Abstract
Autotransporters are secreted virulence factors that comprise three domains: an N-terminal signal peptide, an internal passenger domain, and a C-terminal β-domain. The mechanism of passenger translocation across the outer membrane remains undefined, with four models having been proposed: the “hairpin,” the “threading,” the “multimeric,” and the “Omp85 (YaeT)” models. In an attempt to understand autotransporter biogenesis, we screened the sequences of the serine protease subfamily of autotransporters (SPATEs) for conserved features indicative of a common secretion mechanism. Our analyses revealed a strictly conserved 14-amino-acid motif within the predicted α-helical linker region, upstream of the β-domain of SPATEs. We investigated the function of this motif through a mutagenesis approach using Tsh as a model. Our studies demonstrate that mutations throughout the conserved motif do not block insertion of the β-domain into the outer membrane. However, nonconservative mutations of four hydrophobic (V1099, L1102, G1107, and L1109) and three polar (N1100, K1104, and R1105) residues of the motif severely decrease or even abolish Tsh biogenesis. Further studies showed that these mutations interfere with passenger transport across the outer membrane. Bioinformatical analyses suggest that the critical polar and hydrophobic amino acids localize on opposite sides of the helix that runs through the β-barrel pore. Our data indicate that the conserved motif is important for passenger secretion across the outer membrane and that mutations in certain residues severely affect the secretion process. We discuss how these results fit with the four proposed models for autotransporter secretion and potential applications in antimicrobial and vaccine development.
The autotransporter (AT) or type V secretion pathway is a mechanism employed by gram-negative bacteria for transport of virulence factors to the cell surface or the extracellular environment (9, 12, 19). Substrates of this pathway are large proteins with diverse functions in pathogenesis, classified as toxins, proteases, adhesins, and mediators of intracellular motility or serum resistance (8). ATs comprise three distinct domains: an N-terminal signal sequence, an internal passenger domain that performs the main functions of the protein, and a C-terminal β-domain (9, 12, 19). An α-helical region, known as the linker, is located upstream of the β-domain (12). During the course of secretion, the N-terminal signal peptide targets the protein to the Sec apparatus for secretion across the inner membrane and is then cleaved by a signal peptidase to release the protein into the periplasmic compartment (28). Subsequently the C terminus of the protein inserts into the outer membrane (OM), forming a β-barrel, and the passenger domain is transported to the extracellular environment (14). Once on the bacterial surface, the passenger may either remain attached to the cell or be cleaved and released to the external milieu (12, 14, 19).
Hundreds of ATs have been identified to date by in silico and experimental analyses (39). Based on structural features, ATs are divided into two main subfamilies: the conventional and the trimeric (12, 19). Proteins of the intimin/invasin group are also proposed to be putative ATs; however, there are no experimental data to confirm this hypothesis (12, 19). Conventional ATs contain long C-terminal domains capable of forming a complete β-barrel in the OM. In contrast, trimeric ATs possess very short C-terminal domains and form trimeric β-barrels (3). The serine protease ATs of the Enterobacteriaceae (SPATEs) are a distinct group of conventional ATs, characterized by the conserved proteolytic motif GDSGS within their sequences (4). To date no SPATE has yet been identified in a nonpathogenic bacterium (9). They are implicated in various diseases, and some representative members include Pic from Shigella flexneri (7), Pet from enteroaggregative Escherichia coli (5), and Tsh (Hbp) from avian, human septic, and uropathogenic E. coli strains (6, 23, 26).
Despite the involvement of ATs in pathogenesis, little is known regarding transport of the passenger across the OM (14). Currently, there are four proposed models to describe this critical step of the secretion process. The hairpin and threading models favor transport of the passenger through the OM pore formed by the β-domain of the protein (10, 22, 25). In the hairpin model the passenger is secreted in a C- to N-terminal fashion, whereas in the threading model the N terminus of the passenger is transported first (22). The multimeric model, on the other hand, supports formation of multimeric channels for ATs. In this case, secretion of the passenger domain is believed to occur through a central channel formed by the walls of six or more β-barrels (33). Studies with Neisseria meningitidis have suggested a fourth putative secretion model, in which OM insertion and secretion of ATs involve Omp85 (35), an essential OM accessory factor (36).
In this study we investigated the role of the α-helical linker on AT secretion across the OM. We identified a conserved motif with the consensus sequence EVNNLNKRMGDLRD within the predicted α-helical linker regions of all SPATEs and analyzed its role in protein biogenesis using Tsh as a model. Our data demonstrate that seven amino acids of the motif are essential for protein secretion across the OM. Further analyses suggested that this motif is part of the α-helix spanning the β-barrel pore, and its arrangement confers polarity on the helical structure. Taken together, our findings establish that the 14-amino-acid conserved motif is essential for SPATE secretion across the OM and provide insights into the mechanism of translocation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacteria used in this study were the Escherichia coli K-12 strains UT5600 (OmpT− OmpP−) (17) for extraction of plasmid pYA3418 and expression of a Tsh protein with an alanine instead of serine 259 at the proteolytically active site (TshS259A) from plasmid pYA3432 and XL1-Blue for transformation (Stratagene, La Jolla, CA). Plasmids pYA3418 harboring the tsh gene and pYA3432 carrying a mutated version of the tsh gene that encodes TshS259A have been previously constructed from plasmid pWKS30 (37) as described by Stathopoulos et al. (29). All strains were cultured at 37°C in Luria-Bertani (LB) broth supplemented with ampicillin (100 μg/ml).
Site-directed mutagenesis.
Plasmid pYA3418 was extracted from E. coli UT5600 using the PureLink HQ miniplasmid purification kit (Invitrogen, Carlsbad, CA). Site-directed mutagenesis was performed according to the QuikChange site-directed mutagenesis protocol (Stratagene) using PfuTurbo, a high-fidelity polymerase. The designed primers for all mutations are shown in Table 1. The PCR product was digested with 2 μl of DpnI, purified using the QIAquick PCR purification kit (QIAGEN, Valencia, CA), and subjected to a second DpnI digestion to ensure complete digestion of the methylated parental plasmid. XL1-Blue competent cells were transformed (5 μl of PCR product per 50 μl of cells) by heat shock and screened for ampicillin resistance. All constructs were sequenced at Lone Star Labs (Houston, TX).
