Abstract
Citrobacter rodentium is a natural mouse pathogen related to enteropathogenic and enterohemorrhagic Escherichia coli. We have previously utilized bioluminescence imaging (BLI) to determine the in vivo colonization dynamics of C. rodentium. However, due to the oxygen requirement of the bioluminescence system and the colonic localization of C. rodentium, in vivo localization studies were performed using harvested organs. Here, we report the detection of bioluminescent C. rodentium and commensal E. coli during colonization of the gastrointestinal tract in intact living animals. Bioluminescence was dependent on intact blood circulation, suggesting that the colonic environment is not anaerobic but nanaerobic. In addition, BLI revealed that C. rodentium colonizes the rectum, a site previously unreported for this pathogen.
Citrobacter rodentium belongs to a family of extracellular enteric pathogens that includes the clinically significant enterohemorrhagic Escherichia coli (EHEC) and enteropathogenic E. coli (EPEC) (10). EPEC is a frequent cause of infantile diarrhea in the developing world, while EHEC causes a wide spectrum of illnesses ranging from mild diarrhea to hemorrhagic colitis and hemolytic uremic syndrome. EPEC and EHEC use attaching and effacing (A/E) lesion formation (11), characterized by effacement of the brush border microvilli and intimate bacterial attachment to the apical enterocyte plasma membrane, as an essential step in the colonization and infection of the gastrointestinal mucosa. The capacity to form A/E lesions is encoded mainly on a pathogenicity island termed the locus of enterocyte effacement (LEE), which encodes several transcriptional regulators, the structural components of a type III secretion system (T3SS), the outer membrane adhesin intimin, chaperones, translocators, and effectors proteins (reviewed in reference 8).
EHEC and EPEC are poorly pathogenic in mice. In contrast, C. rodentium is a natural mouse pathogen, related to E. coli, which colonizes the colonic mucosa via A/E lesion formation (reviewed in reference 14). C. rodentium harbors the LEE pathogenicity island, hence providing an in vivo model that permits the robust investigation of pathogen-host interactions under physiological conditions typical of the intestinal environment, with the ability to manipulate both the pathogen and the host. The model has yielded significant novel phenotypes for LEE-encoded proteins not revealed using in vitro infection models and for type III effectors that are encoded on loci different from the LEE (7). In particular, we recently showed that the T3SS effector protein Map is implicated in bacterium-induced diarrhea (17).
Molecular imaging is a rapidly emerging biomedical field which allows the visual representation, characterization, and quantification of biological processes within intact organisms (16). Bioluminescence imaging (BLI) is based on the detection of visible light produced by luciferase-catalyzed reactions which require energy (in the form of FMNH2 and ATP), oxygen, and a specific substrate (13) and so allow the detection of only live, metabolically active cells. Because luciferases are oxygenases, it has been suggested that the requirement for oxygen may limit the use of BLI in anaerobic environments, such as the lumen of the gut (4). Indeed, bacterial cells expressing luciferase were found to be nonluminescent in the gut until exposed to oxygen (3).
To utilize the power of BLI in studying the pathogenesis of C. rodentium, we previously developed a bioluminescent C. rodentium derivative (strain ICC180) which harbors an unpromoted luxCDABE operon from the nematode symbiont Photorhabdus luminescens and a kanamycin resistance cassette, inserted within a homologue of the xylE gene (18), shown to be involved in xylose transport in E. coli K-12 (6). Disruption of this gene in C. rodentium ICC180 was found to have no effect on growth in vitro or colonization dynamics in vivo (18). Because of the requirement for oxygen and the gastrointestinal localization of C. rodentium, we have previously used BLI to visualize C. rodentium ICC180 during infection of the mouse intestinal mucosa using harvested organs (18) and have demonstrated that a few hours after oral inoculation, C. rodentium becomes established within the specialized patch of lymphoid tissue known as the cecal patch, with detectable colonization of the colon occurring subsequently.
Bioluminescence imaging of bacteria within the gastrointestinal tract requires a living host.
