Abstract
Burkholderia pseudomallei is the causative agent of melioidosis and represents a potential bioterrorism threat. In the current studies we have examined gene expression in B. pseudomallei in an animal model of acute melioidosis using whole-genome microarrays. Gene expression profiles were generated by comparing transcriptional levels of B. pseudomallei-expressed genes in infected hamster organs including liver, lung, and spleen following intraperitoneal and intranasal routes of infection to those from bacteria grown in vitro. Differentially expressed genes were similar in infected livers irrespective of the route of infection. Reduced expression of a number of housekeeping genes suggested a lower bacterial growth rate during infection. Energy production during growth in vivo involved specific biochemical pathways such as isomerization of 3-phosphoglycerate, catabolism of d-glucosamine and inositol, and biosynthesis of particular amino acids. In addition, the induction of genes known to be involved in oxidative phosphorylation including ubiquinol oxidase, ferredoxin oxidoreductase, and formate dehydrogenase enzymes suggested the use of alternative pathways for energy production, while the expression of genes coding for ATP-synthase and NADH-dehydrogenase enzymes was reduced. Our studies have identified differentially expressed genes which include potential virulence genes such as those for a putative phospholipase C and a putative two-component regulatory system, and they have also provided a better understanding of bacterial metabolism in response to the host environment during acute melioidosis.
Melioidosis is a major tropical disease prevalent in Southeast Asia and northern Australia (6, 8). This disease was first described in 1912 by Whitmore and Krishnaswami in Rangoon, Myanmar (37). The disease is fatal and is caused by a soil bacterium, Burkholderia pseudomallei. Infection can occur from soil or water contamination of skin abrasions or inhalation of B. pseudomallei from natural sources. Sporadic cases of melioidosis have been reported worldwide (29). These include travelers who have returned from areas of endemicity, especially during the tsunami disaster in Southeast Asian countries in December 2004 (1).
B. pseudomallei is a common cause of human pneumonia and septicemias in areas of endemicity (4). The disease may become dormant, and the infected person may relapse after months, years, or decades (26). In northeastern Thailand, melioidosis accounts for as much as 20% of community-acquired septicemias and causes death in 40% of treated patients (36). At present there are no effective vaccinesagainst B. pseudomallei infection.
Several in vitro studies have demonstrated a better understanding of pathogenesis and virulence of B. pseudomallei in tissue culture models. The bacterium can invade both phagocytic and nonphagocytic cells followed by intracellular multiplication and intercellular spread (15, 16). However, the mechanisms for bacterial survival in host tissues are not fully understood. In addition, a variety of eukaryotic host models for B. pseudomallei infection have been used to study bacterial virulence and to identify virulence determinants. Hosts studied include Caenorhabditis elegans (12, 27) and higher animals (19). The use of animal infection models in combination with bacterial genetics and immunological and antigen purification techniques can teach us much about the pathogenesis of diseases due to microorganisms (38). The hamster model of acute melioidosis was first used to study experimental chemotherapy of melioidosis by Miller and colleagues in 1948 (23), and the model has been used extensively to assess the relative virulence of B. pseudomallei (3, 34). Hamsters are exquisitely sensitive to B. pseudomallei infection and do not display individual variation in susceptibility to B. pseudomallei (10).