TABLE 1.
Sequences of primers used for PCR amplification
| Primer | Sequence (5′-3′)a |
|---|---|
| E1098A-Fwd | GCT ATA ACA ACT TCA TCA CTG CAG TTA ACA ACC TGA ACA AAC GCA TGG |
| E1098A-Rev | CCA TGC GTT TGT TCA GGT TGT TAA CTG CAG TGA TGA AGT TGT TAT AGC |
| V1099R-Fwd | GCT ATA ACA ACT TCA TCA CTG AAC GTA ACA ACC TGA ACA AAC GCA TGG |
| V1099R-Rev | CCA TGC GTT TGT TCA GGT TGT TAC GTT CAG TGA TGA AGT TGT TAT AGC |
| N1100A-Fwd | CTT CAT CAC TGA AGT TGC CAA CCT GAA CAA ACG CAT GGG CG |
| N1100A-Rev | CGC CCA TGC GTT TGT TCA GGT TGG CAA CTT CAG TGA TGA AG |
| N1101A-Fwd | CTT CAT CAC TGA AGT TAA CGC CCT GAA CAA ACG CAT GGG CG |
| N1101A-Rev | CGC CCA TGC GTT TGT TCA GGG CGT TAA CTT CAG TGA TGA AG |
| L1102R-Fwd | CTT CAT CAC TGA AGT TAA CAA CCG GAA CAA ACG CAT GGG CG |
| L1102R-Rev | CGC CCA TGC GTT TGT TCC GGT TGT TAA CTT CAG TGA TGA AG |
| N1103A-Fwd | GAA GTT AAC AAC CTG GCC AAA CGC ATG GGC GAT TTG AGG |
| N1103A-Rev | CCT CAA ATC GCC CAT GCG TTT GGC CAG GTT GTT AAC TTC |
| K1104A-Fwd | CAC TGA AGT TAA CAA CCT GAA CGC ACG CAT GGG CGA TTT GAG GG |
| K1104A-Rev | CCC TCA AAT CGC CCA TGC GTG CGT TCA GGT TGT TAA CTT CAG TG |
| R1105A-Fwd | GTT AAC AAC CTG AAC AAA GCC ATG GGC GAT TTG AGG GAT ATT AAT GGC G |
| R1105A-Rev | CGC CAT TAA TAT CCC TCA AAT CGC CCA TGG CTT TGT TCA GGT TGT TAA C |
| R1105K-Fwd | GTT AAC AAC CTG AAC AAA AAG ATG GGC GAT TTG AGG GAT ATT AAT GGC |
| R1105K-Rev | GCC ATT AAT ATC CCT CAA ATC GCC CAT CTT TTT GTT CAG GTT GTT AAC |
| M1106A-Fwd | CAA CCT GAA CAA ACG CGC GGG CGA TTT GAG GGA TAT TAA TGG CG |
| M1106A-Rev | CGC CAT TAA TAT CCC TCA AAT CGC CCG CGC GTT TGT TCA GGT TG |
| G1107R-Fwd | CCT GAA CAA ACG CAT GCG CGA TTT GAG GGA TAT TAA TGG CGA AGC C |
| G1107R-Rev | GGC TTC GCC ATT AAT ATC CCT CAA ATC GCG CAT GCG TTT GTT CAG G |
| D1108A-Fwd | CCT GAA CAA ACG CAT GGG CGC TTT GAG GGA TAT TAA TGG CG |
| D1108A-Rev | CGC CAT TAA TAT CCC TCA AAG CGC CCA TGC GTT TGT TCA GG |
| L1109R-Fwd | CGC ATG GGC GAT CGG AGG GAT ATT AAT GGC GAA GCC GG |
| L1109R-Rev | CCG GCT TCG CCA TTA ATA TCC CTC CGA TCG CCC ATG CG |
| R1110A-Fwd | CGC ATG GGC GAT TTG GCG GAT ATT AAT GGC GAA GCC GG |
| R1110A-Rev | CCG GCT TCG CCA TTA ATA TCC GCC AAA TCG CCC ATG CG |
| D1111A-Fwd | GC ATG GGC GAT TTG AGG GCT ATT AAT GGC GAA GCC GGT ACG |
| D1111A-Rev | CGT ACC GGC TTC GCC ATT AAT AGC CCT CAA ATC GCC CAT GC |
The mutated nucleotides are in boldface.
Outer membrane preparations.
E. coli XL1-Blue cells transformed with the mutated or wild-type pYA3418 or with the pWKS30 vector were grown overnight at 37°C in LB media containing ampicillin. The cultures were diluted 1:100 in fresh LB media supplemented with ampicillin and incubated at 37°C. At an optical density of 0.6 at 600 nm, expression of Tsh was induced for 3 h by the addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Bacterial cells were harvested by centrifugation at 4,000 rpm (Beckman; J2-21 M centrifuge, rotor JA-10) for 30 min at 4°C and resuspended in 20 mM Tris-HCl, 1 mM EDTA, and 0.1 mM phenylmethylsulfonyl fluoride (PMSF), pH 7.4. The cells were ruptured by passage through a French press twice at 10,000 lb/in2, and unbroken cells were removed by centrifugation at 5,000 rpm (Eppendorf; 5804R, rotor F34638) for 10 min at 4°C. Cell membranes were pelleted at 28,000 rpm (Beckman; L7-55 ultracentrifuge, rotor SW28) for 1 h at 4°C. The inner membrane was solubilized in 0.5% Sarkosyl and 20 mM Tris-HCl, pH 7.4, and separated from the OM by centrifugation at 28,000 rpm for 1 h at 4°C. The pellet containing the OM was resuspended in 20 mM Tris-HCl, pH 7.4.
Protease accessibility assays.