The aim of this study was to determine whether BLI could be used to monitor bacterial loads within the gastrointestinal (GI) environment of living mice. In order to test this, C. rodentium ICC180 was grown overnight at 37°C in LB broth with kanamycin (100 μg ml−1) and orally gavaged into female 6- to 8-week-old C57Bl/6J mice (≈5 × 109 CFU per mouse). Prior to BLI, each animal's abdominal region was depilated to minimize any potential signal impedance by melanin within pigmented fur. Assessment of bioluminescence (photons s−1 cm−2 sr−1) from living animals was measured after gaseous anesthesia with isoflurane by using the IVIS50 system (Xenogen Corporation, Alameda, CA). A photograph (reference image) was taken under low illumination prior to quantification of photons emitted from C. rodentium ICC180 at a binning of 4 over 1 to 10 min using the software program Living Image (Xenogen) as an overlay on Igor (Wavemetrics, Seattle, WA). For anatomical localization, a pseudocolor image representing light intensity (blue, least intense, to red, most intense) was generated using Living Image software and superimposed over the grayscale reference image. Where the bioluminescence from specific tissues was required, mice were killed by cervical dislocation and organs washed with sterile phosphate-buffered saline using a needle and syringe.
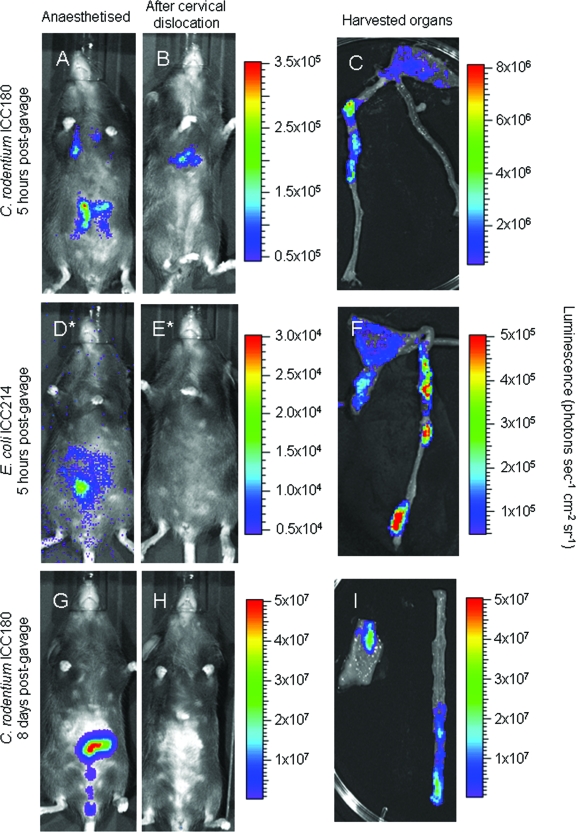
We examined the bioluminescent signal emitted by C. rodentium ICC180 from as early as 5 h postgavage to determine the location of the infection bolus. Interestingly, inoculation by oral gavage often resulted in a small amount of bacteria entering the lungs (Fig. 1A). However, no signal was ever detected from the lungs after day 1 postinfection (p.i.) (Fig. 2B and G), suggesting that the bacteria were efficiently cleared from this site. In addition to the lungs, bioluminescence was also found to be localized within the abdominal region at this early time point (Fig. 1B), and after the removal of tissue, the signal was seen to be localized to the cecum and colon (Fig. 1C). After cervical dislocation of the animal, the signal emanating from the colon and cecum was extinguished within mere minutes (Fig. 1B and H). This is in contrast to the bioluminescent signal detected in the lungs, which remained just as bright after the animal had been killed (Fig. 1B). Cervical dislocation of mice during peak C. rodentium infection (8 days p.i.) also resulted in complete loss of the bioluminescent signal (Fig. 1H); however, when the organs were punctured, light emission was restored (data not shown).
FIG. 1.
BLI in the gastrointestinal tract requires a functional blood supply. Mice were orally gavaged with 109 bacteria, and in vivo bacterial localization was determined by BLI in live animals (A, D, and G), immediately after cervical dislocation (B, E, and H), and on harvested colons and cecums (C, F, and I). Mice were imaged at 5 h postgavage with C. rodentium ICC180 (A to C), at 5 h postgavage with E. coli ICC214 (D to F), or at the peak of C. rodentium ICC180 infection (day 8 postgavage) (G to I) with an integration time of 1 min (except panels marked with  , which required a 10-min integration). Images were acquired using an IVIS50 system and are displayed as pseudocolor images of peak bioluminescence, with variations in color representing light intensity at a given location. Red represents the most intense light emission, while blue corresponds to the weakest signal. The color bar indicates relative signal intensity (as photons s−1 cm−2 sr−1). For each time point, at least three animals were analyzed and a representative animal is shown.
