar. The toxB fragments and flanking IS3 ORFs in pB171 are ORFs 34 to 38.
Base composition.
The mosaic nature of pMAR7 is illustrated by its base composition (Fig. 4A). The average G+C content is 47%, but there are regions with substantially lower values. Altogether, 22 ORFs (20% of pMAR7) in clusters have G+C contents of under 40%, suggesting that these regions originated from sources outside E. coli. For example, the bfp cluster has 37% G+C and the perABC cluster 31% G+C (Fig. 4A). The similar G+C contents and close proximity of these two operons suggest that they were acquired together as a functional unit. The 3-kb region between bfp and per has 55% G+C content and encodes IS fragments, suggesting that the bfp and per operons were separated by IS integration after acquisition. This region may be a hot spot for recent IS integration, which is consistent with a report of substantial differences upstream of the perA promoter in EAF plasmids (21). A higher G+C composition (average 53%) in the three replicon regions suggests that the plasmid maintenance backbone likely served as the target for horizontal acquisition of virulence determinants.
Concluding comments.
Sequence comparison of two prototypic EPEC virulence plasmids revealed an EAF plasmid backbone and nearly identical bfp and per virulence loci. The intact tra region in pMAR7 distinguishes it from pB171, along with 16 pB171 ORFs absent from pMAR7. The tra region is limited to strains of the EPEC1 clonal group, although partial tra sequences were detected in EPEC1, EPEC2, and atypical EPEC strains. The ISEc13 elements flanking the tra genes in pMAR7 suggest that the tra segment might be mobile, possibly acquired by transposition from an F-like plasmid. Subsequently, the region could have mediated the transfer of the entire EAF plasmid to other E. coli strains. Conversely, the IS elements provide targets for homologous recombination by which EAF plasmids might have lost some or all of the tra genes by deletion. This scenario is consistent with the hypothesis of Whittam and McGraw (30) that EPEC1 strains were the reservoir for the emergence of EPEC2. Intact tra genes would have enabled the EAF plasmid to disseminate among diarrheagenic E. coli.
Supplementary Material
Acknowledgments
We thank Isabel Scaletsky, Jane Michalski, and John Chapman for technical assistance; William Reznikoff for helpful discussions; and Michael Chandler for providing IS data.
This study was supported by Public Health Service grants AI-21657 (J.B.K.), AI-44387 (F.R.B. and V.B.), and HSSN26620040040C from NIH/NIAID (ERIC-BRC project) and by the U.S. Army Medical Command (C.B.).
The views expressed here are those of the authors and do not necessarily represent those of the Department of the Army or Department of Defense.
Editor: J. B. Bliska
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Anthony, K. G., W. A. Klimke, J. Manchak, and L. S. Frost. 1999. Comparison of proteins involved in pilus synthesis and mating pair stabilization from the related plasmids F and R100-1: insights into the mechanism of conjugation. J. Bacteriol. 181:5149-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldini, M. M., J. B. Kaper, M. M. Levine, D. C. Candy, and H. W. Moon. 1983. Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J. Pediatr. Gastroenterol. Nutr. 2:534-538. [DOI] [PubMed] [Google Scholar]
- 3.Bieber, D., S. W. Ramer, C.-Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 4.Blank, T. E., H. Zhong, A. L. Bell, T. S. Whittam, and M. S. Donnenberg. 2000. Molecular variation among type IV pilin (bfpA) genes from diverse enteropathogenic Escherichia coli strains. Infect. Immun. 68:7028-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebersbach, G., and K. Gerdes. 2001. The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc. Natl. Acad. Sci. USA 98:15078-15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girón, J. A., A. S. Y. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 9.Glasner, J. D., P. Liss, G. Plunkett III, A. Darling, T. Prasad, M. Rusch, A. Byrnes, M. Gilson, B. Biehl, F. R. Blattner, and N. T. Perna. 2003. ASAP, a systematic annotation package for community analysis of genomes. Nucleic Acids Res. 31:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gómez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jerse, A. E., and J. B. Kaper. 1991. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect. Immun. 59:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 13.Levine, M. M., J. P. Nataro, H. Karch, M. M. Baldini, J. B. Kaper, R. E. Black, M. L. Clements, and A. D. O'Brien. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152:550-559. [DOI] [PubMed] [Google Scholar]
- 14.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manwaring, N. P., R. A. Skurray, and N. Firth. 1999. Nucleotide sequence of the F plasmid leading region. Plasmid 41:219-225. [DOI] [PubMed] [Google Scholar]
- 16.McConnell, M. M., H. Chart, S. M. Scotland, H. R. Smith, G. A. Willshaw, and B. Rowe. 1989. Properties of adherence factor plasmids of enteropathogenic Escherichia coli and the effect of host strain on expression of adherence to HEp-2 cells. J. Gen. Microbiol. 135:1123-1134. [DOI] [PubMed] [Google Scholar]
- 17.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nataro, J. P., M. M. Baldini, J. B. Kaper, R. E. Black, N. Bravo, and M. M. Levine. 1985. Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J. Infect. Dis. 152:560-565. [DOI] [PubMed] [Google Scholar]
- 20.Nataro, J. P. and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okeke, I. N., J. A. Borneman, S. Shin, J. L. Mellies, L. E. Quinn, and J. B. Kaper. 2001. Comparative sequence analysis of the plasmid-encoded regulator of enteropathogenic Escherichia coli strains. Infect. Immun. 69:5553-5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 23.Schneiker, S., B. Kosier, A. Puhler, and W. Selbitschka. 1999. The Sinorhizobium meliloti insertion sequence (IS) element ISRm14 is related to a previously unrecognized IS element located adjacent to the Escherichia coli locus of enterocyte effacement (LEE) pathogenicity island. Curr. Microbiol. 39:274-281. [DOI] [PubMed] [Google Scholar]
- 24.Sohel, I., J. L. Puente, S. W. Ramer, D. Bieber, C.-Y. Wu, and G. K. Schoolnik. 1996. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J. Bacteriol. 178:2613-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone, K. D., H. Z. Zhang, L. K. Carlson, and M. S. Donnenberg. 1996. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol. 20:325-337. [DOI] [PubMed] [Google Scholar]
- 26.Tatsuno, I., M. Horie, H. Abe, T. Miki, K. Makino, H. Shinagawa, H. Taguchi, S. Kamiya, T. Hayashi, and C. Sasakawa. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tobe, T., T. Hayashi, C. G. Han, G. K. Schoolnik, E. Ohtsubo, and C. Sasakawa. 1999. Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect. Immun. 67:5455-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 29.Trabulsi, L. R., R. Keller, and T. A. Tardelli Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whittam, T. S., and E. A. McGraw. 1996. Clonal analysis of EPEC serogroups. Rev. Microbiol. Sao Paulo 27(Suppl. 1):7-16. [Google Scholar]
- 31.Whittam, T. S., M. L. Wolfe, I. K. Wachsmuth, F. Orskov, I. Orskov, and R. A. Wilson. 1993. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect. Immun. 61:1619-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.