Abstract
Antimicrobial peptides, human β-defensin (hBD), and the 18-kDa cationic antimicrobial protein (CAP18) are components of innate immunity. These peptides have antimicrobial activity against bacteria, fungi, and viruses. Actinobacillus actinomycetemcomitans is a gram-negative facultative anaerobe implicated in the initiation of periodontitis. The innate immunity peptides have antibacterial activity against A. actinomycetemcomitans. We investigated the molecular mechanism of human gingival epithelial cells (HGEC) responding to exposure to A. actinomycetemcomitans. HGEC constitutively express hBD1 and inducibly express hBD2, hBD3, and CAP18 on exposure to A. actinomycetemcomitans. The level of expression varies among clinical isolates. In the signaling pathway for hBD2 induction by the bacterial contact, we demonstrate that the mitogen-activated protein (MAP) kinase and not the NF-κB transcription factor pathway is used. We found the outer membrane protein 100 (Omp100; identified by molecular mass) is the component inducing the hBD2 response. Omp100 binds to fibronectin, an extracellular matrix inducing hBD2 via the MAP kinase pathway. Anti-integrin α5β1, antifibronectin, genistein, and PP2 suppress the Omp100-induced expression of hBD2, suggesting that Src kinase is involved through integrin α5β1. The inflammatory cytokines, tumor necrosis factor α (TNF-α), interleukin-1β (IL-1β), IL-6 and IL-8, produced by HGEC on contact with A. actinomycetemcomitans also stimulate expression of hBD2. Further, neutralizing antibody against TNF-α or IL-8 partially inhibits the induction of hBD2 on bacterial contact. Therefore, we found that the induction of the antimicrobial peptides is mediated by a direct response principally through an Omp100-fibronectin interaction, and using secondary stimulation by inflammatory cytokines induced by the bacterial exposure.
Periodontitis is an infectious disease which develops from the interaction of the host and the bacteria in the gingival sulcus (37, 51, 66). Gingival epithelial cells can resist oral pathogenic bacteria as a physical barrier and also produce antimicrobial innate immunity peptides (6). Eukaryotic cells are known to produce many antimicrobial peptides (4, 13). The antimicrobial peptides are defined as containing fewer than 100 amino acids having a broad spectrum of activity against both gram-negative and gram-positive bacteria as well as fungi and viruses (13, 28, 48, 67). Human epithelium produces two major groups of antimicrobial peptides, the β-defensins and the cathelicidin family, including the 18-kDa cationic antimicrobial protein (CAP18, or LL37) (4, 13, 28, 63). The β-defensins are small cysteine-rich cationic antimicrobial peptides (4, 17). They are found in tracheal epithelial cells and in many types of human epithelial cells, including the kidney, urinary tract, oral mucosa, and skin (4, 32, 34, 57). Human β-defensin 1 (hBD1) is constitutively expressed in epithelial cells, whereas hBD2 and hBD3 are inducibly expressed by bacteria, Candida albicans, and inflammatory cytokines (16, 26, 29, 45). These peptides also demonstrate antibacterial activity against the periodontal pathogenic bacteria, Staphylococcus aureus including methicillin-resistant S. aureus and Pseudomonas aeruginosa, although hBD1 and hBD2 have less activity against the gram-positive bacteria (16, 34, 42). hBD1 and hBD2 are salt sensitive whereas hBD3 is not (16). Recently, hBD4 was identified as another salt-sensitive antimicrobial peptide inducibly expressed on bacterial contact (14). CAP18 is a member of the cathelicidin family and is produced in the epithelium and neutrophils (12, 28, 61). CAP18 is salt insensitive and shows a broad spectrum of activity against microorganisms. hBD and CAP18 attract and stimulate immature dendritic cells via the chemokine receptor CCR6 (62, 64).
Actinobacillus actinomycetemcomitans is one of the periodontal pathogenic bacteria implicated in aggressive periodontitis and chronic periodontitis (3, 33, 65). Several virulence factors are identified including lipopolysaccharide (LPS), leukotoxin, cytolethal distending toxin, collagenase, and outer membrane protein (OMP) (2, 9, 22, 27, 44, 52, 53). Studies concerning the interaction between A. actinomycetemcomitans and host cells, especially human gingival epithelial cells (HGEC), are mainly focused on adhesion, invasion by the bacteria, and expression of antimicrobial peptides and inflammatory cytokines in HGEC when cells are exposed to A. actinomycetemcomitans (2, 10, 33, 39, 58). However, there are no reports concerning the bacterial component of A. actinomycetemcomitans that induces antimicrobial peptides. Several groups previously demonstrated that antimicrobial peptides have bactericidal activity against oral bacteria including A. actinomycetemcomitans (21, 31, 36, 42). This suggests that antimicrobial peptides play a role as an immune system against oral bacteria. Therefore, we investigated the expression of antimicrobial peptides in HGEC in response to bacterial contact. We identified the induction molecules on A. actinomycetemcomitans considered to be important for the host-parasite interaction at the molecular level. FliC in Salmonella enterica serovar Enteritidis, protease in Porphyromonas gingivalis, and LPS (Escherichia coli and P. aeruginosa) are the bacterial components presently implicated to induce expression of antimicrobial peptides (8, 30, 40, 59). We studied the A. actinomycetemcomitans cell surface molecules such as LPS or OMPs that may be responsible for induction of antimicrobial peptides. A. actinomycetemcomitans has six major OMPs (identified by their molecular masses), Omp16/18, Omp29, Omp39, Omp64, and Omp100 (23), that may be involved in a variety of factors for virulence including adhesion and invasion into HGEC, serum resistance, and cytokine induction (2). LPS of A. actinomycetemcomitans is one of the major pathogenic factors in periodontal disease. It induces secretion of proinflammatory cytokines and is involved in alveolar bone destruction (19, 60).
Here, we investigate the molecular mechanism that induces antimicrobial peptides after contact with the pathogen, the bacterial surface proteins, or inflammatory cytokines to identify the signaling pathway for hBD2 induction.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains used in this study are listed in Table 1. A. actinomycetemcomitans was cultured in Trypticase soy broth supplemented with 1% (wt/vol) yeast extract in a 5% CO2 atmosphere using the Anaeropack system (Mitsubishi Gas Chemical, Tokyo, Japan). When necessary, kanamycin (25 μg/ml) or spectinomycin (50 μg/ml) was added to the medium.
TABLE 1.
Strains used in this study
| A. actinomycetemcomitans strain | Relevant characteristica | Source or reference |
|---|---|---|
| Y4 | Standard strain (serotype b) | 51 |
| 99 | Clinical isolate | This study |
| 108 | Clinical isolate | This study |
| 126 | Clinical isolate | This study |
| 129 | Clinical isolate | This study |
| 2232 | Clinical isolate | This study |
| 2267 | Clinical isolate | This study |
| 2339 | Clinical isolate | This study |
| IDH781 | Standard strain (serotype d) | 3 |
| KO-76 (RA135) | Spcromp100 knockout | 2 |
| ATCC 29523 | Standard strain (serotype a) | ATCC |
| KO-77 | Kmrspa knockout | 53 |
| SUNYaB75 | Standard strain (serotype a) | SUNYaB |
| KO-78 (SN-T1) | Kmrspa knockout | 53 |
Spc, spectinomycin; Km, kanamycin.
