Abstract
Outer membrane protein P6 is the subject of investigation as a vaccine antigen to prevent infections caused by nontypeable Haemophilus influenzae, which causes otitis media in children and respiratory tract infections in adults with chronic lung disease. P6 induces protective immune responses in animal models and is the target of potentially protective immune responses in humans. P6 is a 16-kDa lipoprotein that shares homology with the peptidoglycan-associated lipoproteins of gram-negative bacteria and is highly conserved among strains of H. influenzae. To characterize the function of P6, an isogenic mutant was constructed by replacing the P6 gene with a chloramphenicol resistance cassette. The P6 mutant showed altered colony morphology and slower growth in vitro than that of the parent strain. By electron microscopy, the P6 mutant cells demonstrated increased size, variability in size, vesicle formation, and fragility compared to the parent cells. The P6 mutant showed hypersensitivity to selected antibiotics with different mechanisms of action, indicating increased accessibility of the agents to their targets. The P6 mutant was more sensitive to complement-mediated killing by normal human serum. Complementation of the mutation in trans completely or partially restored the phenotypes. We concluded that P6 plays a structural role in maintaining the integrity of the outer membrane by anchoring the outer membrane to the cell wall. The observation that the absence of expression of P6 is detrimental to the cell is a highly desirable feature for a vaccine antigen, supporting further investigation of P6 as a vaccine candidate for H. influenzae.
Nontypeable or nonencapsulated Haemophilus influenzae is a common human respiratory tract pathogen, causing disease in both children and adults. The bacterium is an important cause of otitis media, the most common reason for children to receive antibiotic therapy. Based on results of middle-ear-fluid cultures, 25 to 35% of the approximately 25 million episodes of otitis media that occur annually in the United States are caused by H. influenzae (24, 25). The widespread use of pneumococcal conjugate vaccines among infants and children in the past 5 years has led to an increase in the proportion of otitis media cases caused by H. influenzae, particularly among cases of recurrent otitis media (9, 29).
Approximately 20 million adults in the United States have chronic obstructive pulmonary disease (COPD), a debilitating disease which is the fourth leading cause of death nationwide (3, 46). The course of the disease is characterized by intermittent exacerbations, which may lead to physician visits, hospital admissions, respiratory failure requiring mechanical ventilation, and death. Approximately one-half of exacerbations are caused by bacterial infection, and the most common pathogen is H. influenzae (35, 50).
Because of the enormous morbidity associated with H. influenzae otitis media and the morbidity and mortality associated with respiratory tract infections in adults with COPD, H. influenzae is a major focus of vaccine development efforts (16, 36, 37).
Outer membrane protein P6 is a member of the class of outer membrane proteins known as peptidoglycan-associated lipoproteins (10, 28, 44, 45). First discovered in the mid-1980s, P6 is a promising vaccine antigen that has been the subject of extensive study (33, 41, 42). P6 has several features suggesting that the protein may be an effective vaccine antigen. The gene that encodes P6 is present and the protein is expressed in all strains of H. influenzae examined thus far. The nucleotide sequence homology among strains is 97%, and the amino acid sequence homology among strains is 100%, indicating that the protein is highly conserved among strains (43). P6 has epitopes on the bacterial surface, an important characteristic for potentially protective antibodies to bind P6 on the intact bacterial cell.
P6 induces protective immune responses in a variety of animal model systems, including the infant rat model of invasive infection (17, 33, 53), a rat pulmonary clearance model (27), otitis media models in the chinchilla and mouse (12, 18, 48), and nasopharyngeal colonization models (6, 21, 23, 32). P6 is the target of bactericidal antibodies from rats, chinchillas, rabbits, and humans (12, 17, 27, 38). An analysis of antibody responses to P6 in children has provided suggestive evidence that human immune responses to P6 are associated with protection from H. influenzae otitis media (22, 26, 52). Furthermore, T-cell responses to P6 in adults with COPD are associated with relative protection from exacerbations caused by H. influenzae (1). In view of these observations that the highly conserved P6 protein induces potentially protective immune responses in numerous animal models, in vitro systems, and clinical studies, there is great interest in evaluating P6 in clinical trials to assess the extent to which immunization of humans with this antigen will induce protection against H. influenzae infection.
In addition to its potential as a vaccine antigen, P6 is a key mediator in the interaction of H. influenzae with the human host. P6 activates NF-κB through Toll-like receptor 2 signaling (51) and is a potent inducer of proinflammatory cytokines, particularly interleukin 8 and tumor necrosis factor alpha (5). Furthermore, the protein induces the transcription of mucin production genes in middle ear cells (11). These inflammatory effects of P6 parallel those induced by peptidoglycan-associated lipoproteins of other gram-negative bacteria (2, 7, 30). Through these potent effects, P6 is a key virulence factor in mediating the inflammation that is a hallmark of COPD and otitis media.
Little is known about the function of the protein in H. influenzae. P6 shares homology with the Escherichia coli peptidoglycan-associated lipoprotein which appears to play a structural role in anchoring the outer membrane to the cell wall (10, 28, 44, 45). The goal of the present study was to begin to evaluate the function of P6 in H. influenzae. Our approach was to construct a P6-deficient mutant, an undertaking that has eluded us for 2 decades. The mutant and a complemented mutant were studied to assess the function of this putative vaccine antigen in the bacterium.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
H. influenzae strains 49P5H1 and 1479 were isolated from the sputa of adults with COPD. Plasmid pGEM3Zf was obtained from Promega (Madison, WI). Plasmid pSPEC1 was kindly provided by Lauren Bakaletz and Robert Munson (4). H. influenzae was grown on chocolate agar or in brain heart infusion broth supplemented with hemin and NAD at 10 μg/ml (each).
Monoclonal antibodies.
The monoclonal antibody 7F3 recognizes an epitope on outer membrane protein P6 (OMP P6) and was described previously (41, 43). The monoclonal antibody 2C7 recognizes an epitope on OMP P5 (34, 40). Both 7F3 and 2C7 are immunoglobulin G isotypes.
SDS-polyacrylamide gel electrophoresis and immunoblot assays.
Whole-cell lysates were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and Coomassie blue staining by previously described methods (39). Immunoblot assays with monoclonal antibodies were performed as described previously (43).