Bacterial cells were grown under the conditions described above, and after induction of Tsh expression, the cells were harvested by centrifugation and resuspended against 20 mM Tris-HCl, pH 7.4, and 10 mM MgCl2. Incubation with proteinase K (10 μg/ml) was for 1 h at 37°C. The reaction was stopped by addition of 0.1 mM PMSF. After treatment with 1 mM EDTA, the cells were lysed by French press and the OMs were isolated as described above. Cells that were not treated with the corresponding protease served as controls. E. coli UT5600 expressing TshS259A, a protein that is partially retained on the extracellular side of the OM due to the absence of the OM proteases OmpT and OmpP and the S259A mutation in the protein itself, was used as a positive control for the proteinase K activity. To permeabilize the cells, incubation with proteinase K was in 20 mM Tris-HCl and 5 mM EDTA. When OMs were used, 40 μg of total OM proteins was incubated with 2 μg of proteinase K in a total volume of 50 μl of 20 mM Tris-HCl, pH 7.4, and 10 mM MgCl2, at 37°C for 1 h. The reactions were stopped by addition of 0.1 mM PMSF, and 10 μl of each reaction mixture was immediately mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading dye, boiled for 5 min, and resolved on an 8% gel. Reactions with OMs not treated with protease were included as controls. For the purified passenger domain of Tsh (Tshs) 3 μg of protein was incubated with proteinase K (10 μg/ml) in a total volume of 500 μl of 20 mM Tris-HCl, pH 7.4, and 10 mM MgCl2 as described above. Purification of Tshs was performed by ammonium sulfate precipitation and anion-exchange chromatography as described by Kostakioti and Stathopoulos (15).
Electrophoreses and immunoblots.
Supernatants from the cell cultures were screened for secretion of Tsh by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analyses. Culture supernatants (300 μl) were subjected to acetone precipitation, and the proteins were dissolved in SDS-PAGE buffer, separated on an 8% SDS-PAGE gel, and visualized with Coomassie stain. For OMs loading was normalized by estimating equal amounts of the OmpF and OmpA porins (20). For the protease protection assay, equal protein amounts (30 μg) of treated and untreated samples, measured by the bicinchoninic acid assay (Bio-Rad, Hercules, CA), were loaded in each lane. In the case of strain UT5600 expressing TshS259A 50 μg of total protein was loaded for both the treated and untreated samples to account for the smaller amount of Tsh in the OM since some of the protein is secreted into the medium. For immunoblotting, the gels were transferred to a nitrocellulose membrane (Bio-Rad) by semidry electrotransfer. Membranes were blocked with 1% gelatin in Tris-buffered saline overnight at 37°C. For immunodetection of Tsh, membranes were incubated with rabbit Tsh antiserum (diluted 1:3,000) for 1 h 30 min at 37°C. After being washed for 10 min the membranes were probed with horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG; Bio-Rad) at a dilution of 1:20,000, for 1 h at 37°C. The blots were developed by enhanced chemiluminescence (Pierce, Rockford, IL) according to the manufacturer's instructions. SDS-prestained markers (Bio-Rad) were used for estimation of the molecular mass. Densitometric analyses were performed using ImageJ software (http://rsb.info.nih.gov/ij/).
Bioinformatics.
All sequence alignments were performed using MacVector software, version 4.0. For the SPATEs, the whole protein sequence excluding the signal peptide was used in the alignment. Secondary-structure predictions were made based on alignment of the last 299 amino acids of Tsh with the translocator domain of NalP. The model for the translocator domain of Tsh was constructed in Swiss model (www.expasy.org) with NalP as the template. The last 299 amino acids of the protein were used, in accordance to the length of the crystallized NalP translocator domain.
RESULTS
A motif of 14 conserved residues localizes in the helical linker region of SPATEs.
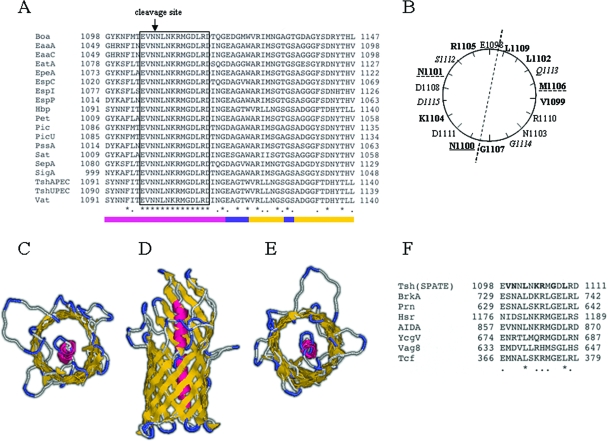
Aiming to understand the mechanism of AT secretion across the OM, we screened the sequences of ATs of the SPATE group for characteristics indicative of a common secretion process. Alignment of the sequences of 19 SPATEs revealed not only a high degree of homology within their C-terminal domains but also an identical 14-amino-acid motif with the consensus sequence EVNNLNKRMGDLRD localizing upstream of their β-domains (Fig. 1A). This sequence represented the longest conserved stretch of amino acids in the sequence alignment of the SPATEs. The remarkable absence of any variation within this sequence in all known SPATE proteins led us to the hypothesis that these amino acids may possess important roles during secretion. Screening of the data bank revealed that this motif is present with some variations in the sequences of other AT proteins as well, including E. coli AIDA and several ATs of Bordetella pertussis (Fig. 1F). Secondary-structure predictions suggested that the motif EVNNLNKRMGDLRD adopts an α-helical configuration. Alignment of the translocator domain (linker region and β-domain) of NalP, the only AT translocator with a solved crystal structure (22), with the corresponding C-terminal sequence of Tsh predicted that the conserved motif lies within the linker region (Fig. 1A). All ATs are predicted to possess an α-helical linker region localizing in the interface between the passenger and the β-domain, with some variations in terms of the exact position of the linker relative to the protein's C-terminal cleavage site (21). Despite the low level of homology between the SPATEs and other ATs that carry a homologue of the 14-amino-acid motif, in all cases the motif is located in the interface between the passenger and the β-domain of the protein. We constructed a model for the translocator domain of Tsh using SWISS model software, which constructs protein models based on an existing structure with the highest degree of homology, and NalP as the template. The constructed model for Tsh consisted of a 12-stranded β-barrel N-terminally connected to the α-helical linker region, in agreement with the NalP structure (Fig. 1C to E). In this model the 14-amino-acid conserved motif localizes within the α-helix that plugs the formed β-barrel pore, suggesting participation in interactions that may lead to translocation of the passenger domain across the OM. Additionally, a helical-wheel projection of the 14 amino acids indicated an asymmetric arrangement of the conserved residues with hydrophobic and polar amino acids clustered on opposite sides of the α-helix (Fig. 1B).