Cell culture.
HGEC were prepared from healthy gingival tissues using a method described previously (56) and grown in MCDB153 (pH 7.4) medium (Sigma Aldrich, Tokyo, Japan) containing 50 μg/ml bovine pituitary extract, 10 μg/ml insulin, 5 μg/ml transferrin, 10 μM 2-mercaptoethanol, 10 μM 2-aminomethanol, 100 units/ml penicillin, 10 nM sodium selenite, 100 μg/ml streptomycin, and 50 ng/ml amphotericin B at 37°C in a 5% CO2 atmosphere.
Preparation of bacterial cells and purification of Omp100.
Exponentially grown A. actinomycetemcomitans strains were harvested and washed with phosphate-buffered saline (PBS) twice. The bacteria were killed by heating cultures at 68°C for 30 min (34). The heat-inactivated bacterial cells were treated with trypsin (10 μg/ml) at 37°C for 30 min and washed with PBS five times. Omp100 was prepared using a method described elsewhere (2, 23).
Coculture of HGEC with bacteria, Omp100, and cytokine.
At 12 h prior to bacterial contact, confluent HGEC in MCDB medium with supplements was replaced with MCDB without supplements. Either the heat-killed bacteria (final concentration, 108 cells/ml), Omp100 (final concentration, 10 ng/ml to 10 μg/ml), or the inflammatory cytokines (interleukin-1β [IL-1β], IL-6, IL-8, and tumor necrosis factor α [TNF-α]; at 1 or 10 ng/ml) (R&D Systems, Inc., Minnesota) were added to the medium. The culture was incubated for no longer than 24 h. This experiment was carried out three times. Reverse transcriptase PCR (RT-PCR) and real-time PCR experiments were performed.
RNA extraction and RT-PCR.
After exposure to bacterial cells, Omp100, or inflammatory cytokines, the total RNA was extracted from the HGEC using ISOGEN (Nippon Gene, Tokyo, Japan) according to the manufacturer's protocol. RT-PCR was performed as described previously (34). One microgram of the total RNA was subject to cDNA synthesis using a first-strand cDNA synthesis kit (Roche, Tokyo, Japan) with a final volume of 20 μl. The cDNA was used as the template DNA for the subsequent PCR. Sense and antisense primer sets for β-defensins, CAP18, and cytokines were described previously (34). The housekeeping gene GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was amplified as a control. The template volumes of the cDNA solutions were 5 μl for hBD1, 20 μl for hBD2, 5 μl for hBD3, 25 μl for CAP18, 2 μl for IL-1β, 10 μl for IL-6, 10 μl for IL-8, 25 μl for TNF-α, and 2 μl for GAPDH.
Quantitative (real-time) RT-PCR.
Quantitative RT-PCR was performed using the ABI 7700 system (Applied Biosystems, Tokyo, Japan) with a Core Reagent kit (Applied Biosystems). The TaqMan probe, sense primers, and antisense primers used in this study were previously described (34).
Inhibition of the signaling pathway of hBD2 induction.
To examine the signaling pathway for hBD2 induction, HGEC were pretreated with NF-κB inhibitor (2 to 50 μM pyrrolidine dithiocarbamate [PDTC]; Sigma Aldrich), mitogen- activated protein (MAP) kinase inhibitors (0.2 to 5 μM SB203580, an inhibitor of p38 MAP kinase; 2 to 50 μM tyrphostin AG126, an inhibitor of tyrosine phosphorylation of Jun N-terminal kinase [JNK]; 0.4 to 10 μM PD98059; and two selective inhibitors of MAP/extracellular signal-regulated kinase [ERK] kinase 1/2 in the p44/42 MAP kinase; Calbiochem, Schwalbach, Germany), anti-human fibronectin antibody (100 ng/ml, Sigma Aldrich), anti-integrin antibody (1:200 dilution; Funakoshi, Tokyo, Japan), PP2 (selective Src tyrosine kinase inhibitor; 0.1 to 1 μM; Sigma Aldrich), PP3 (negative control for the Src tyrosine kinase inhibitor; 0.1 to 1 μM; Sigma Aldrich), and genistein (protein tyrosine kinases inhibitor; 0.1 to 1 μM; Sigma Aldrich). The inhibitors were added 30 min before stimulator contact (bacteria, OMP, or cytokine). Then the HGEC were incubated with each stimulator for 12 h. The inhibitors were present during the experiment.
Fibronectin binding assay.
To identify the Omp100 binding substrate, we performed a binding assay using a flat-bottom plate coated with candidate substrate. Substrates, either fibronectin (Chemicon International, Inc., California), fibrinogen (Chemicon), or bovine serum albumin (BSA; Wako Pure Chemical Industries, Osaka, Japan), were added to 50 mM carbonate buffer (15 mM Na2CO3, 15 mM NaHCO3, 5 mM NaN3) at a final concentration of 1 or 5 μg/ml. One hundred microliters of the substrates was added to 96-well flat plates and incubated for 12 h at 4°C. Then, the plate was washed with wash buffer (150 mM NaCl-0.05% Tween 20) four times and blocked with PBS containing 1% BSA and 0.05% Tween 20 for 1 h at 37°C. The plate was washed once more. A. actinomycetemcomitans cells were pelleted by centrifugation (3,000 × g for 10 min). The pellet was washed and suspended in buffer. The bacterial cell suspension was adjusted to 108 bacterial cells/ml, and 100 μl of the suspension (107 cells) was applied to the wells. After 3 h of bacterial contact, the wells were washed four times with wash buffer, and 10% trypsin was added to remove the adherent bacteria from the plastic dish. After the addition of 100 μl of PBS, the bacterial suspension was diluted appropriately and plated on Trypticase soy broth-yeast extract agar. The plates were incubated for 2 days at 37°C, and the colonies were counted as the adhesive cell number.
Neutralization of the inflammatory cytokine.
Some cytokines have been implicated in antimicrobial peptide production (12, 16, 32, 45). Inflammatory cytokines are expressed in HGEC by bacterial contact (55, 56). To determine the effect of inflammatory cytokines produced by HGEC on the induction of hBD2, HGEC were pretreated for 30 min with monoclonal anti-human IL-8 antibody and/or monoclonal anti-human TNF-α antibody (final concentration, 100 ng/ml; TECHNE Corp., Minnesota) prior to the bacterial contact.
RESULTS
Expression of antimicrobial peptides and cytokines in HGEC stimulated with A. actinomycetemcomitans.
To show the expression of antimicrobial peptides, RT-PCR was performed using the total RNA extracted from the HGEC. The primers specific for hBD1, hBD2, hBD3, and CAP18 were used, and the PCR products of the expected sizes in each reaction mixture were isolated (data not shown). As a negative control, we found no PCR products (no RT reaction) when the total RNA was used as the template (data not shown). We cloned the products in the pGEM-T Easy vector (Promega, Tokyo, Japan), a PCR cloning vector, and determined the DNA sequences that exhibit perfect matches with sequences reported in the database including the sequences for CAP18 (accession no. U19970), hBD1 (U73945), hBD2 (NM_004942), and hBD3 (NM_018661).