Construction of P6 mutant.
A mutant lacking the gene encoding P6 and thus deficient in the expression of P6 was constructed in strain 49P5H1. To accomplish this, a 1,465-bp region upstream of the P6 gene and a 1,260-bp region downstream of the P6 gene were amplified by PCR from genomic DNA of H. influenzae 1479. Primer sequences are noted in Table 1. These amplicons were ligated into pGEM3Zf. A chloramphenicol cassette was amplified from plasmid pACYC184 (New England Biolabs, Beverly, MA) and ligated into the plasmid construct between the fragments upstream and downstream of the P6 gene by using an AccI restriction site. Therefore, the plasmid contained an insert in which a chloramphenicol resistance cassette was flanked by sequences corresponding to regions upstream and downstream of the P6 gene and lacked the P6 gene itself.
TABLE 1.
Oligonucleotide primers used for this study
| Primer | Sequencea | Purpose |
|---|---|---|
| CATacc5′ | CTCTTCTCTCTCTCTGTCGACGACAGCTGATAGAAACAGAAGCCAC | Amplify and clone chloramphenicol resistance cassette |
| CATacc3′ | ATATATATATATATGTCGACAGGCGTAGCACCAGGCGTTTAAGGGCAC | Amplify and clone chloramphenicol resistance cassette |
| CAT5′probe | AGTAAGTTGGCAGCATCACCC | Amplify chloramphenicol resistance cassette probe for Southern blotting |
| CAT3′probe | CATTGAGCAACTGACTGAAATGCC | Amplify chloramphenicol resistance cassette probe for Southern blotting |
| P65′probe | GATGCTGCAGGAAATGGTGCTG | Amplify P6 gene probe for Southern blotting |
| P63′probe | ACGTTCATCAGTGTTACCTTC | Amplify P6 gene probe for Southern blotting |
| pP6spec25′ | TCTCTCAAGCTTCGTTTAGCTTTTGCTTCTTCTCG | Amplify P6 gene and flanking region for pP6Spec2 for complementation |
| pP6spec23′ | TGTGTGTGGAATTCTAAGAATGGTAGGTTGTGAGAG | Amplify P6 gene and flanking region for pP6Spec2 for complementation |
| P6n5′ | ATATATGGTACCGATATTTGTTCAGCTGC | Amplify region upstream of P6 gene for construction of mutant |
| P6n3′acc | ATATATATAGATATGTCGACGATTTCTCCTAAATGAG | Amplify region upstream of P6 gene for construction of mutant |
| P6c5′acc | ATATATATAGATATGTCGACGTATTTCTAATACTTG | Amplify region downstream of P6 gene for construction of mutant |
| P6c3′ | AGTACAAAGCTTGCTGTAACTGCATAACG | Amplify region downstream of P6 gene for construction of mutant |
| P65′ | TGTAGTTCCTCTAACAACGAT | Amplify P6 gene and RT-PCR of P6 transcript |
| P63′ | GTACGCTAACACTGCACGACG | Amplify P6 gene and RT-PCR of P6 transcript |
| tolB5′ | TCGCTGGCGATGAAGGTACGCATTG | RT-PCR of transcript of tolB upstream of P6 gene |
| tolB3′ | CATCATAATCTGCTACACGAA | RT-PCR of transcript of tolB upstream of P6 gene |
| RNAmtrans5′ | TGTTGGAAAATATTCGTATTG | RT-PCR of transcript of HI0380 downstream of P6 gene |
| RNAmtrans3′ | CGTTTTTCAACCGCACTTAGC | RT-PCR of transcript of HI0380 downstream of P6 gene |
Restriction enzyme sites are underlined.
The plasmid was linearized with NdeI and then transformed into strain 49P5H1, which was made competent by the method of Herriott et al. (20), using the transformation protocol of Poje and Redfield (47). Transformants were selected on chocolate agar containing 2 μg/ml of chloramphenicol. A mutant was obtained (49P5H1P6−), and allelic exchange was verified by PCR analysis and Southern blot assay, as detailed in Results.
Complementation was accomplished by using plasmid pSPEC1 (4). A fragment containing the P6 gene and 533 bp upstream and 340 bp downstream of the gene was amplified by PCR from genomic DNA of strain 49P5H1 and ligated into pSPEC1 at HindIII and EcoRI restriction sites. After confirmation of the sequence of the insert of the resulting plasmid (pP6Spec2), H. influenzae 49P5H1P6− was electroporated with pP6Spec2 that had been methylated with CpG methylase (New England Biolabs) in a 0.1-cm cuvette (200 Ω, 2.5 kV, 25 μF). Cells were plated on chocolate agar containing 200 μg/ml of spectinomycin and incubated overnight, and the complemented strain 49P5H1P6−(pP6Spec2) was obtained. Strain 49P5H1P6−(pP6Spec2) was grown in the presence of spectinomycin for all experiments.
Southern blot assay.
Southern blot assays were performed with genomic and plasmid DNAs restricted with EcoRI with a Hoefer TransVac vacuum blotting unit following the manufacturer's instructions (Hoefer, San Francisco, CA). Probes were biotinylated with an NEBlot Phototope kit (New England Biolabs), and blots were developed with a Phototope-Star detection kit (New England Biolabs) according to the manufacturer's instructions.
RT-PCR.
Bacterial RNAs were isolated using a QIAGEN RNeasy kit and a Qiashredder column (QIAGEN, Valencia, CA) following the manufacturer's instructions, with an additional incubation with RNase-free DNase I (Promega) for 30 min at 37°C. Reverse transcriptase PCR (RT-PCR) was performed using a QIAGEN OneStep RT-PCR kit and RNaseOut RNase inhibitor (Invitrogen, Carlsbad, CA). Primers were designed to amplify ∼500-bp fragments of the P6 gene, the upstream gene tolB, and the downstream gene HI0380 (Table 1). In addition, to exclude the possibility of contaminating DNA, parallel reactions with TaqI DNA polymerase (HotMaster mix; Eppendorf, Hamburg, Germany) were performed. Following amplification, samples were electrophoresed in 1.5% agarose gels and stained with ethidium bromide.
Transmission electron microscopy.