, which required a 10-min integration). Images were acquired using an IVIS50 system and are displayed as pseudocolor images of peak bioluminescence, with variations in color representing light intensity at a given location. Red represents the most intense light emission, while blue corresponds to the weakest signal. The color bar indicates relative signal intensity (as photons s−1 cm−2 sr−1). For each time point, at least three animals were analyzed and a representative animal is shown.
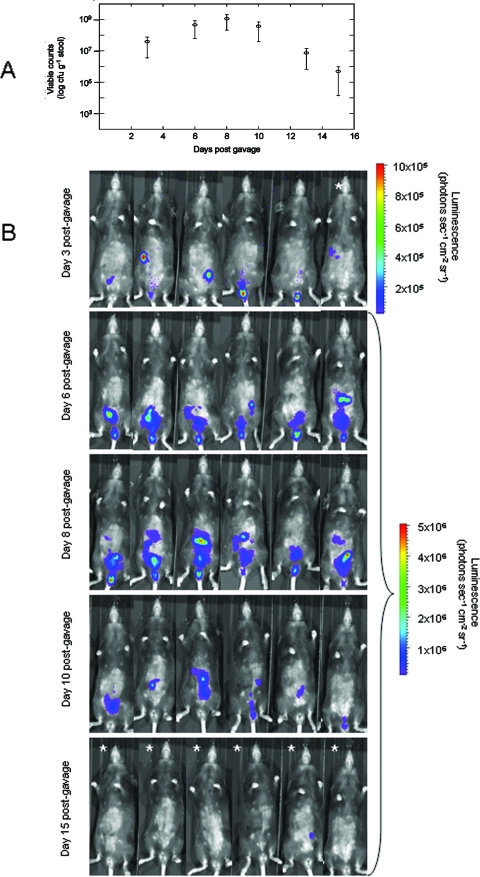
FIG. 2.
In vivo growth dynamics of luminescent C. rodentium ICC180. Mice were orally gavaged with 109 bacteria, and in vivo growth dynamics were determined by viable counts (given as CFU per gram of stool) (A) and BLI (B). Images were acquired using an IVIS50 system and are displayed as pseudocolor images of peak bioluminescence, with variations in color representing light intensity at a given location. Red represents the most intense light emission, while blue corresponds to the weakest signal. The color bar indicates relative signal intensity (as photons s−1 cm−2 sr−1). Mice were imaged at various time points postgavage with an integration time of 1 min. If no luminescence was detected at 1 min, then a 10-min exposure was used (indicated by  ).
).
We were interested in determining whether the ability of C. rodentium to form an intimate attachment with the epithelial cell was required for the detection of bioluminescence in vivo. To do this, we isolated a commensal E. coli strain from a C57Bl/6J mouse by plating cecal contents and stool onto MacConkey agar. Isolated bacteria were then identified using the API20E strip system (bioMérieux UK Ltd., Basingstoke, United Kingdom), and an E. coli strain was selected for further study. This strain was used to generate a spontaneous nalidixic acid-resistant mutant, which was then rendered bioluminescent as previously described (18) to generate strain ICC214. As with C. rodentium ICC180, we also detected a signal from the cecum 5 h after oral gavage of mice with 109 CFU of E. coli ICC214, which is unable to form A/E lesions (Fig. 1D and F), suggesting that intimate bacterium-cell attachment is not required for the detection of bioluminescence within the GI tract.
Determination of the colonization dynamics of C. rodentium in the GI tract of living mice.
As we were able to detect bacteria within the GI tract of living animals, we were interested in following the colonization dynamics of C. rodentium from oral gavage to clearance in vivo. Our results demonstrate that bioluminescence can be detected from within anesthetized mice at 3 days postgavage (Fig. 2B), requiring a 1- to 10-min exposure time to detect signals of 105 photons s−1 cm−2 sr−1. Within the majority of animals, this signal was localized to a single focus within the abdominal region which we have previously identified to be the cecum and cecal patch (18). In addition, a number of animals exhibited a signal from the region of the rectum, a localization previously unreported for C. rodentium colonization.