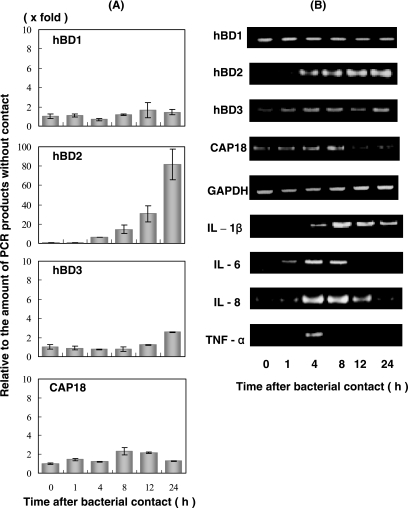
We investigated the mRNA expression of antimicrobial peptides in HGEC stimulated with heat-killed A. actinomycetemcomitans using a time course experiment (Fig. 1A and B). The concentration of hBD1 mRNA was relatively high compared to the mRNA of other antimicrobial peptides without bacterial contact, and hBD2 mRNA was not found in HGEC without bacterial contact, indicating that it is inducible. The concentration of hBD2 mRNA gradually increased in HGEC during the first 4 h following exposure to A. actinomycetemcomitans Y4 (fivefold greater). After 8 h, the quantity of hBD2 mRNA was 15-fold greater than at 0 h, and the expression further increased until 24 h. The concentration of hBD3 mRNA did not alter for 12 h, and then increased to 2.6-fold after 24 h. The peak concentration of CAP18 mRNA was at 8 h (twofold greater) after bacterial contact and then gradually decreased, whereas the concentration of hBD1 mRNA was not affected by exposure to A. actinomycetemcomitans cells. We also investigated the expression of the cytokines IL-1β, IL-6, IL-8, and TNF-α in HGEC exposed to A. actinomycetemcomitans cells (Fig. 1B). IL-1β mRNA gradually increased at 8 h after the initial bacterial contact and then gradually decreased. IL-6, IL-8, and TNF-α mRNA were found 4 h after bacterial contact, and each mRNA decreased after 8 h.
FIG. 1.
Expression of β-defensins, CAP18, and cytokine mRNA in HGEC stimulated with heat-inactivated A. actinomycetemcomitans Y4 cells. The total RNA was extracted from cells after exposure to A. actinomycetemcomitans at various times and then used for real-time PCR (A) and RT-PCR (B) performed as described in Materials and Methods. The results of the real-time PCR are expressed as a ratio comparing the value at time zero to the value at the data time point. The values represent the means ± standard deviations of triplicate experiments.
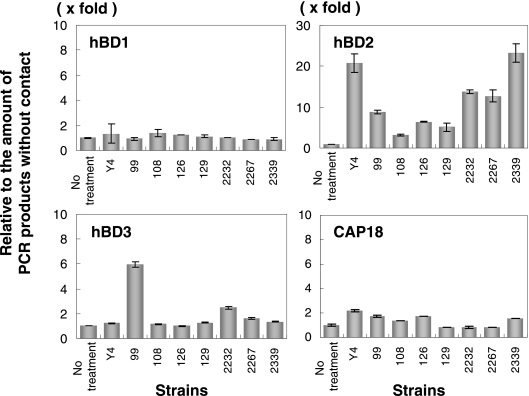
Expression of antimicrobial peptides using stimulation with A. actinomycetemcomitans clinical isolates.
We investigated the expression of four antimicrobial peptides in HGEC stimulated with seven clinical isolates and compared the levels in HGEC without bacterial contact. We found that there was a difference among the strains with regard to the amount of expression of induced antimicrobial peptide mRNAs. Inducible hBD2 mRNA was observed (4- to 25-fold) in all strains tested, while hBD3 or CAP18 mRNA was observed in some strains (hBD3, 1.5- to 6-fold; CAP18, 1- to 2-fold) at 12 h (Fig. 2). hBD1 mRNA concentration was not affected by bacterial exposure. There was no correlation in the inducibility of hBD2, hBD3, or CAP18 mRNA of each strain; A. actinomycetemcomitans 2339 induced hBD2 mRNA strongly but did not induce hBD3 or CAP18, while A. actinomycetemcomitans 99 induced hBD2 weakly but induced hBD3 strongly.
FIG. 2.
Expression of hBD1, hBD2, hBD3, and CAP18 mRNA in HGEC stimulated with heat-inactivated clinically isolated A. actinomycetemcomitans cells. The total RNA was extracted from cells after exposure to A. actinomycetemcomitans for 12 h and used for real-time PCR performed as described in Materials and Methods. The results of the real-time PCR are expressed as a ratio comparing the value at time zero to the value at the data time point. The values represent the means ± standard deviations of triplicate experiments.
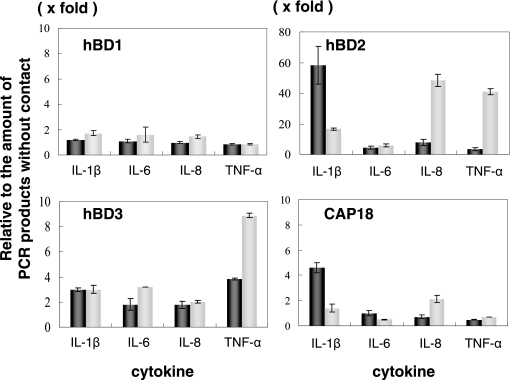
Inflammatory cytokines induce expression of antimicrobial peptides in HGEC.
We determined the effect of inflammatory cytokines (IL-1β, IL-6, IL-8, and TNF-α) on the expression of antimicrobial peptides (Fig. 3). Expression of hBD1 mRNA was not affected by the cytokines, but hBD2 mRNA was greatly induced by IL-1β, IL-8, and TNF-α (4- to 60-fold). Expression of hBD3 mRNA was also induced by all cytokines (two- to ninefold). And CAP18 mRNA was increased by IL-1β and IL-8 (1.5- to 4-fold). The expression of hBD2 and CAP18 induced with IL-1β shows that the concentration of mRNA increased to a higher concentration with 1 ng/ml IL-1β than with 10 ng/ml. This was because the 10 ng/ml IL-1β induced a faster response, and expression was in decline by the next sampling point.
FIG. 3.
Expression of hBD1, hBD2, hBD3, and CAP18 mRNA in HGEC stimulated with IL-1β, IL-6, IL-8, and TNF-α. The total RNA was extracted from the cells after the addition of several cytokines for 12 h and then used for real-time PCR performed as described in Materials and Methods. Black bars, 1 ng/ml; gray bars, 10 ng/ml. The results of the real-time PCR are expressed as a ratio comparing the value at time zero to the value at the data time point. The values represent the means ± standard deviations of triplicate experiments.
Identification of A. actinomycetemcomitans induction molecules leading to hBD2 expression.