Bacteria were grown to logarithmic phase in broth and harvested by centrifugation. Pellets were resuspended in phosphate-buffered saline (PBS) and washed three times by centrifugation. Bacterial cells were fixed by suspension in 2.5% glutaraldehyde in PBS and incubation for 2 h at 4°C. After being washed three times by centrifugation and then resuspended, cells were stained by suspension in 1.5% osmium tetroxide and incubation for 1 h at 4°C. After the cells were washed three times in PBS, they were dehydrated in graded ethanol followed by acetone. Samples were embedded in Araldite/Embed (Electron Microscopy Sciences, Hatfield, PA), thin sections were cut and mounted on coated specimen grids, and sections were stained with saturated aqueous uranyl acetate-lead citrate. Sections were examined using a JEOL 100CX-II transmission electron microscope in the Electron Microscopy Laboratory in the School of Medicine and Biomedical Sciences at the University at Buffalo.
MICs.
MIC determinations were performed in the laboratory of Gary Doern at the University of Iowa by broth microdilution as described previously (19).
Disk diffusion assays.
Assays to assess the susceptibility of strains to a panel of hydrophobic agents and to selected antimicrobial agents were performed as described by Luke et al. (31). Bacteria were suspended to an optical density at 600 nm of 0.2, and aliquots of 0.1 ml were spread onto chocolate agar plates. Sterile blank paper disks (Becton Dickinson, Cockeysville, MD) were saturated with the various agents and placed on the agar plates in triplicate, and the plates were incubated overnight at 36°C in 5% CO2. Sensitivity to each of the agents was determined by measuring the diameter of the zone of inhibition in two axes, and the mean value was recorded. Values were analyzed using pairwise comparison in a mixed model.
Bactericidal assays.
The sensitivity of strains to killing by normal human serum was assessed by bactericidal assays performed in 96-well plates. Strains were grown to logarithmic phase in broth and diluted in 10% bovine serum albumin in Hanks' balanced salt solution so that the same number of viable bacteria was placed into each well, as determined by colony counts (final concentration in the assay well, ∼104 CFU per ml). It was necessary to use different dilutions of bacteria for the different strains (parent strain, P6 mutant, and complemented mutant) based on the optical density of each broth culture to have the same number of viable bacteria in each reaction. Serum from a healthy donor was added at various dilutions, and colony counts were determined in duplicate after incubation for 30 min at 36°C. The percent kill for each serum dilution was calculated by dividing the number of colonies after 30 min of incubation with serum by the number of colonies in control wells incubated for 30 min in the absence of serum. Colony counts were performed at time zero and at 30 min for each assay to ensure that bacteria remained viable under the conditions of the assay.
RESULTS
Construction and characterization of a P6 mutant.
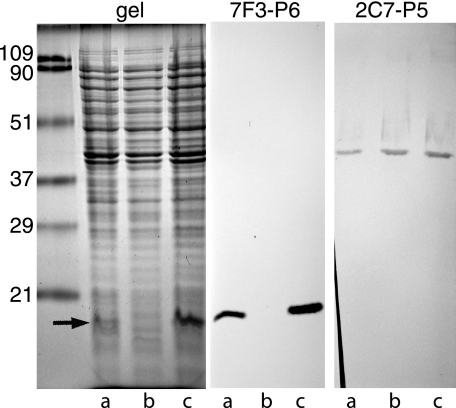
To create a mutant that completely lacks the gene encoding OMP P6, a construct that contained a chloramphenicol cassette in place of the P6 gene in the center of a fragment of DNA with the sequences flanking the P6 gene in H. influenzae was constructed. Homologous recombination with the construct generated a mutant in which P6 was absent. The mutation was complemented by cloning the P6 gene and its flanking sequences into a plasmid that replicates in H. influenzae. Figure 1 shows an SDS gel of a whole-cell lysate in which the P6 band separates as a somewhat wavy band characteristic of a lipoprotein in the parent strain 49P5H1 (lanes a) but is absent in the mutant strain (lanes b). The complemented mutant shows the presence of a prominent P6 band (lanes c). Immunoblot assays with a monoclonal antibody that recognizes an epitope on OMP P6 confirmed that P6 is not expressed in the mutant but is expressed in the complemented mutant (Fig. 1).
FIG. 1.
Coomassie blue-stained SDS gel (left panel) and immunoblot assays of whole bacterial cell lysates (middle and right panels). Lanes a, parent strain; lanes b, P6 mutant; lanes c, complemented P6 mutant. The center panel was probed with the monoclonal antibody 7F3, which recognizes an epitope on outer membrane protein P6. The right panel was probed with the monoclonal antibody 2C7, which recognizes an epitope on outer membrane protein P5. The arrow denotes the location of P6. Molecular mass markers are noted on the left, in kilodaltons.
To confirm the insertion of the chloramphenicol cassette into the mutant, DNA from the parent, mutant, and complemented mutant strains was subjected to analysis by PCR, using primers corresponding to sequences in the chloramphenicol cassette, the P6 gene, and the region flanking the P6 gene. All PCRs yielded amplicons of the predicted sizes (data not shown).
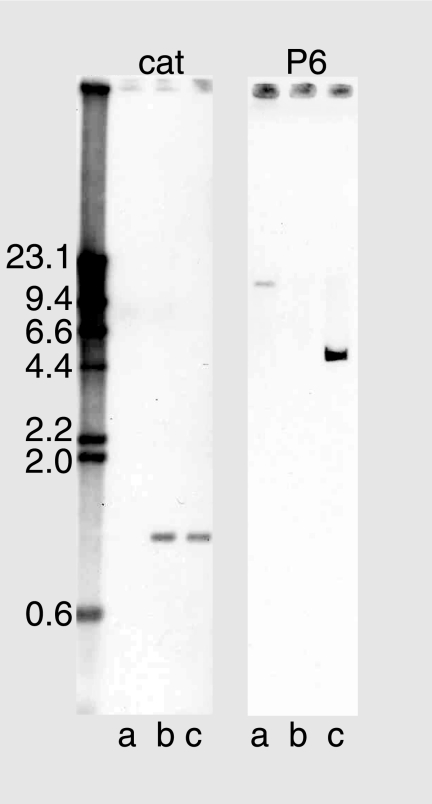
Southern blot analysis was performed to further characterize the mutant. DNA isolated from the parent, mutant, and complemented mutant strains was restricted with EcoRI and subjected to agarose gel electrophoresis. After transfer, probing of the DNA with a 200-bp fragment of the chloramphenicol cassette showed single bands for the mutant and the complemented mutant, as expected (Fig. 2, left panel). Probing the DNA with a 200-bp fragment of the P6 gene showed a band of between 9.4 and 23.1 kb for the parent and no band for the mutant, confirming the absence of the P6 gene in the mutant (Fig. 2, right panel). The complemented mutant yielded a band of approximately 5 kb as a result of detection of the P6 gene on the pP6Spec2 plasmid (Fig. 2, right panel, lane c).