FIG. 1.
Identification and structural characterization of the 14-amino-acid conserved motif upstream of the β-domains of all SPATEs. (A) Alignment of the sequences of 19 SPATEs showing the conserved motif. Identical residues are marked with asterisks and homologous residues with dots. The identical motif is within the boxed region. The bar indicates secondary-structure predictions based on bioinformatic analyses of Tsh using the NalP translocator domain as a template. Color coding: α-helix in pink, extracellular loops and periplasmic turns in blue, and β-strands in yellow. (B) Schematic representation of the residues of the conserved motif depicted on a helical wheel. The hydrophobic and polar residues that affect outer membrane transport are in boldface, residues that interfere with protein maturation are underlined with a solid line, and residues that have minor effects on protein biogenesis are underlined with a broken line. Residues in italics are not part of the conserved motif. (C) Top view of the model constructed for the translocator domain of Tsh using SWISS model software. Tertiary-structure predictions were based on the crystal structure of NalP. (D) Side view of the Tsh model showing the 12-strand β-barrel with the α-helix within the pore. (E) Bottom view of the Tsh model showing the periplasmic turns. (F) Identification of the conserved motif in non-SPATE autotransporters and alignment with the SPATE motif. The NCBI protein data bank was screened using the C-terminal 299 amino acids of Tsh and narrowed by querying the 14-amino-acid motif. Seven of the identified proteins along with their accession numbers are BrkA, Prn, Vag8, and Tcf from Bordetella pertussis (AAA51646, A32560, AAC31247, and CAD12826, respectively); Hsr from Helicobacter mustelae (AAC36865); AIDA from E. coli F11 (ZP00725747); and YcgV from E. coli (NP415720).
Specific hydrophobic and polar residues of the conserved α-helical motif are critical for secretion of Tsh.
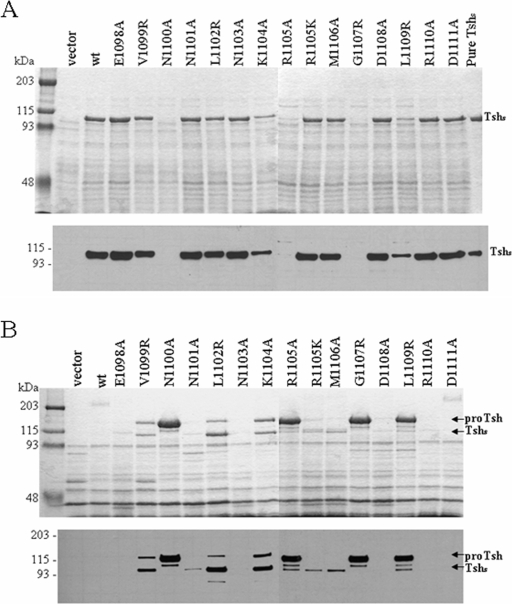
To test the significance of the conserved α-helical motif in the secretion of SPATEs, we constructed a series of mutations within the motif of the Tsh AT (residues 1098 to 1111 of the precursor protein) and examined their effects in Tsh biogenesis. Each mutant maintained the whole tsh gene intact but contained a mutation in one of the residues composing the EVNNLNKRMGDLRD motif. All constructs were screened for secretion of Tsh in the supernatant by SDS-PAGE (Fig. 2A, top) and immunoblot analyses (Fig. 2A, bottom) with an antiserum raised against Tshs. This antiserum detects the free passenger domain (∼106 kDa) and the unprocessed form of Tsh (∼140 kDa). Our data reveal that nonconservative substitution of residues K1104 and L1109 significantly decreases the release of Tshs in the external medium, whereas nonconservative mutations of residues N1100, R1105, and G1107 result in complete absence of Tshs from the culture supernatant. In contrast, conservative mutation (R to K) of residue R1105 does not seem to affect secretion. No effects on secretion were observed in the case of nonconservative mutations in the rest of the amino acids of the motif. Our results demonstrate that specific polar and hydrophobic residues of the helical motif are essential for Tsh biogenesis.
FIG. 2.
Mutations in the conserved α-helical motif affect biogenesis of Tsh. (A) SDS-PAGE profile (top panel) and Western blot analyses (bottom panel) of culture supernatants. Samples were concentrated by acetone precipitation. (B) SDS-PAGE profile (top panel) and Western blot analyses (bottom panel) of OMs isolated from E. coli XL1-Blue expressing the different constructs. The gels were stained with Coomassie blue. Polyclonal antiserum against Tshs was used for immunoblot analyses. Lanes are labeled according to the specific mutation that each construct carries within the unprocessed Tsh. Vector indicates the absence of tsh, wt indicates the presence of the wild-type tsh gene cloned into vector pWKS30, and Tshs is the secreted domain purified by anion-exchange chromatography. ProTsh, the 140-kDa unprocessed form of Tsh.
Mutations of five hydrophobic and four polar residues of the motif lead to accumulation of the protein in the OM.