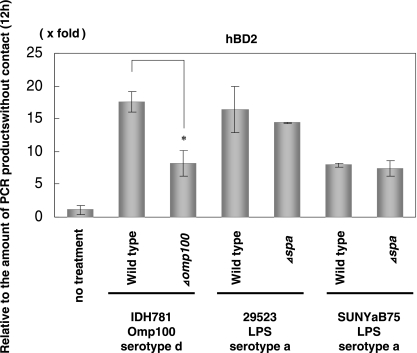
A. actinomycetemcomitans whole cells induce expression of hBD2 mRNA, indicating that a bacterial surface factor(s) may be associated with hBD2 induction. We compared the expression of hBD2 mRNA in HGEC stimulated with live A. actinomycetemcomitans, heat-killed cells, and trypsin-treated cells (data not shown). The concentration of hBD2 mRNA stimulated by heat-killed cells and by trypsin-treated cells was 27% and 75%, respectively, lower than the mRNA stimulated by living cells.
Two major cell surface components, serotype-specific polysaccharide antigen (SPA) and Omp100, are reported to induce some inflammatory cytokines and activate the immune response (2, 22, 60). Therefore, we investigated the induction of hBD2 mRNA in HGEC by A. actinomycetemcomitans mutants lacking SPA or Omp100 (Fig. 4). The induction by the Omp100 mutant was suppressed by 50% compared to the wild type (A. actinomycetemcomitans IDH781), whereas there was no difference in the induction level of hBD2 between the wild type (A. actinomycetemcomitans ATCC 29523 and SUNYaB75) and the SPA mutant.
FIG. 4.
Effect of Omp100 and SPA on the expression of hDB2. Heat-inactivated A. actinomycetemcomitans strains were added to HGEC and incubated for 12 h. The total RNA was extracted from cells after contact with bacteria for 12 h and used for real-time PCR. The results of the real-time PCR are expressed as a ratio comparing the value at time zero to the value at the data time point. The values represent the means and ± standard deviations of triplicate experiments.
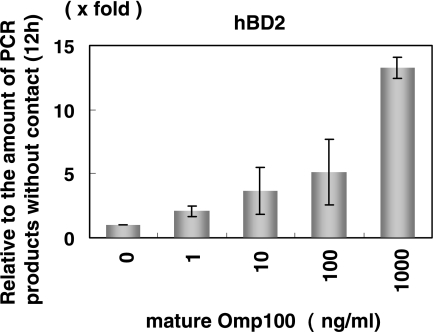
Because the Omp100 deletion decreased hBD2 induction, we investigated the expression of hBD2 in HGEC using purified Omp100 (Fig. 5). The purified Omp100 induced hBD2 mRNA in a dose-dependent manner (1 ng/ml to 1,000 ng/ml).
FIG. 5.
Effect of purified A. actinomycetemcomitans Y4 Omp100 on the expression of hDB2 in HGEC. Purified Omp100 was incubated with HGEC at various concentrations for 12 h. After the total RNA extraction, real-time PCR was performed. The results of the real-time PCR are expressed as a ratio comparing the value at time zero to the value at the data time point. The values represent the means ± standard deviations of triplicate experiments.
Signaling pathway for hBD2 expression in HGEC.
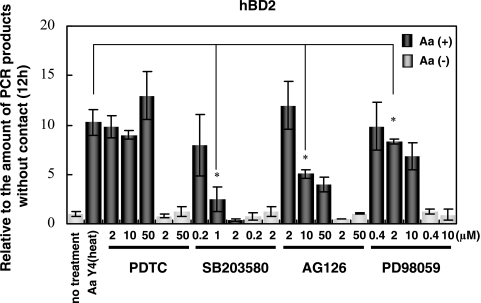
To investigate whether hBD2 is induced through the NF-κB or the MAP kinase pathway, HGEC were pretreated with various doses of PDTC, SB203580, AG126, or PD98059 and then stimulated with A. actinomycetemcomitans Y4 for 12 h. hBD2 induction was not inhibited by PDTC, whereas the MAP kinase inhibitor SB203580, AG126, and PD98059 inhibited hBD2 induction in a dose-dependent manner (Fig. 6). SB203580 completely inhibited the induction of hBD2.
FIG. 6.
Effect of inhibitors on hBD2 mRNA induction by A. actinomycetemcomitans cells. HGEC were pretreated with PDTC (NF-κB inhibitor; 2 to 50 μM), SB203580 (p38 inhibitor; 0.2 to 5 μM), AG126 (JNK inhibitor, 2 to 50 μM), or PD98059 (ERK inhibitor; 0.4 to 10 μM) for 1 h and then stimulated with heat-inactivated A. actinomycetemcomitans Y4 for 12 h. The total RNA was extracted and analyzed using real-time PCR. Gray bar, without bacterial contact; black bar, with bacterial contact. The results of the real-time PCR are expressed as a ratio in comparison to the value at time zero, and the values represent the means ± standard deviations of triplicate cultures. *, differs significantly (P < 0.01) from cells stimulated with A. actinomycetemcomitans Y4 alone.
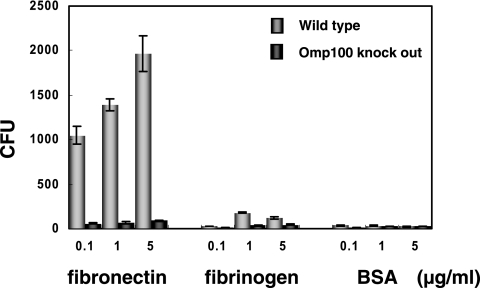
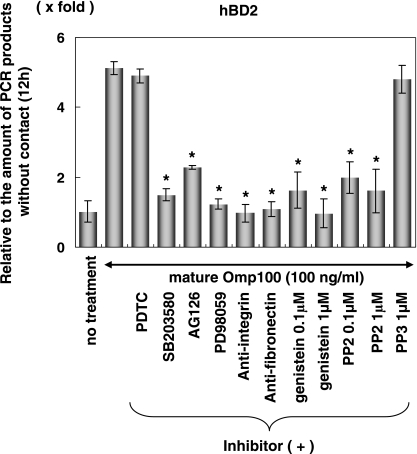
Previous studies showed that A. actinomycetemcomitans cells adhere and invade HGEC (2), and we found that Omp100 was the major adhesin to HGEC. The YadA protein of Yersinia enterocolitica, a homologue of Omp100, binds to fibronectin and activates tyrosine kinase (46, 54). To find the target molecule for Omp100, we performed a binding assay of A. actinomycetemcomitans and its Omp100 mutant to fibronectin, fibrinogen, and BSA. A. actinomycetemcomitans IDH781 (wild type) selectively bound fibronectin, whereas the Omp100 knockout mutant did not bind to this matrix (Fig. 7). To investigate the Omp100 signaling pathway, we studied hBD2 mRNA expression by Omp100 using MAP or NF-κB inhibitors. We found that hBD2 expression induced by Omp100 was inhibited with MAP kinase inhibitors but not with NF-κB inhibitor. Previously, the staphylococcal fibronectin binding protein was shown to be a transducing signal to the cell via the integrin α5β1 and to activate focal adhesion kinase (FAK) (1, 50). We looked at integrin α5β1-FAK to determine if it was involved in the signaling pathway of Omp100-dependent hBD2 expression. We used anti-human fibronectin antibody, anti-human integrin α5β1 antibody, PP2 (selective Src tyrosine kinase inhibitor), and genistein (protein tyrosine kinases inhibitor) to determine if there was an inhibitory effect on Omp100 induction. Figure 8 shows that hBD2 RNA expression was inhibited by the antibodies and by genistein and PP2. This suggests that Omp100 activates tyrosine kinase FAK through the fibronectin-integrin α5β1 pathway.