FIG. 2.
Southern blot assay. DNAs from the parent strain (lanes a), the P6 mutant (lanes b), and the complemented P6 mutant (lanes c) were digested with EcoRI. Panels were probed with ∼200-bp biotinylated PCR fragments of the chloramphenicol resistance cassette (left) and the P6 gene (right). Molecular size markers are noted in kilobases on the left.
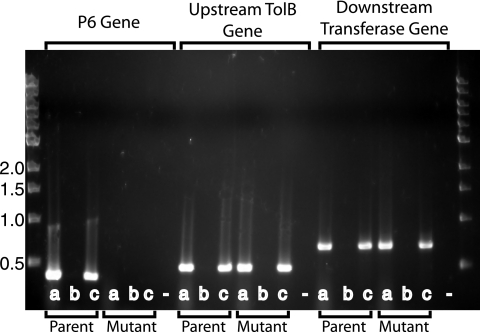
To assess whether replacing the P6 gene with a chloramphenicol resistance cassette in the mutant altered transcription of the genes upstream and downstream of the P6 gene, RT-PCR was performed. Primers that amplify an ∼500-bp fragment of tolB, which is located immediately upstream of the P6 gene, were designed. A strong transcriptional terminator is present downstream of the P6 gene (49), and the next open reading frame, corresponding to HI0380, encoding a hypothetical tRNA/rRNA methyltransferase, is located 430 bp downstream of the P6 gene. Primers corresponding to an ∼600-bp fragment of HI0380 were designed. Transcription of both the upstream tolB and downstream HI0380 genes in the P6 mutant appeared to be indistinguishable from that in the parent strain (Fig. 3). Control assays confirmed that the purified RNA was free of contaminating DNA and that, as expected, transcription of the P6 gene in the mutant was absent. These results demonstrate that, based on semiquantitative RT-PCR, deletion of the P6 gene in the mutant did not alter the transcription of the genes immediately upstream and downstream of the P6 gene.
FIG. 3.
Results of RT-PCR with the P6 mutant and its parent strain, 49P5H1, as noted at the bottom of the gel. Primers used in reactions corresponding to the P6 gene, the upstream tolB gene, and the downstream hypothetical tRNA/rRNA methyltransferase gene are noted at the top of the gel. Lanes a, purified RNA amplified with reverse transcriptase; lanes b, purified RNA amplified with TaqI polymerase to exclude DNA contamination; lanes c, purified DNA amplified with TaqI polymerase; lanes −, distilled water with reverse transcriptase as a negative control. Molecular size markers are noted in kilobases on the left.
Colony morphology and growth characteristics.
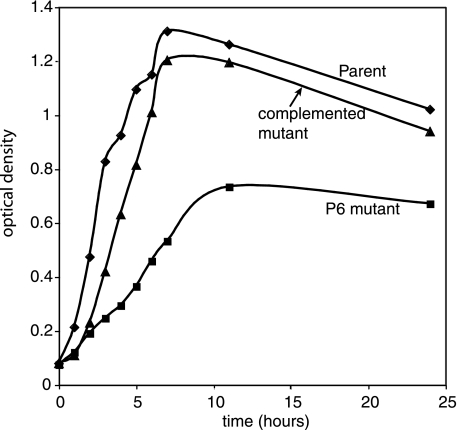
P6 mutants demonstrated altered colony morphology on chocolate agar compared to the parent strain. Mutant colonies had approximately one-half the diameter of colonies of the parent strain after overnight incubation, and the mutant colonies demonstrated a “stickier” consistency when probed with a loop. The mutant strain had a tendency to show clumping when suspended in buffers, in contrast to the parent strain. The complemented mutant had an intermediate phenotype for these features. The mutant showed slower growth in broth and reached a lower cell density in broth than did the parent (Fig. 4).
FIG. 4.
Growth curves for H. influenzae strains.
Cell morphology.
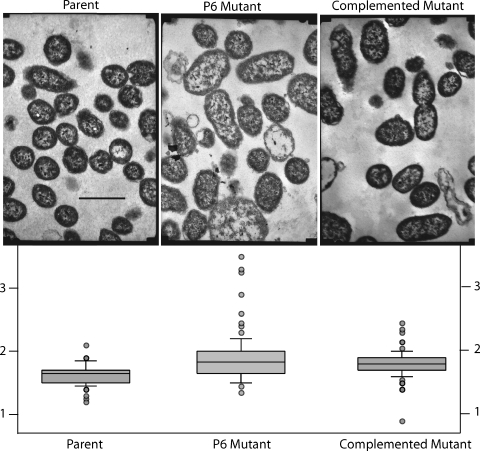
Transmission electron microscopy showed that compared to those of the parent strain, cells of the P6 mutant were larger and showed more size variation from cell to cell (Fig. 5). In addition, more fragments of cells and empty cell envelopes were apparent in mutant sections than in parent sections, suggesting that the cells were more fragile during growth and during processing for electron microscopy. The complemented mutant had an intermediate phenotype with regard to cell size, size variation from cell to cell, and the presence of cell fragments (Fig. 5).
FIG. 5.
Electron micrographs of parent strain 49P5H1, P6 mutant, and complemented mutant. Bar, 1 μm. The graphs indicate the smallest diameters of 100 cells of each strain. The boxes denote 25 and 75% quartiles around the means, and the bars denote 10 and 90% values. Cell diameters lying outside the 10 and 90% values are noted by circles. The y axis shows the cell diameters measured on prints at a magnification of 20,000, in cm.