In the present studies, we used microarray techniques to compare differential gene expression in B. pseudomallei during infection of Syrian hamsters to gene expression in B. pseudomallei grown in vitro. Our studies have identified potential virulence determinant genes as well as providing interesting information regarding the bacterial metabolic response to the host environment.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. To isolate bacterial RNA from the broth culture, B. pseudomallei 1026b was grown in Luria-Bertani (LB) broth at 37°C with shaking at 250 rpm. The bacterial cells were harvested when the cell growth reached an optical density at 600 nm of 0.9, equivalent to mid-exponential-phase growth.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| B. pseudomallei | ||
| 1026b | Clinical isolate; Gmr Smr Pxr Tps | 11 |
| DD503 | 1026b derivative; Δ(amrR-oprA) rpsL (Smr) AGs Tcs | 24 |
| ATL0378 | DD503 derivative; ΔBPSL0378 Gmr | This study |
| ATL1876 | DD503 derivative; ΔBPSL1876 Gmr | This study |
| ATL2025 | DD503 derivative; ΔBPSL2025 Gmr | This study |
| ATL2515 | DD503 derivative; ΔBPSL2515 Gmr | This study |
| ATL3050 | DD503 derivative; ΔBPSL3050 Gmr | This study |
| ATL3320 | DD503 derivative; ΔBPSL3320 Gmr | This study |
| ATS0067 | DD503 derivative; ΔBPSS0067 Gmr | This study |
| ATS0464 | DD503 derivative; ΔBPSS0464 Gmr | This study |
| ATS1388 | DD503 derivative; ΔBPSS1388 Gmr | This study |
| ATS1489 | DD503 derivative; ΔBPSS1489 Gmr | This study |
| ATS1530 | DD503 derivative; ΔBPSS1530 Gmr | This study |
| ATCPL2025 | ATL2025 carrying pCPL2025; Gmr Tmpr | This study |
| ATCPS0067 | ATS0067 carrying pCPS0067; Gmr Tmpr | This study |
| E. coli | ||
| TOP10 | General cloning and blue-white screening. | Invitrogen |
| SM10λpir | E. coli cloning strain | 30 |
| S17-1 | E. coli cloning strain | 30 |
| Plasmids | ||
| pCR2.1-TOPO | 3.9-kb TA cloning vector; pMB1 oriR Apr Kmr | Invitrogen |
| pGSV3-lux | Mobilizable suicide vector containing lux operon from pSC26; oriT Gmr | 25 |
| pUCP28T | Shuttle vectors derived from pUC18/19; Tmpr | 35 |
| PCPL2025 | (BamHI-XbaI)-pUCP28T::full BPSL2025; Tmpr | This study |
| PCPS0067 | (BamHI-XbaI)-pUCP28T::full BPSS0067; Tmpr | This study |
Animal studies and routes of infection.
Two different routes of infection were used in this study, intraperitoneal (i.p.) and intranasal (i.n.). Syrian hamsters (female, 6 to 8 weeks of age) were used throughout the studies.
(i) Intraperitoneal route of infection.
Five hamsters were injected with a 100-μl suspension of organisms grown in LB. Serial dilutions were made in phosphate-buffered saline to obtain an infective dose 10 times higher than the known 50% lethal dose (LD50) of B. pseudomallei 1026b (11). Serial dilution and plate counts were used to confirm the dose. All hamsters were severely ill at 40 h after the inoculation. At this time, all hamsters were euthanized, and the infected organs were immediately collected. The infected lungs, spleens, and livers were preserved in RNAlater solution (Ambion) at −70°C until required.
(ii) Intranasal route of infection.
Five hamsters were infected with B. pseudomallei 1026b via the intranasal route. After the hamsters were anesthetized with 8 mg of ketamine, a 20-μl suspension of organisms grown in LB was intranasally inoculated into one nostril of each hamster. The infective dose confirmed by serial dilution and plate count was approximately 106 CFU. All hamsters were ill at 40 h after the inoculation. The hamsters were euthanized, and the infected organs including the livers, lungs, and spleens were collected as described for the intraperitoneal inoculation experiments and preserved in RNAlater solution (Ambion) at −70°C until required.
RNA isolation. (i) Bacterial total RNA from broth culture.
Bacterial cells were harvested from 20 ml broth culture in LB at an optical density at 600 nm of 0.9. Prior to harvest of the cells, 1% (vol/vol) of RNAlater was added to the culture and maintained at 4°C. The bacterial cells were harvested by centrifugation at 8,000 × g at 4°C for 5 min. The cell pellets were treated with 1 ml of Trizol solution (Invitrogen), mixed well, and maintained at room temperature for 5 min. Two hundred microliters of chloroform was added and mixed for 20 seconds. The mixture was maintained at room temperature for 10 min. The tube was centrifuged at 10,000 × g at 4°C for 15 min, and the aqueous phase was transferred to a fresh tube containing 1 ml of isopropanol, mixed well, and maintained at room temperature for 10 min. The tube was centrifuged at 10,000 × g at 4°C for 15 min. At this step, the total RNA pellet was precipitated. The supernatant was carefully discarded, and the pellet was washed with 1 ml of 75% ethanol by centrifugation at 10,000 × g at 4°C for 5 min. The RNA pellet was dried at room temperature. The pellet was resuspended in 30 μl of the RNase-free distilled water and treated with DNase I (DNA-free; Ambion Inc.). RNA concentration was quantified by spectrophotometry, and samples were electrophoresed in a formaldehyde agarose gel to assess the quality of RNA.