FIG. 7.
Binding assay of A. actinomycetemcomitans strains expressing Omp100 and knockout mutant. The binding assay was performed as described in Materials and Methods. Gray bar, wild type; black bar, Omp100 knockout mutant. Values represent the means ± standard deviations of triplicate wells.
FIG. 8.
Effect of inhibitors on hBD2 mRNA expression by Omp100. HGEC were pretreated with PDTC (NF-κB inhibitor; 2 μM), SB203580 (p38 inhibitor; 0.2 μM), AG126 (JNK inhibitor; 2 μM), PD98059 (ERK inhibitor; 0.4 μM), anti-human fibronectin affinity-isolated antibody, anti-integrin affinity-isolated antibody, PP2 (selective Src tyrosine kinase inhibitor), PP3 (negative control for the Src tyrosine kinase inhibitor), and genistein (protein tyrosine kinases inhibitor) with each concentration for 1 h and then stimulated with purified Omp100 (100 ng/ml) for 12 h. The total RNA was extracted and analyzed using real-time PCR. The results of the real-time PCR are expressed as a ratio comparing the value at time zero to the value at the data time point. The values represent the means and ± standard deviations of triplicate experiments. *, differs significantly (P < 0.01) from cells stimulated with Omp100 alone.
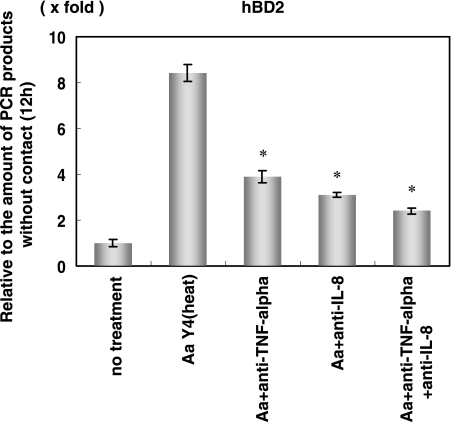
Effect of anticytokine antibody on hBD2 expression in HGEC after 12 h of bacterial contact.
A. actinomycetemcomitans exposure induces the expression of inflammatory cytokines (IL-1β, IL-6, IL-8, and TNF-α) after 1 to 4 h (Fig. 1). Among these inflammatory cytokines, IL-1β, IL-8, and TNF-α induce hBD2 mRNA 8 to 24 h after addition (Fig. 3). Therefore, we investigated whether cytokines induced by A. actinomycetemcomitans contact affect the hBD2 expression as autocrine (Fig. 9). We used neutralizing antibody against IL-8 or TNF-α. Anti-TNF-α antibody or anti-IL-8 antibody decreased the expression of hBD2 to approximately 52% to 64% after 12 h of bacterial exposure. Using anti-TNF-α antibody and anti-IL-8 antibody together, the expression of hBD2 mRNA was further decreased (75% inhibition).
FIG. 9.
Effect of anti-TNF-α and anti-IL-8 on hBD2 mRNA induction. HGEC were pretreated with monoclonal anti-human TNF-α antibody and monoclonal anti-human IL-8 antibody (final concentration, 100 ng/ml) for 1 h and then stimulated with heat-inactivated A. actinomycetemcomitans Y4 for 12 h. The total RNA was extracted and analyzed using real-time PCR. The results of the real-time PCR are expressed as a ratio comparing the value at time zero to the value at the data time point. The values represent the means and ± standard deviations of triplicate experiments. *, differs significantly (P < 0.01) from cells stimulated with A. actinomycetemcomitans Y4 alone.
DISCUSSION
hBD1 is reported to be constitutively expressed in some primary cultured cells, and this expression is not affected by bacterial contact and thus is constitutive (6, 26, 34). In contrast, the expression of hBD2, hBD3, and CAP18 is induced in epithelial, skin, gingival, and intestinal tract cells by bacterial components or by cytokines (5, 16, 34, 41, 59). Our results using HGEC are consistent with these findings (Fig. 1). The signal for hBD2 expression using bacterial stimuli is reported by two different pathways, NF-κB and MAP kinase, in different cell lines (18, 25, 40). In human gingival epithelial cells, a cell wall extract from Fusobacterium nucleatum induced hBD2 expression via MAP kinase (25), while S. enterica serovar Enteritidis FliC (flagella filament protein) and P. aeruginosa induced the expression via NF-κB pathway in Caco-2 human carcinoma cells and in human respiratory epithelium, respectively (18, 40). Here, we demonstrate that A. actinomycetemcomitans and Omp100 induce hBD2 expression in HGEC not via the NF-κB pathway but via the MAP kinase pathway (Fig. 6 and 8).
We found that Omp100 is a major component of A. actinomycetemcomitans in inducing the expression of hBD2. Omp100 is one of six major OMPs of A. actinomycetemcomitans that show a variety of virulence, adhesion, invasion, serum resistance, and cytokine induction functions (2). We demonstrate that Omp100 binds fibronectin and that this triggers hBD2 expression (Fig. 4 and 7). Fibronectin-binding proteins are found in many bacterial species including S. aureus, Streptococcus pyogenes, and Y. enterocolitica (1, 24, 50, 54). The proteins mediate adhesion to host cells or to substrate materials or invasion into the host cell to establish colonization (11). Fibronectin is an adhesive glycoprotein that binds to α5β1 integrin through the RGD binding domain (38, 50). The α5β1 integrin signal induces activation of FAK and MAP kinase. FAK in association with Src activates downstream effector molecules either directly or through activation of phosphatidylinositol 3 kinase (7, 15, 35, 49). Our inhibition assay suggests that Omp100 activates fibronectin-integrin α5β1 and is possibly associated with FAK and Src, which are involved in activation of MAP kinase (Fig. 8). Pattern recognition receptors, such as Toll-like receptors and nucleotide-binding oligomerization domains, are considered to play an important role in the response against microorganisms (20, 43). It was reported that LPS from P. aeruginosa is recognized by Toll-like receptor 4 following activation of the NF-κB pathway and induction of hBD2 in the human lung epithelial cell line A549 (30). Our preliminary investigations indicate that low concentrations of LPS (<100 ng/ml) did not increase hBD2 expression in HGEC (data not shown), and as shown in Fig. 4, SPA deletion mutants still induced some amount of hBD2 mRNA expression with the wild type. These differences might be due to the differences in species specificity of LPS. hBD2 expression induced by whole cells of A. actinomycetemcomitans was inhibited by MAP kinase inhibitor but not by NF-κB inhibitor. Therefore, these results suggest that hBD2 expression induced by whole A. actinomycetemcomitans cell exposure in HGEC is mainly mediated by Omp100 activating the MAP kinase pathway, but not NF-κB pathway, through the α5β1 integrin signal.