To assess quantitatively the size differences apparent by visual inspection of electron micrographs, the smallest diameters of 100 cells each of the parent strain, the P6 mutant, and the complemented mutant were measured at a magnification of 20,000. Analysis using Aabel software (Tulsa, OK) is shown in the graphs below the electron micrographs in Fig. 5. The mean diameters of cells (in cm, measured on prints of cells at a magnification of 20,000) of the parent strain (1.63), the mutant (1.89), and the complemented strain (1.79) were different from one another (P < 0.001; analysis of variance). Inspection of the graphs also reveals that cells of the P6 mutant show greater size variation than do those of the parent strain (Fig. 5).
Antimicrobial susceptibility.
The MICs of the parent strain, the P6 mutant, and the complemented mutant were determined for a panel of antimicrobial agents. No significant difference in MIC was observed among the three strains for ampicillin, amoxicillin-clavulanic acid, cefpodoxime, ceftriaxone, moxifloxacin, levofloxacin, azithromycin, clarithromycin, telithromycin, tetracycline, and meropenem. Differences in MICs between parent and mutant strains were observed for three cephalosporins, and these results are shown in Table 2, along with results for selected β-lactam antibiotics. The P6 mutant was 8- to 16-fold more sensitive than the parent strain to cefdinir, cefuroxime, and cefprozil. The phenotype was restored to within one dilution of the parent strain by complementation of the P6 mutation in the case of cefdinir and cefuroxime and was partially restored in the case of cefprozil.
TABLE 2.
MICs for H. influenzae strains of selected β-lactam antibiotics
| Strain | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| Ampicillin | Cefpodoxime | Cefdinir | Cefuroxime | Cefprozil | |
| Parent | 0.5 | 0.12 | 1.0 | 8.0 | 16 |
| P6 mutant | 0.5 | 0.12 | 0.12 | 1.0 | 1.0 |
| Complemented mutant | 0.5 | 0.12 | 0.5 | 4.0 | 4.0 |
Disk diffusion assays were performed to assess the relative susceptibilities of the parent strain, the P6 mutant, and the complemented mutant to the antimicrobial agents novobiocin and polymyxin B. The P6 mutant was significantly more susceptible to inhibition of growth by both novobiocin and polymyxin B (Table 3). Complementation of the mutation completely restored the parent phenotype to the mutant with regard to novobiocin susceptibility and partially restored the polymyxin B phenotype.
TABLE 3.
Results of disk diffusion assays with H. influenzae strains
| Strain | Zone of inhibition (mm) (mean± SD)
|
|
|---|---|---|
| Novobiocin | Polymyxin B | |
| Parent | 19.5 ± 0.5 | 16.0 ± 0.6 |
| P6 mutant | 25.0 ± 0.6a | 20.0 ± 0.6a |
| Complemented mutant | 20.0 ± 0.6b | 17.8 ± 0.8c |
P < 0.0001 (P6 mutant versus parent strain).
P = 0.22 (complemented mutant versus parent strain).
P = 0.002 (complemented mutant versus parent strain).
Disk diffusion assays were also performed to assess the sensitivity profiles of the strains for a panel of hydrophobic agents, including Triton X-100, sodium deoxycholate, Tween 20, and sodium dodecyl sulfate. No reproducible differences were observed between the parent strain and the P6 mutant in disk diffusion assays with these detergents.
Serum sensitivity.
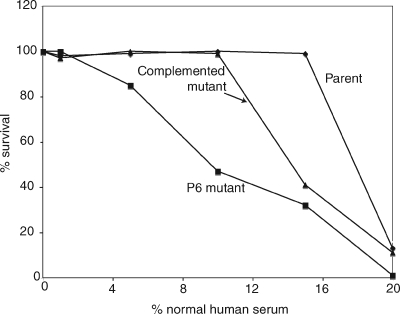
The effect of P6 on sensitivity to complement-mediated killing by human serum was assessed by performing bactericidal assays with dilutions of normal human serum. The P6 mutant was more sensitive to killing by normal human serum than the parent strain, as shown in Fig. 6. The complemented mutant demonstrated an intermediate phenotype with regard to serum sensitivity. The assay was performed multiple times, and the results consistently showed that the P6 mutant was more sensitive to killing by normal human serum than the parent strain.
FIG. 6.
Results of serum bactericidal assays with normal human serum used against H. influenzae strains, as noted. The x axis shows final concentrations of serum in bactericidal reaction mixtures, and the y axis shows percent survival after 30 min.
DISCUSSION
A mutant deficient in expression of the peptidoglycan-associated lipoprotein outer membrane protein P6 was constructed by deleting the entire P6 gene and replacing it with a chloramphenicol resistance cassette. Analysis of the mutant revealed several phenotypic changes compared to the parent strain, including (i) slower growth in broth culture; (ii) altered morphology, including a larger cell size and increased variability in cell size; (iii) decreased cell wall integrity, as indicated by vesicle formation and fragmentation of cells; (iv) increased susceptibility to selected antimicrobial agents; and (v) increased susceptibility to complement-mediated killing by human serum. Complementation of the P6 mutation partially or completely restored these phenotypes. Based on these observations, we concluded that P6 plays a structural function in the bacterial cell by virtue of its role in cell growth, cell shape, and integrity of the cell wall.
Complementation of the mutation in trans resulted in either full or partial restoration of the parental phenotype for each of the characteristics studied. Furthermore, transcription of the genes upstream and downstream of the P6 gene was unaltered in the mutant compared to the parent strain (Fig. 3). These observations support the conclusion that deletion of the P6 gene was responsible for the phenotypes observed in the mutant. The most likely explanation for incomplete complementation in some experiments is that transcription, translation, and processing to the outer membrane are not as efficient for the plasmid construct as for the chromosomal gene.
The P6 mutant demonstrated hypersensitivity to several antibiotics, including polymyxin B, novobiocin, and selected cephalosporins. The mechanisms of action of these antibiotics differ from one another. The target for novobiocin is DNA gyrase, and the targets for cephalosporins are penicillin binding proteins on the cytoplasmic membrane. We speculate that the loss of P6 increases the accessibility of the targets for these agents by alteration of the outer membrane architecture, accounting for the increased antibiotic susceptibility. Polymyxin B binds the outer membrane component lipooligosaccharide, suggesting that alteration in the outer membrane structure by the loss of P6 results in the increased susceptibility to this agent.