In the wild-type strain almost all Tsh is released into the extracellular environment as a 106-kDa protein and only a small amount of protein remains covalently attached to the OM (29). Therefore, the inability of some of the mutants to secrete the protein into the external medium can be interpreted in three ways: either protein insertion into the OM is blocked, secretion across the membrane is inefficient, or cleavage and release of the passenger are abolished. To distinguish between these three possibilities, we screened all our constructs for the presence of Tsh in the OM. All mutants were grown under identical conditions and lysed by French press, and their OMs were isolated and examined by SDS-PAGE (Fig. 2B, top) and Western blot analyses (Fig. 2B, bottom). Cells carrying the empty vector or the wild-type Tsh were used as negative controls. Our results indicate accumulation of the unprocessed 140-kDa form of Tsh in the OMs of all strains carrying a mutation in any of the hydrophobic amino acids of the motif, with the exception of M1106. In addition, the 140-kDa band was detected in the OMs of constructs carrying a nonconservative mutation in the charged residues K1104 and R1105 and the polar residue N1100. This band was absent from all other mutants, in agreement with the ability of these strains to secrete the protein into the external medium with an efficiency similar to that of the wild-type strain (Fig. 2A). Furthermore, increased amounts of the processed 106-kDa Tshs were observed in the OM as a result of mutations in residues V1099, L1102, K1104, R1105, and L1109 of the motif. Some Tshs was also detected in the OM due to mutations N1101 and M1106, which seem to have minor effects in Tsh biogenesis. These data indicate that mutations in the conserved motif do not block insertion of the β-domain into the OM but interfere with transport of the passenger across the membrane and/or its proteolytic processing and maturation.
Mutations in seven residues of the motif severely affect passenger transport across the OM.
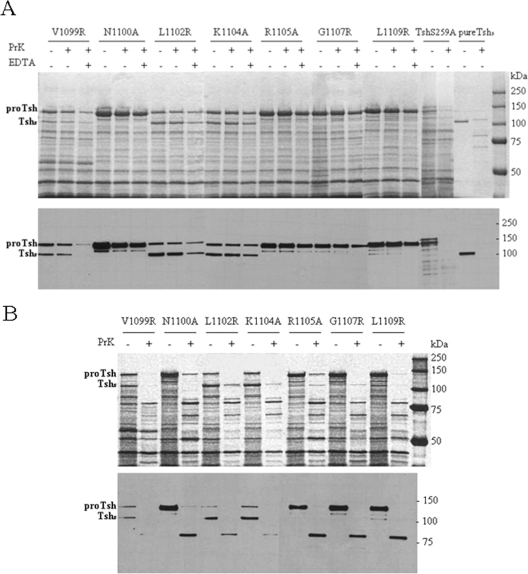
We used a protease protection assay to examine whether the OM-accumulated protein localizes in the external or periplasmic side of the OM. Such localization would indicate whether the corresponding residues of the motif are important in the initial or later steps of OM secretion. For this purpose, equal volumes of whole cells of all mutants with major effects in Tsh biogenesis were treated with the same amount of proteinase K under identical conditions. Proteinase K was used at a concentration of 10 μg/ml as higher concentrations of the enzyme resulted in nonspecific degradation of most OM proteins. The reaction was stopped by addition of PMSF, and OMs were isolated and examined for cleavage of unprocessed Tsh. Since cleavage was expected within the passenger domain had it been transported extracellularly, the digestion profile of purified Tshs was used for comparison (Fig. 3A). A surface-localized form of Tsh expressed by UT5600 was used as an additional positive control. Our analyses showed that the 140-kDa band remains intact in all mutants with the exception of construct N1100A, where there is a 30% ± 3% decrease in the corresponding band intensity, as determined by densitometric analyses. A 5% ± 0.4% decrease in the 140-kDa band is also observed in construct V1099R (Fig. 3A). As expected almost complete degradation of the surface-localized TshS259A was observed in the presence of the same amount of proteinase K. When incubation with proteinase K proceeds in the presence of the permeabilizing agent EDTA, a small degree of cleavage of the 140-kDa protein is observed in all mutants. Similar results were obtained when trypsin was used instead of proteinase K under the same conditions (data not shown). These observations argue against surface localization of the protein. To further confirm these results, we treated isolated OMs of these constructs with proteinase K and looked for cleavage of the unprocessed form of Tsh. Our data reveal that under these conditions the 140-kDa protein is almost completely digested in all mutants (Fig. 3B). This was also the case for the OM-accumulated 106-kDa Tshs (Fig. 3A and B). Taken together these observations demonstrate that mutations in residues V1099, L1102, K1104, R1105, G1107, and L1109 interfere with the early stages of AT transport across the OM, whereas substitution of residue N1100 affects transport efficiency and subsequent processing of the 140-kDa protein.
FIG. 3.
Mutations in specific hydrophobic and polar residues of the motif block passenger secretion across the OM. (A) SDS-PAGE (top panel) and Western blot (bottom panel) analyses of isolated OMs. Whole cells were treated with proteinase K (PrK; 10 μg/ml) in the presence and absence of EDTA. Cells that were not treated with externally added protease were used as controls. TshS259A expressed from E. coli UT5600 represents a surface-localized form of Tsh and was used as a control for assay efficiency. (B) SDS-PAGE (top panel) and Western blot (bottom panel) analyses of isolated OMs treated with proteinase K (10 μg/ml). ProTsh, the 140-kDa unprocessed form of Tsh.