All A. actinomycetemcomitans clinical isolates tested in this study induced hBD2 expression; however, the concentration of mRNA produced varied among strains (Fig. 2). Because we demonstrated that Omp100 is a major component of the bacteria in inducing hBD2 expression, we determined the concentration of Omp100 protein in each strain by immunoblotting and found no differences among strains (data not shown). Also, deletion of Omp100 did not result in complete inhibition of hBD2 expression (Fig. 4). These results suggest that other factors of the bacterial cell surface component are involved in hBD2 expression. Also, other OMPs other than Omp100 may be involved in hBD2 induction. Further study is necessary to determine the various modes of hBD2 induction with different clinical isolates.
Cytokines have been implicated in the induction of antimicrobial peptides including TNF-α for hBD3 production, IL-1β and TNF-α for hBD2, and IL-6 for CAP18 (12, 16, 32, 45). In HGEC, we had the same findings (Fig. 3). In addition, we found that IL-1β, IL-8, IL-6, and ΤNF-α were induced in HGEC by exposure to A. actinomycetemcomitans (Fig. 1). As shown in Fig. 9, cytokines as autocrine signals are associated to induce hBD2 production. We found hBD2 expression after 8 h of the addition of cytokines (data not shown), while hBD2 expression by A. actinomycetemcomitans whole cells or by Omp100 was seen after 4 h. Therefore, we concluded that the induction of hBD2 expression occurs by direct bacterial contact to HGEC and later by inflammatory cytokine in an autocrine response. But the induction is not completely inhibited by these neutralization antibodies, so there might be other factors such as IL-1β or IL-6 that may induce expression of hBD2. It is reported that IL-8/CXCL8 works as the chemokine for migration of polymorphonuclear leukocytes into tissues (47). So not only activation of innate immunity but also induction of adaptive immunity are found by A. actinomycetemcomitans contacts.
Omp100 has many complex functions; as a fibronectin binding protein, it is believed to be a major virulence factor to adhere and/or to invade into host cells (2), and it may also induce antimicrobial peptides, factors in innate immunity. This single molecule plays an important role in the balance of the host/parasite relationship, having two conflicting effects on host cells. In these two aspects, we think that the advantage of binding to the cells for the bacteria is to colonize and/or escape from the host immune system by invading host cells, while the disadvantage is to induce several chemical factors involved in the immune system. We previously demonstrated that the susceptibilities to antimicrobial peptides are different among A. actinomycetemcomitans strains (42). Here, we show that the induction level of hBD2 expression varies among strains. The balance between resistance of A. actinomycetemcomitans to antimicrobial peptides and its potential to induce the peptides may influence the infectivity of the bacteria. Further study will be needed to determine if the degree of response by antimicrobial peptide induction against A. actinomycetemcomitans contributes to its infection in the host cell as well as to determine all of the effects of the molecule Omp100 during infection.
Acknowledgments
Part of this study was carried out in the Research Center for Molecular Medicine, Faculty of Medicine, Hiroshima University. We thank Yoshio Nakano for SPA mutants. We also thank Jim Nelson for editorial assistance.
H.K. and M.S. were supported by a Grant-in-Aid for Scientific Research by the Ministry of Education, Science, Sports and Culture. M.S. was supported by a Grant-in-Aid for Scientific Research on Priority Areas (C) by the Ministry of Education, Science, Sports and Culture.
Editor: F. C. Fang
REFERENCES
- 1.Agerer, F., S. Lux, M. A., M. Rohde, K. Ohlsen, and C. R. Hauck. 2005. Cellular invasion by Staphylococcus aureus reveals a functional link between focal adhesion kinase and cortactin in integrin-mediated internalization. J. Cell Sci. 118:2189-2200. [DOI] [PubMed] [Google Scholar]
- 2.Asakawa, R., H. Komatsuzawa, T. Kawai, S. Yamada, R. B. Goncalves, S. Izumi, T. Fujiwara, Y. Nakano, N. Suzuki, Y. Uchida, K. Ouhara, H. Shiba, M. A. Taubman, H. Kurihara, and M. Sugai. 2003. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 50:1125-1139. [DOI] [PubMed] [Google Scholar]
- 3.Asikainen, S., C. Chen, M. Saarela, L. Saxen, and J. Slots. 1997. Clonal specificity of Actinobacillus actinomycetemcomitans in destructive periodontal disease. Clin. Infect. Dis. 25(Suppl. 2):S227-S229. [DOI] [PubMed] [Google Scholar]
- 4.Bals, R. 2000. Epithelial antimicrobial peptides in host defense against infection. Respir. Res. 1:141-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bals, R., X. Wang, Z. Wu, T. Freeman, V. Bafna, M. Zasloff, and J. M. Wilson. 1998. Human beta-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J. Clin. Investig. 102:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissell, J., S. Joly, G. K. Johnson, C. C. Organ, D. Dawson, P. B. J. McCray, and J. M. Guthmiller. 2004. Expression of beta-defensins in gingival health and in periodontal disease. J. Oral Pathol. Med. 33:278-285. [DOI] [PubMed] [Google Scholar]
- 7.Black, D. S., L. G. Montagna, S. Zitsmann, and J. B. Bliska. 1998. Identification of an amino-terminal substrate binding domain in the Yersinia tyrosine phosphatase that is required for efficient recognition of focal adhesion targets. Mol. Microbiol. 29:1263-1274. [DOI] [PubMed] [Google Scholar]
- 8.Chung, W. O., S. R. Hansen, D. Rao, and B. A. Dale. 2004. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J. Immunol. 173:5165-5170. [DOI] [PubMed] [Google Scholar]
- 9.Farida, R., M. Wilson, and L. Ivanyi. 1986. Serum IgG antibodies to lipopolysaccharides in various forms of periodontal disease in man. Arch. Oral Biol. 31:711-715. [DOI] [PubMed] [Google Scholar]
- 10.Feucht, E. C., C. L. DeSanti, and A. Weinberg. 2003. Selective induction of human beta-defensin mRNAs by Actinobacillus actinomycetemcomitans in primary and immortalized oral epithelial cells. Oral Microbiol. Immunol. 18:359-363. [DOI] [PubMed] [Google Scholar]
- 11.Foster, T. J., and M. Höök. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 12.Frohm, M., B. Agerberth, G. Ahangari, M. Ståhle-Bäckdahl, S. Liden, H. Wigzell, and G. H. Gudmundsson. 1997. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J. Biol. Chem. 272:15258-15263. [DOI] [PubMed] [Google Scholar]
- 13.Ganz, T., and R. I. Lehrer. 1995. Defensins. Pharmacol. Ther. 66:191-205. [DOI] [PubMed] [Google Scholar]