The increased sensitivity to complement-mediated killing by human serum of the P6 mutant observed in the present study is also likely a result of altered outer membrane architecture. The structural changes resulting from the absence of P6 may cause the bacterial cell to be killed more readily by insertion of the membrane attack complex. Alternatively, the absence of P6 may expose new epitopes to serum antibodies directed at other surface antigens.
P6 is a peptidoglycan-associated lipoprotein (PAL) with homology to the PALs of E. coli and other gram-negative bacteria. The PAL gene is part of a complex of genes that encode five proteins (TolQ, TolR, TolA, TolB, and PAL) that form a bridge linking the cytoplasmic and outer membranes in E. coli and other gram-negative bacteria (8, 13, 14). In H. influenzae, the tolQRAB genes are also located upstream of the P6 gene (49). Analysis of the P6 mutant that was developed and characterized in the present study provided evidence that P6 plays a similar role for H. influenzae in anchoring the outer membrane to the peptidoglycan.
Fortney et al. (15) developed a mutant of Haemophilus ducreyi that is deficient in the 18-kDa homologue of P6 of H. influenzae. The H. ducreyi mutant also showed altered colony morphology and hypersensitivity to selected antibiotics, but in contrast to the P6 mutant of H. influenzae, the H. ducreyi mutant had a similar growth rate in broth to that of the parent strain. However, wild-type H. ducreyi is quite fastidious in its growth in broth, and perhaps this characteristic of slow growth obscures the detection of a difference. It is interesting that a PAL mutant of H. ducreyi showed a reduced level of pathogenicity in a human challenge model of H. ducreyi infection, suggesting that PAL facilitates pustule formation.
The construction of a P6 mutant of H. influenzae eluded our efforts for many years. The successful construction of the mutant resulted largely from “trial and error,” and although we are not certain of the relative importance of various factors, several approaches likely contributed. These included using a highly transformable strain, ensuring that the transforming DNA contained an H. influenzae-specific uptake sequence, using a chloramphenicol resistance cassette in favor of a kanamycin resistance cassette, using chocolate agar rather than supplemented brain heart infusion agar to select transformants, and screening a large number of transformants to identify a mutant.
P6 continues to be the focus of significant interest as a vaccine antigen for nontypeable H. influenzae. The observation that deleting P6 is detrimental to the bacterium has important implications in further evaluating P6 as a potential vaccine antigen. A mechanism used by bacteria to evade immune responses to antigens is to turn off or down regulate the expression of antigens that are targets of immune responses. The results of the present study show that P6 plays a critical role in the bacterium, indicating that the expression of P6 is important for its viability. This observation is consistent with the previously documented observation of a high degree of sequence conservation among strains (100% conserved at the amino acid level for strains studied thus far), further supporting the concept that P6 is critical for the bacterium. This characteristic is highly desirable for a vaccine antigen.
Acknowledgments
This work was supported by NIH grant AI19641 from the National Institute of Allergy and Infectious Diseases and by the Department of Veterans Affairs.
We thank Robert Munson and Lauren Bakaletz for plasmid pSPEC1, Gary Doern for determining MICs, Ted Szczesny for assistance with electron microscopy, and Xueya Cai for assistance with statistical analysis.
Editor: D. L. Burns
REFERENCES
- 1.Abe, Y., T. F. Murphy, S. Sethi, H. S. Faden, J. Dmochowski, Y. Harabuchi, and Y. M. Thanavala. 2002. Lymphocyte proliferative response to P6 of Haemophilus influenzae is associated with relative protection from exacerbations of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 165:967-971. [DOI] [PubMed] [Google Scholar]
- 2.Aliprantis, A. O., R. B. Yang, M. R. Mark, S. Suggett, B. Devaux, J. D. Radolf, G. R. Klimpel, P. Godowski, and A. Zychlinsky. 1999. Cell activation and apoptosis by bacterial lipoproteins through Toll-like receptor-2. Science 285:736-739. [DOI] [PubMed] [Google Scholar]
- 3.American Thoracic Society. 1995. Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 152:S77-S121. [PubMed] [Google Scholar]
- 4.Bakaletz, L. O., B. D. Baker, J. A. Jurcisek, A. Harrison, L. A. Novotny, J. E. Bookwalter, R. Mungur, and R. S. Munson, Jr. 2005. Demonstration of type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect. Immun. 73:1635-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berenson, C. S., T. F. Murphy, C. T. Wrona, and S. Sethi. 2005. Outer membrane protein P6 of nontypeable Haemophilus influenzae is a potent and selective inducer of human macrophage proinflammatory cytokines. Infect. Immun. 73:2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertot, G. M., P. D. Becker, C. A. Guzman, and S. Grinstein. 2004. Intranasal vaccination with recombinant P6 protein and adamantylamide dipeptide as mucosal adjuvant confers efficient protection against otitis media and lung infection by nontypeable Haemophilus influenzae. J. Infect. Dis. 189:1304-1312. [DOI] [PubMed] [Google Scholar]
- 7.Brightbill, H. D., D. H. Libraty, S. R. Krutzik, R. B. Yang, J. T. Belisle, J. R. Bleharski, M. Maitland, M. V. Norgard, S. E. Plevy, S. T. Smale, P. J. Brennan, B. R. Bloom, P. J. Godowski, and R. L. Modlin. 1999. Host defense mechanisms triggered by microbial lipoproteins through Toll-like receptors. Science 285:732-736. [DOI] [PubMed] [Google Scholar]