DISCUSSION
In this study we investigated the role of the linker region in AT secretion across the OM in the SPATE group. We identified a 14-amino-acid conserved motif with the consensus sequence EVNNLNKRMGDLRD upstream of the β-domains of all SPATEs. Bioinformatical analyses using Tsh suggested that this motif (residues 1098 to 1111 of the unprocessed protein) is part of the α-helical linker region that runs through the C-terminal β-barrel pore. Consideration that the linker region has been previously implicated in AT secretion made our observations even more intriguing. More specifically, sequential deletions of the linker region have been shown to decrease the translocation efficiency of heterologous proteins (16, 30), truncated natural AT passenger domains (13, 21), or natural passenger domains carrying deletions and epitope insertions (34). However, the exact role and mechanism of function of the linker in the secretion process are undefined. Our analyses supported the view that arrangement of the 14 conserved residues in the α-helix confers polarity to the helical structure, suggesting that each side of the helix participates in different interactions. Site-directed mutagenesis, targeting one residue of the conserved motif at a time, revealed that, although none of the 14 amino acids is essential for protein insertion into the OM, 9 residues are involved in Tsh biogenesis (Table 2). More specifically secretion of Tshs into the supernatant was decreased as a result of nonconservative mutations in residues K1104 and L1109 and abolished in the case of residues N1100, R1105, and G1107. The observed phenotypes were due to inefficient protein translocation through the OM, which led to accumulation of the ∼140-kDa unprocessed form of Tsh in the membrane. Unprocessed Tsh was also observed in the OM as a result of mutations V1099R and L1102R, which had no detectable effects on passenger release. In addition, processed 106-kDa Tshs was detected in the OM due to mutations V1099R, L1102R, K1104A, R1105A, and L1109R. Small amounts of Tshs were present in the OM due to mutations N1101A, R1105K, and M1106A, which had a minor impact in Tsh biogenesis. Treatment of intact and permeabilized whole cells with proteinase K demonstrated that, in the 140-kDa OM-accumulated Tsh, the passenger domain is found on the periplasmic side of the membrane in mutants L1102R, K1104A, R1105A, G1107R, and L1109R. This was also the case for the processed Tshs. Some amounts of passenger are shown to localize extracellularly in mutant N1100A, where the mutation has a dual effect, interfering not only with passenger secretion but also with its maturation as well. In agreement with these data, Navarro-Garcia et al. reported that mutation of the corresponding residue (N1019) in Pet affects cleavage and release of the protein to the external milieu in the double mutant N1018G-N1091I, although confocal microscopy indicated the presence of Pet at the cell surface (18). Our findings were further supported by the extensive degradation of the 140-kDa band when isolated OMs were treated with proteinase K under identical conditions. The smaller degree of cleavage observed in the case of permeabilized cells may be the result of interactions with periplasmic chaperones. Interactions with periplasmic chaperones that protect the AT from proteolytic degradation have been previously reported for IcsA and the IgA1 protease (2, 32). Taken together, these results show that the conserved α-helical motif is important for secretion of the passenger domain across the OM and that mutations in specific residues either block passenger translocation in its early steps or interfere with maturation after surface exposure.
TABLE 2.
Summary of the effects that mutations in the conserved α-helical motif have on Tsh biogenesis
| Mutation | Secretion in the supernatantb | Presencec of Tsh in the OM
|
Cleavaged of the 140-kDa band by PrKa
|
||
|---|---|---|---|---|---|
| 106 kDa | 140 kDa | Whole cells | OM | ||
| None (wild type) | + | − | − | NA | NA |
| E1098A | + | − | − | NA | NA |
| V1099R | + | + | + | − | + |
| N1100A | − | − | ++ | + | + |
| N1101A | + | + | − | NA | NA |
| L1102R | + | ++ | + | − | + |
| N1103A | + | − | − | NA | NA |
| K1104A | Decreased | ++ | + | − | + |
| R1105A | − | + | ++ | − | + |
| R1105K | + | + | − | NA | NA |
| M1106A | + | + | − | NA | NA |
| G1107R | − | − | ++ | − | + |
| D1108A | + | − | − | NA | NA |
| L1109R | Decreased | + | ++ | − | + |
| R1110A | + | − | − | NA | NA |
| D1111A | + | − | − | NA | NA |
PrK, proteinase K.
+, secretion; −, no secretion.
++, increased amount; +, present; −, absent.
+, cleavage; −, no cleavage; NA, not applicable.
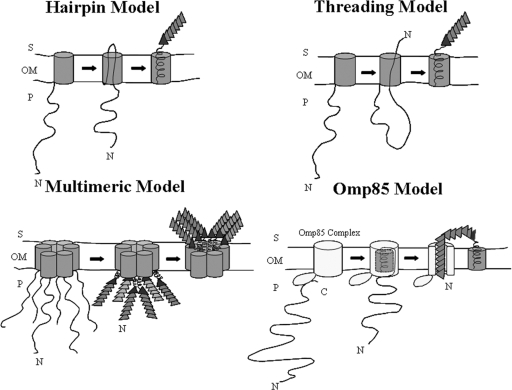
Currently four different models have been proposed to describe the mechanism for secretion of ATs across the OM: the hairpin, the threading, the multimeric, and the Omp85 (YaeT) models (Fig. 4). The hairpin and threading secretion models suggest that AT secretion occurs through the OM β-barrel pore formed by the C-terminal protein domain. An α-helix at the N terminus of the β-domain obstructs the pore after secretion. In both models the passenger domain adopts its native conformation extracellularly. Their main difference lies in the way transport of the passenger domain is initiated. In the hairpin model the helical domain at the translocator-passenger interface inserts first into the β-barrel, in an extended mode, and drives transport of the unfolded passenger domain to the cell surface. In the threading model, secretion of the passenger domain proceeds in a N- to C-terminal manner, with the helical domain at the translocator-passenger boundary entering the channel last. This model suggests the presence of only one polypeptide at a time within the pore, and therefore the transported protein could be partially folded (22). The multimeric model, supported by the studies of Veiga and colleagues with IgA1 protease, suggests secretion through a central pore formed by the walls of at least six β-barrel monomers assembled in a ring-shaped complex (33). Such a pore is large enough to allow secretion of folded passenger domains (31, 32). The Omp85 model is based on observations of Voulhoux et al. indicating participation of the Omp85 complex in AT secretion (35). The Omp85 complex is a heteroligomeric protein complex with four currently identified components in E. coli: Omp85/YaeT, YfgL, YfiO, and NlpB (24, 38). According to this model, secretion of ATs may occur through a pore formed in the OM by Omp85 and not the pore of the translocator domain itself (22). This could become possible after interaction of the more conserved C terminus of the AT with the Omp85 complex at the periplasmic side of the OM. Such an interaction may trigger insertion of the translocator domain into the Omp85 pore coupled to folding into a β-barrel that encompasses an α-helix. Once the passenger domain is transported extracellularly, the Omp85 complex may release the AT translocator domain into the OM (1). This model makes AT secretion more homologous to two-partner secretion (12, 19).