(ii) Bacterial total RNA from infected tissues.
The infected hamsters' organs preserved in RNAlater were thawed at room temperature. RNA isolation from each organ was performed independently using the same protocol. Briefly, the RNAlater solution was removed from the container and then 2, 3, and 5 ml of Trizol were added to the samples of spleen, lung, and liver, respectively. Samples were homogenized with a Brinkman Polytron homogenizer, and the homogenized samples were maintained at room temperature for 5 min. The samples were divided, and 1 ml each was placed into microcentrifuge tubes. To each tube was added 200 μl of chloroform, and tubes were maintained at room temperature for 10 min. The tubes were centrifuged at 10,000 × g at 4°C for 15 min. After centrifugation, the aqueous phase was transferred to a fresh tube containing 1 ml of isopropanol, mixed well, and maintained at room temperature for 10 min. The following steps were preceded by the same procedure as that for the bacterial total RNA isolation described above, but the product from this step was a mixture of total RNA from eukaryotic cells and bacterial cells. After removal of contaminating DNA with DNase I, the total bacterial RNA was purified from the mixture using MicrobEnrich (Ambion) per the manufacturer's instructions. Briefly, 100 μg (volume no larger than 30 μl) of the mixed RNA was combined with 300 μl of binding buffer and gently mixed. Forty microliters of the Capture Oligo Mix (containing oligonucleotides specific for eukaryotic RNA) was added to the mixture and then heated at 70°C for 10 min. This heat denaturation helped to facilitate maximal hybridization of the capture oligonucleotide to the mammalian RNA. The mixture was then incubated at 37°C for 1 h. In the meantime, the Oligo Magbeads (containing sequences for specific binding to the Capture Oligo Mix and conjugated with magnetic beads) was prepared by being washed with nuclease-free water and resuspended in the binding buffer using 500 μl of Oligo Magbeads per 100 μg of input RNA. After incubation, the RNA mixture and the Oligo Magbeads were combined and further incubated at 37°C for 10 min. The Oligo Magbeads containing the eukaryotic RNA were captured to one side of the tube's inner wall using the magnetic stand. The supernatant containing the total bacterial RNA was transferred to a fresh tube, further precipitated in ethanol, and centrifuged. The total bacterial RNA was resuspended in nuclease-free water. The total bacterial RNA purified from this technique yielded approximately 10% by weight of the input RNA.
(iii) Bacterial mRNA purification.
Bacterial mRNA purification was used to purify the bacterial mRNA from the total bacterial RNA from both sources described above using MICROBExpress (Ambion Inc.) per the manufacturer's instructions. Briefly, 2 to 10 μg (volume no larger than 15 μl) of the bacterial total RNA was mixed with 200 μl of binding buffer. Four microliters of Capture Oligo mix was added to the mixture and incubated at 70°C for 10 min. This heat denaturation helped to facilitate maximal hybridization of the rRNA to the capture oligonucleotides. The tube was incubated further at 37°C for 15 min. In the meantime, 50 μl of Oligo Magbeads (containing sequences for specific binding to the capture oligonucleotides and conjugated with magnetic beads) was prepared by being washed with nuclease-free water, resuspended with the binding buffer, and incubated at 37°C until required. Fifty microliters of the prepared Oligo Magbeads was added to the mixture and incubated further at 37°C for 15 min. This step allowed the hybridization of Oligo Magbeads and allowed the capture oligonucleotide with rRNA to be formed. The Oligo Magbeads containing the rRNA were captured to one side of the tube's inner wall using the magnetic stand. The supernatant containing the purified bacterial mRNA was transferred to a fresh tube, further precipitated in ethanol, and centrifuged. The mRNA was resuspended in nuclease-free water. The mRNA purified from this technique yielded approximately 10% and 80% by weight of the input total RNA previously isolated from LB culture and from the MicrobEnrich purified total RNA of the infected organs, respectively.
Examination of bacterial RNA by a quantitative RT-PCR.
All bacterial total RNA and mRNA obtained after purification were analyzed by reverse transcription-PCR (RT-PCR) to quantify the presence of the 16S and 23S rRNA. One-step RT-PCR was used as previously described (33). PCR primers for the 16S and 23S rRNA are shown in Table 2.