- 14.García, J. R., A. Krause, S. Schulz, F. J. Rodríguez-Jimenez, E. Klüver, K. Adermann, U. Forssmann, A. Frimpong-Boateng, R. Bals, and W. G. Forssmann. 2001. Human beta-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 15:1819-1821. [PubMed] [Google Scholar]
- 15.Giancotti, F. G., and E. Ruoslahti. 1999. Integrin signaling. Science 285:1028-1032. [DOI] [PubMed] [Google Scholar]
- 16.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 17.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 1997. A peptide antibiotic from human skin. Nature 387:861. [DOI] [PubMed] [Google Scholar]
- 18.Harder, J., U. Meyer-Hoffert, L. M. Teran, L. Schwichtenberg, J. Bartels, S. Maune, and J. M. Schröder. 2000. Mucoid Pseudomonas aeruginosa, TNF-alpha, and IL-1beta, but not IL-6, induce human beta-defensin-2 in respiratory epithelia. Am. J. Respir. Cell Mol. Biol. 22:714-721. [DOI] [PubMed] [Google Scholar]
- 19.Hong, C. C., M. Shimomura-Shimizu, M. Muroi, and K. Tanamoto. 2004. Effect of endocrine disrupting chemicals on lipopolysaccharide-induced tumor necrosis factor-alpha and nitric oxide production by mouse macrophages. Biol. Pharm. Bull. 27:1136-1139. [DOI] [PubMed] [Google Scholar]
- 20.Inohara, N., M. Chamaillard, C. McDonald, and G. Nunez. 2004. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. [DOI] [PubMed]
- 21.Joly, S., C. Maze, P. B. McCray, Jr., and J. M. Guthmiller. 2004. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J. Clin. Microbiol. 42:1024-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiley, P., and S. C. Holt. 1980. Characterization of the lipopolysaccharide from Actinobacillus actinomycetemcomitans Y4 and N27. Infect. Immun. 30:862-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsuzawa, H., R. Asakawa, T. Kawai, K. Ochiai, T. Fujiwara, M. A. Taubman, M. Ohara, H. Kurihara, and M. Sugai. 2002. Identification of six major outer membrane proteins from Actinobacillus actinomycetemcomitans. Gene 288:195-201. [DOI] [PubMed] [Google Scholar]
- 24.Kreikemeyer, B., S. Oehmcke, M. Nakata, R. Hoffrogge, and A. Podbeiski. 2004. Streptococcus pyogenes fibronectin-binding protein F2: expression profile, binding characteristics, and impact on eukaryotic cell interactions. J. Biol. Chem. 279:15850-15859. [DOI] [PubMed] [Google Scholar]
- 25.Krisanaprakornkit, S., J. R. Kimball, and B. A. Dale. 2002. Regulation of human beta-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways but not the NF-kappaB transcription factor family. J. Immunol. 168:316-324. [DOI] [PubMed] [Google Scholar]
- 26.Krisanaprakornkit, S., A. Weinberg, C. N. Perez, and B. A. Dale. 1998. Expression of the peptide antibiotic human beta-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect. Immun. 66:4222-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lally, E. T., I. R. Kieba, D. R. Demuth, J. Rosenbloom, E. E. Golub, N. S. Taichman, and C. W. Gibson. 1989. Identification and expression of the Actinobacillus actinomycetemcomitans leukotoxin gene. Biochem. Biophys. Res. Commun. 159:256-262. [DOI] [PubMed] [Google Scholar]
- 28.Lehrer, R. I., and T. Ganz. 1999. Antimicrobial peptides in mammalian and insect host defence. Curr. Opin. Immunol. 11:23-27. [DOI] [PubMed] [Google Scholar]
- 29.Liu, A. Y., D. Destoumieux, A. V. Wong, C. H. Park, E. V. Valore, L. Liu, and T. Ganz. 2002. Human beta-defensin-2 production in keratinocytes is regulated by interleukin-1, bacteria, and the state of differentiation. J. Investig. Dermatol. 118:275-281. [DOI] [PubMed] [Google Scholar]
- 30.MacRedmond, R., C. Greene, C. C. Taggart, N. McElvaney, and S. O'Neill. 2005. Respiratory epithelial cells require Toll-like receptor 4 for induction of human beta-defensin 2 by lipopolysaccharide. Respir. Res. 6:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maisetta, G., G. Batoni, S. Esin, F. Luperini, M. Pardini, D. Bottai, W. Florio, M. R. Giuca, M. Gabriele, and M. Campa. 2003. Activity of human beta-defensin 3 alone or combined with other antimicrobial agents against oral bacteria. Antimicrob. Agents Chemother. 47:3349-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews, M., H. P. Jia, J. M. Guthmiller, G. Losh, S. Graham, G. K. Johnson, B. F. Tack, and P. B. J. McCray. 1999. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 67:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayer, D. H., and P. M. Fives-Taylor. 1997. The role of Actinobacillus actinomycetemcomitans in the pathogenesis of periodontal disease. Trend Microbiol. 5:224-228. [DOI] [PubMed] [Google Scholar]
- 34.Midorikawa, K., K. Ouhara, H. Komatsuzawa, T. Kawai, S. Yamada, T. Fujiwara, K. Yamazaki, K. Sayama, M. A. Taubman, H. Kurihara, K. Hashimoto, and M. Sugai. 2003. Staphylococcus aureus susceptibility to innate antimicrobial peptides, beta-defensins and CAP18, expressed by human keratinocytes. Infect. Immun. 71:3730-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Milliano, M. T., and B. A. Luxton. 2003. Initial signaling of the fibronectin receptor (alpha5beta1 integrin) in hepatic stellate cells is independent of tyrosine phosphorylation. J. Hepatol. 39:32-37. [DOI] [PubMed] [Google Scholar]
- 36.Mineshiba, F., S. Takashiba, J. Mineshiba, K. Matsuura, S. Kokeguchi, and Y. Murayama. 2003. Antibacterial activity of synthetic human B defensin-2 against periodontal bacteria. J. Int. Acad. Periodontol. 5:35-40. [PubMed] [Google Scholar]
- 37.Moore, W. E., and L. V. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000. 5:66-77. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa, I., A. Amano, H. Inaba, S. Kawai, and S. Hamada. 2005. Inhibitory effects of Porphyromonas gingivalis fimbriae on interactions between extracellular matrix proteins and cellular integrins. Microbes. Infect. 7:157-163. [DOI] [PubMed] [Google Scholar]
- 39.Noguchi, T., H. Shiba, H. Komatsuzawa, N. Mizuno, Y. Uchida, K. Ouhara, R. Asakawa, S. Kudo, H. Kawaguchi, M. Sugai, and H. Kurihara. 2003. Syntheses of prostaglandin E2 and E-cadherin and gene expression of beta-defensin-2 by human gingival epithelial cells in response to Actinobacillus actinomycetemcomitans. Inflammation 27:341-349. [DOI] [PubMed] [Google Scholar]
- 40.Ogushi, K., A. Wada, T. Niidome, N. Mori, K. Oishi, T. Nagatake, A. Takahashi, H. Asakura, S. Makino, H. Hojo, Y. Nakahara, M. Ohsaki, T. Hataleyama, H. Aoyagi, H. Kurazono, J. Moss, and J. Hirayama. 2001. Salmonella enteritidis FliC (flagella filament protein) induces human beta-defensin-2 mRNA production by Caco-2 cells. J. Biol. Chem. 276:30521-30526. [DOI] [PubMed] [Google Scholar]