FIG. 4.
Proposed models for AT secretion. The folded N-terminal passenger domains are depicted as triangles and the C-terminal domains as cylinders anchored in the OM. The folding initiator domain at the C terminus of the passenger is indicated with a darker triangle. Omp85 represents an OM hetero-oligomeric protein complex composed of at least four currently identified components in E. coli: Omp85/YaeT, YfgL, YfiO, and NlpB. S, cell surface; P periplasm.
Taking into account the proposed models for AT secretion across the OM, our results support transport of the passenger from the C to the N terminus. If secretion occurred in an N terminus-to-C terminus mode, as presented by the threading model, the conserved α-helical linker motif would enter the pore last, and thus mutations in the conserved residues would not have abolished translocation of Tshs to the bacterial surface. This could only be the case if the conserved linker motif folded in an α-helix while still in the periplasm and participated in interactions that drove secretion of the passenger through the pore prior to its own transport. However, this scenario cannot explain the accumulation of processed Tshs in the OM, as the cleavage site would localize intracellularly, being inaccessible to extracellular proteases. Furthermore, the monomeric nature of purified Tsh, as determined by blue native gel electrophoresis, gel filtration, and cross-linking analyses, argues against a multimeric model with a central secretion pore (11). These findings are in agreement with other studies indicating that the EspP, AIDA, and NalP ATs are monomeric (17, 22, 27). Moreover, the multimeric model poses several questions as to how a hydrophilic pore could be formed by the hydrophobic interface of the β-barrel monomers and how the OM lipids could be excluded from such a pore. In addition, given that secretion of six folded passenger domains proceeds through the same pore and almost at the same time, passenger crowding could present a real problem. Thus passenger secretion may occur via either the hairpin or the Omp85 model. In the case of the hairpin model, the secreted protein has to be unfolded, in accordance with the NalP pore dimensions (10 by 12.5 Å). Therefore, the important residues of the conserved motif may interact with residues in the barrel wall, and these interactions prevent the formation of the helix before the passenger has passed by. If due to the mutations the interactions do not take place, the helix will form and the pore will be blocked, which will prevent passenger translocation. It is possible that the cleavage site is more easily accessible to the corresponding protease when the protein is transported in the unfolded form. This would mean that processing takes place before the helix is formed, which would be consistent with the idea that the linker region is initially present within the barrel in a delineated fashion. In this way the polypeptide would be long enough to expose the processing site at the cell surface. Only after processing does the C-terminal part fold back in the barrel to form the helical plug. On the other hand, the conserved motif could adopt its α-helical conformation prior to passenger transport and participate in a folded mode in interactions that pull the passenger to the cell surface. This scenario is supported by observations of folded periplasmic AT intermediates for IcsA, IgA protease, and EspP (2, 27, 32). The presence of a folded α-helix inside the pore makes it unlikely that secretion occurs through the β-barrel of the translocator domain, since the NalP pore dimensions support the presence of barely two extended polypeptides at a time within the channel (22). In this case, passenger transport may occur through the pore of an accessory factor such as Omp85, and SPATE secretion across the OM would better fit with the Omp85 model. The fact that this motif is conserved with slight variations in other ATs, such as B. pertussis BrkA and Prn (Fig. 1F), enhances the importance of our study, suggesting that our findings may apply in the secretion mechanisms of other conventional ATs as well.
The identification and characterization of this 14-amino-acid conserved motif within the α-helical linker region of all SPATEs reported to date may also lead to the development of new tools for prevention and treatment of numerous infectious diseases. Each SPATE is among the predominant secreted proteins and also the most immunogenic proteins of their host pathogen (9). Since SPATEs are found only in pathogenic bacteria (9), this highly conserved motif may be an ideal antigen for vaccine development. In addition, the critical role of certain amino acids of the motif in passenger secretion makes it an ideal target for design of antimicrobials that could interfere with interactions required for export and biogenesis of the secreted virulence factor. Future studies will be performed to explore these possibilities.
Acknowledgments
We thank Jan Tommassen at Utrecht University for helpful discussions and critical review of the manuscript and Anne Delcour at the University of Houston for critical review of the manuscript.
This work was supported by two grants (E-1548) by the Robert Welch Foundation.