TABLE 2.
PCR primers used in this study
| Gene | Primer name | Nucleotide sequence, 5′ to 3′ | Purpose |
|---|---|---|---|
| BPSL0378 | L0378-for | CAAAGGCTCCCGATGAGCGA | Internal PCR |
| L0378-rev | AAATGTCGGCATCGAGCGCC | Internal PCR | |
| Up-L0378-for | GAAATTCGTGTCGAGCTGCG | Merodiploid confirmation | |
| BPSL1876 | L1876-for | GGTCGGCGTCTACCGGAT | Internal PCR |
| L1876-rev | TCGAGGAACGTGAACGCGGG | Internal PCR | |
| Up-L1876-for | CCCAACGGAGTCCGCTCGT | Merodiploid confirmation | |
| BPSL2025 | L2025-for | ACCGCGACCTGAAGCGGCT | Internal PCR |
| L2025-rev | GATCGACACGACCCGCCC | Internal PCR | |
| Up-L2025-for | TCGCGTTCCTGACGCTGCC | Merodiploid confirmation | |
| CPL2025-for | CCATAAGCGGAACGCCGCAG | Complementation of ΔBPSL2025 | |
| CPL2025-rev | TCGATACGTCGCCCATGCCC | Complementation of ΔBPSL2025 | |
| BPSL2515 | L2515-for | ATCAGGGCGAGGTTGAGGTT | Internal PCR |
| L2515-rev | ATGCGATGTCGGTGATGTG | Internal PCR | |
| Up-L2515-for | CGAACAGGTGCTCGCGTG | Merodiploid confirmation | |
| BPSL3050 | L3050-for | CCAACGAGGGCTACAACCGC | Internal PCR |
| L3050-rev | GCACGCTCGCCGACGTGA | Internal PCR | |
| Up-L3050-for | AATTCGGCAGGATCAGCG | Merodiploid confirmation | |
| BPSL3320 | L3320-for | GCCCTCGACGGGCAATTCGT | Internal PCR |
| L3320-rev | TTGCGACCATGCGACGCTGC | Internal PCR | |
| Up-L3320-for | GTCGACTTCGGCAACCAGCG | Merodiploid confirmation | |
| BPSS0067 | S0067-for | GATACCCGCCTTCGTCATCT | Internal PCR |
| S0067-rev | CAGCGTGCCGAAGTAGTTG | Internal PCR | |
| Up-S0067-for | GATACCCGCCTTCGTCATCT | Merodiploid confirmation | |
| CPS0067-for | CTCGATGTGCAACGCCGCC | Complementation of ΔBPSS0067 | |
| CPS0067-rev | CCATTGTCGTGCGCGTCAGG | Complementation of ΔBPSS0067 | |
| BPSS0464 | S0464-for | GGTCAGGCTTCTCGCTGAAG | Internal PCR |
| S0464-rev | TGATGAAGAGCGTCGCGAGC | Internal PCR | |
| Up-S0464-for | GGCTATCCGATGGCGTACTA | Merodiploid confirmation | |
| BPSS1388 | S1388-for | GCGAATCTCGCGAAAGCGGA | Internal PCR |
| S1388-rev | CCAGTCTCTTGGCGACCTTC | Internal PCR | |
| Up-S1388-for | TGTTCCGGCAACTCGAAACG | Merodiploid confirmation | |
| BPSS1489 | S1489-for | GCGACGGCAACCGATTCGCT | Internal PCR |
| S1489-rev | ACTGAATGCCGAGCAGCGCG | Internal PCR | |
| Up- S1489-for | GCGACGGCAACCGATTCGCT | Merodiploid confirmation | |
| BPSS1530 | S1530-for | AAGGAACTGAAGGCTCGGGC | Internal PCR |
| S1530-rev | TCCATTTCGGCTCGATGCCG | Internal PCR | |
| Up-S1530-for | AAGAACCAGATCGACGGCGC | Merodiploid confirmation | |
| luxC | luxC-rev | ACGAATGTATGTCCTGCGTCTTGAG | pGSV3-lux fusion and merodiploid confirmation |
| 16S rRNA | 16S rRNA-for | TGGCTAATACCCGGAGTGGA | RT-PCR |
| 16S rRNA-rev | CGGTTTGTCACCGGCAGTCT | RT-PCR | |
| 23S rRNA | 23S rRNA-for | GCCAGCTAAGGTCCCCAAAT | RT-PCR |
| 23S rRNA-rev | TGCAGGTCGGAACTTACCCG | RT-PCR |
Microarray analysis.