Following adaptation within the cecum, both bioluminescence data and viable counts demonstrate that the challenge bacterial population increases in number, reaching a plateau of between 108 and 109 CFU g−1 stool by day 8 p.i. (Fig. 2). At this time, the bioluminescent signal was localized throughout the abdominal region, corresponding to colonization of the cecum and colon, requiring a 1-min exposure time to detect signals of 106 photons s−1 cm−2 sr−1. In this model, mice begin to clear the infection from day 10 p.i. and this corresponded with a decrease in detectable bioluminescence until day 15 p.i., when a low signal was detectable in only one animal and required a 10-min exposure to be visualized (Fig. 2B).
In addition to information regarding localization, it is also possible to quantify the bioluminescence signal using Living Image software. Interestingly, despite the presence of both intimately attached C. rodentium strains and those being shed from the cecum and colon, the bioluminescent signal detected in vivo was found to be slightly less than that for the harvested organs containing only attached bacteria (2.20 × 108 compared to 2.75 × 108 photons s−1) (Fig. 1D and F). Indeed, the sensitivity of BLI is dependent on several factors, including the availability of cofactors, the distance the photons must travel through tissue, and potential signal impedance (such as absorption of light by oxyhemoglobin and deoxyhemoglobin, or by melanin within pigmented skin and fur). Clearly, some or all of these factors will be responsible for this difference in sensitivity in vivo compared with ex vivo.
Colonization of the rectum by C. rodentium.
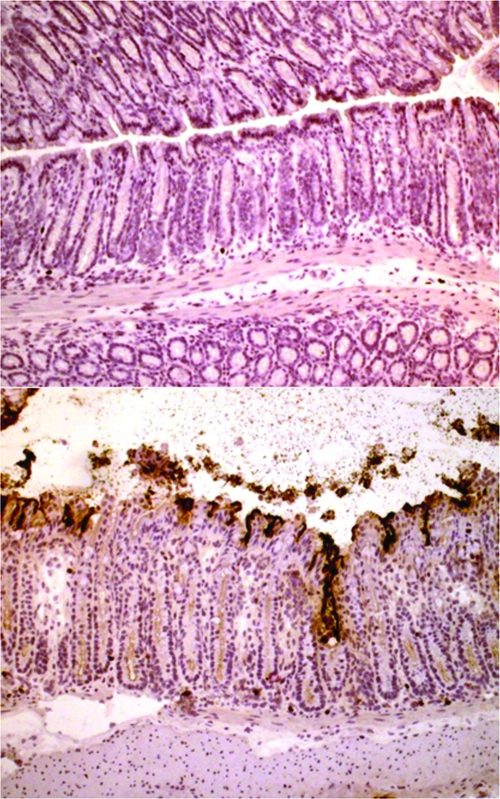
Using BLI, we observed a number of infected animals exhibiting a bioluminescent signal from the region of the rectum, a localization previously unreported for C. rodentium colonization. Analysis of tissue sections by immunohistochemistry demonstrated obvious and heavy colonization of the rectum by C. rodentium (Fig. 3) until about 1 mm from the anal margin. Furthermore, hyperplasia of the rectal crypts was clearly observed.
FIG. 3.
Colonization of the rectum by C. rodentium. Frozen sections of rectum harvested from uninfected mice and mice at 6 days post-C. rodentium infection were cut and stained for immunohistochemistry with a rabbit antibacterial antibody as described previously (9). A section showing representative rectal tissue from an uninfected mouse is given in the top panel, while that from a C. rodentium-infected animal is given in the bottom panel. Bacteria are clearly evident, adhering to the surface epithelium and crypts of the rectum with resulting hyperplasia (bottom panel). Magnification, ×84.
In this paper, we have reported, for the first time, the use of BLI in following the infection dynamics of C. rodentium in the GI tract of live animals. We have shown that, as we reported using harvested organs from orally challenged mice (18), at early time points, C. rodentium colonizes the cecal patch. However, the current study shows that C. rodentium also targets the rectum. Involvement of the rectum in C. rodentium infection has not been reported before; significantly, it was recently shown that EHEC O157:H7 targets the distal bovine rectum (15).