- 8.Cascales, E., and R. Lloubes. 2004. Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol. Microbiol. 51:873-885. [DOI] [PubMed] [Google Scholar]
- 9.Casey, J. R., and M. E. Pichichero. 2004. Changes in frequency and pathogens causing acute otitis media in 1995-2003. Pediatr. Infect. Dis. J. 23:824-828. [DOI] [PubMed] [Google Scholar]
- 10.Chen, R., and U. Henning. 1987. Nucleotide sequence of the gene for the peptidoglycan-associated lipoprotein of Escherichia coli K12. Eur. J. Biochem. 163:73-77. [DOI] [PubMed] [Google Scholar]
- 11.Chen, R., J. H. Lim, H. Jono, X. X. Gu, Y. S. Kim, C. B. Basbaum, T. F. Murphy, and J. D. Li. 2004. Nontypeable Haemophilus influenzae lipoprotein P6 induces MUC5AC mucin transcription via TLR2-TAK1-dependent p38 MAPK-AP1 and IKKbeta-IkappaBalpha-NF-kappaB signaling pathways. Biochem. Biophys. Res. Commun. 324:1087-1094. [DOI] [PubMed] [Google Scholar]
- 12.DeMaria, T. F., D. M. Murwin, and E. R. Leake. 1996. Immunization with outer membrane protein P6 from nontypeable Haemophilus influenzae induces bactericidal antibody and affords protection in the chinchilla model of otitis media. Infect. Immun. 64:5187-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis, J. J., E. R. Lafontaine, and P. A. Sokol. 1996. Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa. J. Bacteriol. 178:7059-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubuisson, J. F., A. Vianney, N. Hugouvieux-Cotte-Pattat, and J. C. Lazzaroni. 2005. Tol-Pal proteins are critical cell envelope components of Erwinia chrysanthemi affecting cell morphology and virulence. Microbiology 151:3337-3347. [DOI] [PubMed] [Google Scholar]
- 15.Fortney, K. R., R. S. Young, M. E. Bauer, B. P. Katz, A. F. Hood, R. S. Munson, Jr., and S. M. Spinola. 2000. Expression of peptidoglycan-associated lipoprotein is required for virulence in the human model of Haemophilus ducreyi infection. Infect. Immun. 68:6441-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giebink, G. S., Y. Kurono, L. O. Bakaletz, J. M. Kyd, S. J. Barenkamp, T. F. Murphy, B. Green, P. L. Ogra, X. X. Gu, J. A. Patel, T. Heikkinen, S. I. Pelton, M. Hotomi, and P. Karma. 2005. Recent advances in otitis media. 6. Vaccine. Ann. Otol. Rhinol. Laryngol. 194(Suppl.):86-103. [PubMed] [Google Scholar]
- 17.Green, B. A., T. Quinn-Dey, and G. W. Zlotnick. 1987. Biologic activities of antibody to a peptidoglycan-associated lipoprotein of Haemophilus influenzae against multiple clinical isolates of H. influenzae type b. Infect. Immun. 55:2878-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green, B. A., M. E. Vazquez, G. W. Zlotnick, G. Quigley-Reape, J. D. Swarts, I. Green, J. L. Cowell, C. D. Bluestone, and W. J. Doyle. 1993. Evaluation of mixtures of purified Haemophilus influenzae outer membrane proteins in protection against challenge with nontypeable H. influenzae in the chinchilla otitis media model. Infect. Immun. 61:1950-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heilmann, K. P., C. L. Rice, A. L. Miller, N. J. Miller, S. E. Beekmann, M. A. Pfaller, S. S. Richter, and G. V. Doern. 2005. Decreasing prevalence of beta-lactamase production among respiratory tract isolates of Haemophilus influenzae in the United States. Antimicrob. Agents Chemother. 49:2561-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herriott, R. M., E. M. Meyer, and M. Vogt. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hotomi, M., T. Saito, and N. Yamanaka. 1998. Specific mucosal immunity and enhanced nasopharyngeal clearance of nontypeable Haemophilus influenzae after intranasal immunization with outer membrane protein P6 and cholera toxin. Vaccine 16:1950-1956. [DOI] [PubMed] [Google Scholar]
- 22.Hotomi, M., N. Yamanaka, T. Saito, J. Shimada, M. Suzumoto, M. Suetake, and H. Faden. 1999. Antibody responses to the outer membrane protein P6 of non-typeable Haemophilus influenzae and pneumococcal capsular polysaccharides in otitis-prone children. Acta Otolaryngol. (Stockholm) 119:703-707. [DOI] [PubMed] [Google Scholar]
- 23.Hotomi, M., N. Yamanaka, J. Shimada, M. Suzumoto, Y. Ikeda, A. Sakai, J. Arai, and B. Green. 2002. Intranasal immunization with recombinant outer membrane protein P6 induces specific immune responses against nontypeable Haemophilus influenzae. Int. J. Pediatr. Otorhinolaryngol. 65:109-116. [DOI] [PubMed] [Google Scholar]
- 24.Kilpi, T., E. Herva, T. Kaijalainen, R. Syrjanen, and A. K. Takala. 2001. Bacteriology of acute otitis media in a cohort of Finnish children followed for the first two years of life. Pediatr. Infect. Dis. J. 20:654-662. [DOI] [PubMed] [Google Scholar]
- 25.Klein, J. O. 1994. Otitis media. Clin. Infect. Dis. 19:823-833. [DOI] [PubMed] [Google Scholar]
- 26.Kodama, H., H. Faden, Y. Harabuchi, A. Kataura, J. M. Bernstein, and L. Brodsky. 1999. Cellular immune response of adenoidal and tonsillar lymphocytes to the P6 outer membrane protein of non-typeable Haemophilus influenzae and its relation to otitis media. Acta Otolaryngol. (Stockholm) 119:377-383. [DOI] [PubMed] [Google Scholar]
- 27.Kyd, J. M., M. L. Dunkley, and A. W. Cripps. 1995. Enhanced respiratory clearance of nontypeable Haemophilus influenzae following mucosal immunization with P6 in a rat model. Infect. Immun. 63:2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazzaroni, J. C., and R. Portalier. 1992. The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL). Mol. Microbiol. 6:735-742. [DOI] [PubMed] [Google Scholar]
- 29.Leibovitz, E., M. R. Jacobs, and R. Dagan. 2004. Haemophilus influenzae: a significant pathogen in acute otitis media. Pediatr. Infect. Dis. J. 23:1142-1152. [PubMed] [Google Scholar]