- 41.O'Neil, D. A., E. M. Porter, D. Elewaut, G. M. Anderson, L. Eckmann, T. Ganz, and M. F. Kagnoff. 1999. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 163:6718-6724. [PubMed] [Google Scholar]
- 42.Ouhara, K., H. Komatsuzawa, S. Yamada, H. Shiba, T. Fujiwara, M. Ohara, K. Sayama, K. Hashimoto, H. Kurihara, and M. Sugai. 2005. Susceptibilities of periodontopathogenic and cariogenic bacteria to antimicrobial peptides, beta-defensins and LL37, produced by human epithelial cells. J. Antimicrob. Chemother. 55:888-896. [DOI] [PubMed] [Google Scholar]
- 43.Philpott, D. J., and S. E. Girardin. 2004. The role of Toll-like receptors and Nod proteins in bacterial infection. Mol. Immunol. 41:1099-1108. [DOI] [PubMed] [Google Scholar]
- 44.Robertson, P. B., M. Lantz, P. T. Marucha, K. S. Kornman, C. L. Trummel, and S. C. Holts. 1982. Collagenolytic activity associated with Bacteroides species and Actinobacillus actinomycetemcomitans. J. Periodont. Res. 17:275-283. [DOI] [PubMed] [Google Scholar]
- 45.Schroder, J. M., and J. Harder.1999. Human beta-defensin-2. Int. J. Biochem. Cell. Biol. 31:645-651. [DOI] [PubMed] [Google Scholar]
- 46.Schulze-Koops, H., H. Burkhardt, J. Heesemann, T. Kirsch, B. Swoboda, C. Bull, S. Goodman, and F. Emmrich. 1993. Outer membrane protein YadA of enteropathogenic yersiniae mediates specific binding to cellular but not plasma fibronectin. Infect. Immun. 61:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scimone, M. L., V. P. Lutzky, S. I. Zittermann, P. Maffia, C. Jancic, F. Buzzola, A. C. Issekutz, and H. E. Chuluyan. 2005. Migration of polymorphonuclear leucocytes is influenced by dendritic cells. Immunology 114:375-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selsted, M. E., S. S. Harwig, T. Ganz, J. W. Schilling, and R. I. Lehrer. 1985. Primary structures of three human neutrophil defensins. J. Clin. Investig. 76:1436-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shinji, H., K. Seki, A. Tajima, A. Uchida, and S. Masuda. 2003. Fibronectin bound to the surface of Staphylococcus aureus induces association of very late antigen 5 and intracellular signaling factors with macrophage cytoskeleton. Infect. Immun. 71:140-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinha, B., P. P. Francois, O. Nusse, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann, and K. H. Krause. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin binding to integrin α5β1. Cell Microbiol. 1:101-117. [DOI] [PubMed] [Google Scholar]
- 51.Socransky, S. S., and A. D. Haffajee. 1994. Implications of periodontal microbiology for the treatment of periodontal infections. Compend. Suppl. 1994:S684-S685, S688-S693; S714-S717. [PubMed] [Google Scholar]
- 52.Sugai, M., T. Kawamoto, S. Y. Peres, Y. Ueno, H. Komatsuzawa, T. Fujiwara, H. Kurihara, H. Suginaka, and E. Oswald. 1998. The cell cycle-specific growth-inhibitory factor produced by Actinobacillus actinomycetemcomitans is a cytolethal distending toxin. Infect. Immun. 66:5008-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki, N., Y. Nakano, Y. Yoshida, H. Nakao, Y. Yamashita, and T. Koga. 2000. Genetic analysis of the gene cluster for the synthesis of serotype a-specific polysaccharide antigen in Actinobacillus actinomycetemcomitans. Biochim. Biophys. Acta 1517:135-138. [DOI] [PubMed] [Google Scholar]
- 54.Tertti, R., M. Skurnik, T. Vartio, and P. Kuusela. 1992. Adhesion protein YadA of Yersinia species mediates binding of bacteria to fibronectin. Infect. Immun. 60:3021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uchida, Y., H. Shiba, H. Komatsuzawa, C. Hirono, A. Ashikaga, T. Fujita, H. Kawaguchi, M. Sugai, Y. Shiba, and H. Kurihara. 2005. Irsogladine malate influences the response of gap junctional intercellular communication and IL-8 of human gingival epithelial cells following periodontopathogenic bacterial challenge. Biochem. Biophys. Res. Commun. 333:502-507. [DOI] [PubMed] [Google Scholar]
- 56.Uchida, Y., H. Shiba, H. Komatsuzawa, T. Takemoto, M. Sakata, T. Fujita, H. Kawaguchi, M. Sugai, and H. Kurihara. 2001. Expression of IL-1 beta and IL-8 by human gingival epithelial cells in response to Actinobacillus actinomycetemcomitans. Cytokine 14:152-161. [DOI] [PubMed] [Google Scholar]
- 57.Valore, E. V., C. H. Park, A. J. Quayle, K. R. Wiles, P. B. J. McCray, and T. Ganz. 1998. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J. Clin. Investig. 101:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vankeerberghen, A., H. Nuytten, K. Dierickx, M. Quirynen, J. J. Cassiman, and H. Cuppens. 2005. Differential induction of human beta-defensin expression by periodontal commensals and pathogens in periodontal pocket epithelial cells. J. Periodontol. 76:1293-1303. [DOI] [PubMed] [Google Scholar]
- 59.Vora, P., A. Youdim, L. S. Thomas, M. Fukata, S. Y. Tesfay, K. Lukasek, K. S. Michelsen, A. Wada, T. Hirayama, M. Arditi, and M. T. Abreu. 2004. Beta-defensin-2 expression is regulated by TLR signaling in intestinal epithelial cells. J. Immunol. 173:5398-5405. [DOI] [PubMed] [Google Scholar]
- 60.Wada, N., H. Maeda, Y. Yoshimine, and A. Akamine. 2004. Lipopolysaccharide stimulates expression of osteoprotegerin and receptor activator of NF-kappa B ligand in periodontal ligament fibroblasts through the induction of interleukin-1 beta and tumor necrosis factor-alpha. Bone 35:629-635. [DOI] [PubMed] [Google Scholar]
- 61.Woo, J. S., J. Y. Jeong, Y. J. Hwang, S. W. Chae, S. J. Hwang, and H. M. Lee. 2003. Expression of cathelicidin in human salivary glands. Arch. Otolaryngol. Head Neck Surg. 129:211-214. [DOI] [PubMed] [Google Scholar]
- 62.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schröder, J. M. Wang, O. M. Howard, and J. J. Oppenheim. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-529. [DOI] [PubMed] [Google Scholar]
- 63.Yang, D., O. Chertov, and J. J. Oppenheim. 2001. Participation of mammalian defensins and cathelicidin (LL-37). J. Leukoc. Biol. 69:691-697. [PubMed] [Google Scholar]
- 64.Yang, D., O. Chertov, and J. J. Oppenheim. 2001. The role of mammalian antimicrobial peptides and proteins in awakening of innate host defenses and adaptive immunity. Cell Mol. Life Sci. 58:978-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]
- 66.Zambon, J. J. 1996. Periodontal diseases: microbial factors. Ann. Periodontol. 1:879-925. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, L., W. Yu, T. He, J. Yu, R. E. Caffrey, E. A. Dalmasso, S. Fu, T. Pham, J. Mei, J. J. Ho, W. Zhang, P. Lopez, and D. D. Ho. 2002. Contribution of human alpha-defensin 1, 2, and 3 to the anti-HIV-1 activity of CD8 antiviral factor. Science 298:995-1000. [DOI] [PubMed] [Google Scholar]