The GI tract is the largest and most complex environment in the mammalian host, covering several diverse macroenvironments, including the stomach, the small intestine, and the colon. Within these macroenvironments are several microenvironments in which bacteria can live, including the lumen, the mucus layer overlying the epithelium, mucus within intestinal crypts, and the surface of the mucosal epithelial cells.
A recent report described the analysis of >5,000 16S rRNA gene sequences isolated from the murine cecum (12). Researchers found that 64% of the sequences could not be assigned to known genera, and only 7% represented previously cultured species. The two most abundant bacterial divisions were the Firmicutes and the Bacteroidetes (12). The presence of strict anaerobes residing within the GI tract has lead to the long-held belief that this environment is anaerobic and, as such, unsuitable for the use of BLI. However, we have clearly demonstrated that there is sufficient oxygen present within the murine colon and cecum to allow the generation of detectable light by C. rodentium ICC180 and commensal E. coli (ICC214). Furthermore, light production was seen only in live animals, suggesting the requirement for a circulating blood supply to the colon and cecum to provide sufficient oxygen.
It was recently reported that the common GI inhabitant Bacteroides fragilis, previously believed to be a strict anaerobe, can grow in nanomolar concentrations of O2 (300 nM) (1). B. fragilis was found to encode a cytochrome bd oxidase (CydA) essential for O2 consumption. Indeed, many species of Bacteroides have now been shown to grow in nanomolar concentrations of O2, and homologues of cydA have been identified in the genomes of many prokaryotes classified as strict anaerobes. The authors suggested a new term, nanaerobes, for such organisms which can benefit from, yet do not require, O2 for growth. Furthermore, it has been reported that luminescence can be detected from marine bioluminescent bacteria under O2 concentrations as low as 10 nM (2). These finding suggest that regions of the murine GI tract should be referred to as nanaerobic.
Previously, bioluminescent Salmonella enterica serovar Typhimurium was found to be luminescent in the cecum but nonluminescent in the small intestine until exposed to oxygen (3). The researchers suggested that the presence of a signal from within the cecum was due to an intimate coupling of bacteria and epithelial cells, thus allowing the bacteria access to oxygen. However, we detected a signal from the cecum at a time when an intimate coupling is unlikely to have occurred for C. rodentium ICC180. Indeed, our data suggest that after oral gavage of mice with 109 CFU ICC180, the majority of bacteria do not colonize the animals but instead pass straight through the gastrointestinal tract. While a signal is detectable from within animals at 5 h postgavage, by 24 h postgavage, no detectable signal remains (data not shown), suggesting that fewer than 103 CFU in a spot (the detection limits of the machine for ICC180) (18) have attached to the cecal patch. It is not until 3 days postgavage that sufficient C. rodentium strains are present for a signal to be detectable from within the cecum. Our conclusions are supported by the fact that a signal can be recorded from the cecum following oral inoculation with a bioluminescent commensal E. coli strain (ICC214) which is unable to form such an intimate attachment. Instead, we suggest that sufficient levels of oxygen are present within the lumen of some regions of the gastrointestinal tract. Indeed, cervical dislocation resulted in loss of bioluminescence, suggesting that the nanaerobic environment in the GI tract is maintained by diffusion of O2 from blood vessels into the lumen.
The C. rodentium model permits robust investigation of pathogen-host interactions under physiological conditions typical of the intestinal environment, with the ability to manipulate both the pathogen and the host. Indeed, defined mutants can easily be made in C. rodentium ICC180 by using the lambda red one-step method (5), and we are currently investigating the roles of a number of T3SS effectors in colonization using BLI. The finding that BLI technology can be utilized for studying events occurring in the GI tract of living animals is important, as it has the potential to have a significant impact on the reduction of the numbers of animals required for such experiments. In our previous studies, the requirement to image organs ex vivo resulted in the use of at least three mice per time point. However, using BLI on live animals, this number can be drastically reduced as group sizes of five to six animals are sufficient to follow the colonization dynamics for an entire experiment. In addition, better-quality data are obtained by following the same animals throughout the course of an experiment.