- 30.Liang, M. D., A. Bagchi, H. S. Warren, M. M. Tehan, J. A. Trigilio, L. K. Beasley-Topliffe, B. L. Tesini, J. C. Lazzaroni, M. J. Fenton, and J. Hellman. 2005. Bacterial peptidoglycan-associated lipoprotein: a naturally occurring Toll-like receptor 2 agonist that is shed into serum and has synergy with lipopolysaccharide. J. Infect. Dis. 191:939-948. [DOI] [PubMed] [Google Scholar]
- 31.Luke, N. R., S. Allen, B. W. Gibson, and A. A. Campagnari. 2003. Identification of a 3-deoxy-d-manno-octulosonic acid biosynthetic operon in Moraxella catarrhalis and analysis of a KdsA-deficient isogenic mutant. Infect. Immun. 71:6426-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason, K. W., D. Zhu, C. A. Scheuer, J. C. McMichael, G. W. Zlotnick, and B. A. Green. 2004. Reduction of nasal colonization of nontypeable Haemophilus influenzae following intranasal immunization with rLP4/rLP6/UspA2 proteins combined with aqueous formulation of RC529. Vaccine 22:3449-3456. [DOI] [PubMed] [Google Scholar]
- 33.Munson, R. S., Jr., and D. M. Granoff. 1985. Purification and partial characterization of outer membrane proteins P5 and P6 from Haemophilus influenzae type b. Infect. Immun. 49:544-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munson, R. S., Jr., S. Grass, and R. West. 1993. Molecular cloning and sequence of the gene for outer membrane protein P5 of Haemophilus influenzae. Infect. Immun. 61:4017-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, T. F. 2003. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis. 16:129-134. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, T. F. 2005. Vaccine development for non-typeable Haemophilus influenzae and Moraxella catarrhalis: progress and challenges. Expert Rev. Vaccines 4:843-853. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, T. F., L. O. Bakaletz, J. M. Kyd, B. Watson, and D. L. Klein. 2005. Vaccines for otitis media: proposals for overcoming obstacles to progress. Vaccine 23:2696-2702. [DOI] [PubMed] [Google Scholar]
- 38.Murphy, T. F., L. C. Bartos, P. A. Rice, M. B. Nelson, K. C. Dudas, and M. A. Apicella. 1986. Identification of a 16,600-dalton outer membrane protein on nontypable Haemophilus influenzae as a target for human serum bactericidal antibody. J. Clin. Investig. 78:1020-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy, T. F., K. C. Dudas, J. M. Mylotte, and M. A. Apicella. 1983. A subtyping system for nontypable Haemophilus influenzae based on outer-membrane proteins. J. Infect. Dis. 147:838-846. [DOI] [PubMed] [Google Scholar]
- 40.Murphy, T. F., and C. Kirkham. 2002. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol. 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murphy, T. F., M. B. Nelson, K. C. Dudas, J. M. Mylotte, and M. A. Apicella. 1985. Identification of a specific epitope of Haemophilus influenzae on a 16,600-dalton outer membrane protein. J. Infect. Dis. 152:1300-1307. [DOI] [PubMed] [Google Scholar]
- 42.Nelson, M. B., M. A. Apicella, T. F. Murphy, H. Vankeulen, L. D. Spotila, and D. Rekosh. 1988. Cloning and sequencing of Haemophilus influenzae outer membrane protein P6. Infect. Immun. 56:128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson, M. B., R. S. Munson, Jr., M. A. Apicella, D. J. Sikkema, J. P. Molleston, and T. F. Murphy. 1991. Molecular conservation of the P6 outer membrane protein among strains of Haemophilus influenzae: analysis of antigenic determinants, gene sequences, and restriction fragment length polymorphisms. Infect. Immun. 59:2658-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parsons, L. M., F. Lin, and J. Orban. 2006. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry 45:2122-2128. [DOI] [PubMed] [Google Scholar]
- 45.Parsons, L. M., and J. Orban. 2005. NMR assignment of the periplasmic domain of peptidoglycan-associated lipoprotein (Pal) from Haemophilus influenzae. J. Biomol. NMR 32:93. [DOI] [PubMed] [Google Scholar]
- 46.Pauwels, R. A., A. S. Buist, P. M. Calverley, C. R. Jenkins, and S. S. Hurd. 2001. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) workshop summary. Am. J. Respir. Crit. Care Med. 163:1256-1276. [DOI] [PubMed] [Google Scholar]
- 47.Poje, G., and R. J. Redfield. 2003. Transformation of Haemophilus influenzae, p. 57-70. In M. Herbert, D. Wood, and E. Moxon (ed.), Haemophilus influenzae protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 48.Sabirov, A., S. Kodama, T. Hirano, M. Suzuki, and G. Mogi. 2001. Intranasal immunization enhances clearance of nontypeable Haemophilus influenzae and reduces stimulation of tumor necrosis factor alpha production in the murine model of otitis media. Infect. Immun. 69:2964-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sen, K., D. J. Sikkema, and T. F. Murphy. 1996. Isolation and characterization of the Haemophilus influenzae tolQ, tolR, tolA and tolB genes. Gene 178:75-81. [DOI] [PubMed] [Google Scholar]
- 50.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000. A state of the art review. Clin. Microbiol. Rev. 14:336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shuto, T., H. Xu, B. Wang, J. Han, H. Kai, X. X. Gu, T. F. Murphy, D. J. Lim, and J. D. Li. 2001. Activation of NF-kappa B by nontypeable Haemophilus influenzae is mediated by Toll-like receptor 2-TAK1-dependent NIK-IKK alpha/beta-I kappa B alpha and MKK3/6-p38 MAP kinase signaling pathways in epithelial cells. Proc. Natl. Acad. Sci. USA 98:8774-8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamanaka, N., and H. Faden. 1993. Antibody response to outer membrane protein of nontypeable Haemophilus influenzae in otitis-prone children. J. Pediatr. 122:212-218. [DOI] [PubMed] [Google Scholar]
- 53.Yang, Y.-P., R. S. Munson, Jr., S. Grass, P. Chong, R. E. Harkness, L. Gisonni, O. James, Y. Kwok, and M. H. Klein. 1998. Effect of lipid modification on the physicochemical, structural, antigenic and immunoprotective properties of Haemophilus influenzae outer membrane protein P6. Vaccine 15:976-987. [DOI] [PubMed] [Google Scholar]