Editor: J. B. Bliska
REFERENCES
- 1.Bos, M. P., and J. Tommassen. 2004. Biogenesis of the gram-negative bacterial outer membrane. Curr. Opin. Microbiol. 7:610-616. [DOI] [PubMed] [Google Scholar]
- 2.Brandon, L. D., and M. B. Goldberg. 2001. Periplasmic transit and disulfide bond formation of the autotransported Shigella protein IcsA. J. Bacteriol. 183:951-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cotter, S. E., N. K. Surana, and J. W. St Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13:199-205. [DOI] [PubMed] [Google Scholar]
- 4.Dutta, P. R., R. Cappello, F. Navarro-Garcia, and J. P. Nataro. 2002. Functional comparison of serine protease autotransporters of Enterobacteriaceae. Infect. Immun. 70:7105-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eslava, C., F. Navarro-Garcia, J. R. Czeczulin, I. R. Henderson, A. Cravioto, and J. P. Nataro. 1998. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect. Immun. 66:3155-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heimer, S. R., D. A. Rasko, C. V. Lockatell, D. E. Johnson, and H. L. Mobley. 2004. Autotransporter genes pic and tsh are associated with Escherichia coli strains that cause acute pyelonephritis and are expressed during urinary tract infection. Infect. Immun. 72:593-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson, I. R., J. Czeczulin, C. Eslava, F. Noriega, and J. P. Nataro. 1999. Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 11.Hritonenko, V., M. Kostakioti, and C. Stathopoulos. Quaternary structure of a SPATE autotransporter protein. Mol. Membr. Biol., in press. [DOI] [PubMed]
- 12.Jacob-Dubuisson, F., R. Fernandez, and L. Coutte. 2004. Protein secretion through autotransporter and two-partner pathways. Biochim. Biophys. Acta 1694:235-257. [DOI] [PubMed] [Google Scholar]
- 13.Klauser, T., J. Kramer, K. Otzelberger, J. Pohlner, and T. F. Meyer. 1993. Characterization of the Neisseria Iga beta-core. The essential unit for outer membrane targeting and extracellular protein secretion. J. Mol. Biol. 234:579-593. [DOI] [PubMed] [Google Scholar]
- 14.Kostakioti, M., C. L. Newman, D. G. Thanassi, and C. Stathopoulos. 2005. Mechanisms of protein export across the bacterial outer membrane. J. Bacteriol. 187:4306-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostakioti, M., and C. Stathopoulos. 2004. Functional analysis of the Tsh autotransporter of an avian pathogenic Escherichia coli strain. Infect. Immun. 72:5548-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maurer, J., J. Jose, and T. F. Meyer. 1999. Characterization of the essential transport function of the AIDA-I autotransporter and evidence supporting structural predictions. J. Bacteriol. 181:7014-7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller, D., I. Benz, D. Tapadar, C. Buddenborg, L. Greune, and M. A. Schmidt. 2005. Arrangement of the translocator of the autotransporter adhesin involved in diffuse adherence on the bacterial surface. Infect. Immun. 73:3851-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navarro-Garcia, F., A. Canizalez-Roman, J. Luna, C. Sears, and J. P. Nataro. 2001. Plasmid-encoded toxin of enteroaggregative Escherichia coli is internalized by epithelial cells. Infect. Immun. 69:1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman, L. C., and C. Stathopoulos. 2004. Autotransporter and Two-partner secretion: delivery of large-size virulence factors by gram-negative bacterial pathogens. Crit. Rev. Microbiol. 30:275-286. [DOI] [PubMed] [Google Scholar]
- 20.Nikaido, H., and M. Vaara. 1987. Outer membrane, p. 10-11. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik., M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 21.Oliver, D. C., G. Huang, and R. C. Fernandez. 2003. Identification of secretion determinants of the Bordetella pertussis BrkA autotransporter. J. Bacteriol. 185:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oomen, C. J., P. Van Ulsen, P. Van Gelder, M. Feijen, J. Tommassen, and P. Gros. 2004. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23:1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otto, B. R., R. Sijbrandi, J. Luirink, B. Oudega, J. G. Heddle, K. Mizutani, S. Y. Park, and J. R. Tame. 2005. Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J. Biol. Chem. 280:17339-17345. [DOI] [PubMed] [Google Scholar]
- 24.Paschen, S. A., W. Neupert, and D. Rapaport. 2005. Biogenesis of beta-barrel membrane proteins of mitochondria. Trends Biochem. Sci. 30:575-582. [DOI] [PubMed] [Google Scholar]
- 25.Pohlner, J., R. Halter, K. Beyereuther, and T. F. Meyer. 1987. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325:458-462. [DOI] [PubMed] [Google Scholar]
- 26.Provence, D. L., and R. Curtiss III. 1994. Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect. Immun. 62:1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skillman, K. M., T. J. Barnard, J. H. Peterson, R. Ghirlando, and H. D. Bernstein. 2005. Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Mol. Microbiol. 58:945-958. [DOI] [PubMed] [Google Scholar]
- 28.Stathopoulos, C., D. R. Hendrixson, D. G. Thanassi, S. J. Hultgren, J. W. St Geme III, and R. Curtiss III. 2000. Secretion of virulence determinants by the general secretory pathway in gram-negative pathogens: an evolving story. Microbes Infect. 2:1061-1072. [DOI] [PubMed] [Google Scholar]
- 29.Stathopoulos, C., D. L. Provence, and R. Curtiss III. 1999. Characterization of the avian pathogenic Escherichia coli hemagglutinin Tsh, a member of the immunoglobulin A protease-type family of autotransporters. Infect. Immun. 67:772-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki, T., M. C. Lett, and C. Sasakawa. 1995. Extracellular transport of VirG protein in Shigella. J. Biol. Chem. 270:30874-30880. [DOI] [PubMed] [Google Scholar]
- 31.Veiga, E., V. de Lorenzo, and L. A. Fernandez. 1999. Probing secretion and translocation of a beta-autotransporter using a reporter single-chain Fv as a cognate passenger domain. Mol. Microbiol. 33:1232-1243. [DOI] [PubMed] [Google Scholar]
- 32.Veiga, E., V. de Lorenzo, and L. A. Fernandez. 2004. Structural tolerance of bacterial autotransporters for folded passenger protein domains. Mol. Microbiol. 52:1069-1080. [DOI] [PubMed] [Google Scholar]
- 33.Veiga, E., E. Sugawara, H. Nikaido, V. de Lorenzo, and L. A. Fernandez. 2002. Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO J. 21:2122-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velarde, J. J., and J. P. Nataro. 2004. Hydrophobic residues of the autotransporter EspP linker domain are important for outer membrane translocation of its passenger. J. Biol. Chem. 279:31495-31504. [DOI] [PubMed] [Google Scholar]
- 35.Voulhoux, R., M. P. Bos, J. Geurtsen, M. Mols, and J. Tommassen. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299:262-265. [DOI] [PubMed] [Google Scholar]
- 36.Voulhoux, R., and J. Tommassen. 2004. Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res. Microbiol. 155:129-135. [DOI] [PubMed] [Google Scholar]
- 37.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 38.Wu, T., J. Malinverni, N. Ruiz, S. Kim, T. J. Silhavy, and D. Kahne. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235-245. [DOI] [PubMed] [Google Scholar]
- 39.Yen, Y. T., and C. Stathopoulos. Identification of autotransporter proteins secreted by type V secretion systems in gram-negative bacteria. In M. Van der Giejen (ed.), Methods in molecular biology: protein targeting protocols, in press. Humana Press, Totowa, N.J. [DOI] [PubMed]