In conclusion, we have shown that (i) the colonic environment is nanaerobic, (ii) it is possible to determine the colonization dynamics of C. rodentium infection in the GI tract of live animals using BLI, (iii) detection of bioluminescent C. rodentium in vivo is dependent on a functioning blood supply, and (iv) C. rodentium targets both the cecum and the distal rectum at early time points postchallenge from which the bacteria spread to colonize the colon. The mechanism by which A/E pathogens colonize the rectum is not known, but C. rodentium provides a practical infection model to investigate this phenomenon experimentally.
Acknowledgments
We would like to thank Alan Huett and Francis Girard for helpful discussions.
This work was supported by the Wellcome Trust (grant 071006/Z/03/Z).
Editor: J. T. Barbieri
REFERENCES
- 1.Baughn, A. D., and M. H. Malamy. 2004. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature 427:441-444. [DOI] [PubMed] [Google Scholar]
- 2.Bourgois, J. J., F. E. Sluse, F. Baguet, and J. Mallefet. 2001. Kinetics of light emission and oxygen consumption by bioluminescent bacteria. J. Bioenerg. Biomembr. 33:353-363. [DOI] [PubMed] [Google Scholar]
- 3.Contag, C. H., P. R. Contag, J. I. Mullins, S. D. Spilman, D. K. Stevenson, and D. A. Benaron. 1995. Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol. 18:593-603. [DOI] [PubMed] [Google Scholar]
- 4.Contag, C. H., and M. H. Bachmann. 2002. Advances in in vivo bioluminescence imaging of gene expression. Annu. Rev. Biomed. Eng. 4:235-260. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and W. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, E. O., and P. J. Henderson. 1987. The cloning and DNA sequence of the gene xylE for xylose-proton symport in Escherichia coli K12. J. Biol. Chem. 262:13928-13932. [PubMed] [Google Scholar]
- 7.Deng, W., J. L. Puente, S. Gruenheid, Y. Li, B. A. Vallance, A. Vazquez, J. Barba, J. A. Ibarra, P. O'Donnell, P. Metalnikov, K. Ashman, S. Lee, D. Goode, T. Pawson, and B. B. Finlay. 2004. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc. Natl. Acad. Sci. USA 101:3597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garmendia, J., G. Frankel, and V. F. Crepin. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 73:2573-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonçalves, N. S., M. Ghaem-Maghami, G. Monteleone, G. Frankel, G. Dougan, D. J. M. Lewis, C. P. Simmons, and T. T. MacDonald. 2001. Critical role for tumor necrosis factor alpha in controlling the number of lumenal pathogenic bacteria and immunopathology in infectious colitis. Infect. Immun. 69:6651-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 11.Knutton, S., D. R. Lloyd, and A. S. McNeish. 1987. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect. Immun. 55:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meighen, E. A. 1994. Genetics of bacterial bioluminescence. Annu. Rev. Genet. 28:117-139. [DOI] [PubMed] [Google Scholar]
- 14.Mundy, R., T. T. MacDonald, G. Dougan, G. Frankel, and S. Wiles. 2005. Citrobacter rodentium: of mice and man. Cell. Microbiol. 7:1697-1706. [DOI] [PubMed] [Google Scholar]
- 15.Naylor, S. W., J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, I. J. McKendrick, D. G. Smith, and D. L. Gally. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piwnica-Worms, D., D. P. Schuster, and J. R. Garbow. 2004. Molecular imaging of host-pathogen interactions in intact small animals. Cell. Microbiol. 6:319-331. [DOI] [PubMed] [Google Scholar]
- 17.Simpson, N., R. Shaw, V. F. Crepin, R. Mundy, A. J. FitzGerald, N. Cummings, A. Straatman-Iwanowska, I. Connerton, S. Knutton, and G. Frankel. 2006. The enteropathogenic Escherichia coli type III secretion system effector Map binds EBP50/NHERF1: implication for cell signalling and diarrhoea. Mol. Microbiol. 60:349-363. [DOI] [PubMed] [Google Scholar]
- 18.Wiles, S., S. Clare, J. Harker, A. Huett, D. Young, G. Dougan, and G. Frankel. 2004. Organ-specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell. Microbiol. 6:963-972. [DOI] [PubMed] [Google Scholar]