Microarray analyses were performed as previously described (33) with minor modifications. Briefly, equal amounts of 3 to 5 μg of bacterial mRNA obtained from both sources (LB culture and infected organ) were annealed with 10 μg of the random hexamer pd(N)6 (Amersham Bioscience) by incubation at 70°C for 10 min and maintained on ice. The cDNA was generated in a reverse transcription reaction using 100 U of StrataScript reverse transcriptase, 2 μl of 10× StrataScript reaction buffer, 1 μl of 20× deoxynucleoside triphosphate mix with an amino-allyl-dUTP, 1.5 μl of 0.1 M dithiothreitol, and 0.5 μl of 40 U μl−1 of RNase block (Stratagene Inc.). The reaction mixture was incubated at 48°C for 1 h 30 min. The amino-allyl cDNAs from both samples were labeled differently with 5 μl of Cy3 or Cy5 (Cy3-, Cy5-monofunctional dyes; Amersham Bioscience) at room temperature for 30 min in the dark. The uncoupled dye was removed from the fluorescent dye-labeled cDNA by using a microspin cup and DNA binding solution (Stratagene Inc.), and the volume wasreduced to approximately 3 μl by vacuum centrifugation. Each Cy3- and Cy5-labeled cDNA was diluted with hybridization solution containing 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate. Both hybridization solutions were combined and mixed carefully by pipetting and then heated at 95°C for 3 min. Volumes of the hybridization were 50 μl. The hybridization solution was left to cool at room temperature and mixed thoroughly by centrifugation. The hybridization solution was applied onto the microarray and covered with a plastic coverslip. The slide was incubated at 50°C in a slide hybridization chamber (Boekel Scientific) for 18 h. Following incubation, the slide was washed with four solutions: solution 1, 4× SSC, 0.2% (wt/vol) sodium dodecyl sulfate for 5 min at 50°C, twice; solution 2, 0.2× SSC for 1 min; and solution 3, 0.1× SSC for 1 min each at room temperature. The slide was spin dried in a centrifuge at 550 rpm for 5 min and scanned immediately. Scanning was performed with a Virtek ChipReader (Virtek Biotech, Canada). The fluorescence signals from both dyes were quantified using QuantArray microarray analysis software (Packard Bioscience). Normalization and data analysis were performed with GeneTraffic software (Iobion Informatics). Data obtained from at least 24 hybridizations of two independent RNA preparations of each type of infected organ including flip-dye replications were used in each analysis. Normalization of data was performed on each hybridization using LOWESS (Locally Weighted Scatter Plot Smoother), a curve-fitting algorithm.
Calculation and statistical analysis.
To select significantly expressed genes from the whole-genome microarray data, basic calculation and statistics including differences (n-fold), mean log2 ratio, standard deviation (SD), coefficient of variance (COV), and Student's t test were used in the analysis. Hierarchical clustering and K-mean clustering analyses were performed using TIGR-MeV software (www.tigr.org).
Microarray data repository.
Raw microarray data from this study are available at ftp://136.159.173.80/. Further information is available upon request.
Construction of insertion mutants.
Insertion mutations were constructed in 11 up-regulated genes including BPSL0378 (MarR family regulatory protein), BPSL1876 (putative phospholipase), BPSL2025 (putative two-component regulatory system), BPSL2515 (rpsA; 30S ribosomal protein S1), BPSL3320 (fliD; flagellar hook-associated protein), BPSL3050 (trpE; anthranilate synthase component I), BPSS0067 (putative phospholipase C precursor), BPSS0464 (potI, putrescine transport system permease protein), BPSS1388 (putative membrane protein), BPSS1489 (putative exported protein), and BPSS1530 (bprA, putative H-NS-like regulatory protein).
A previously described lux-based suicide vector, pGSV3-lux, was used in this study (25). Briefly, an internal region of approximately 300 to 600 bp of those 11 selected genes was amplified by PCR. The PCR products were cloned with pCR2.1-TOPO (Invitrogen). The recombinant plasmid was digested with EcoRI, and a small fragment (the insert) was purified and ligated into EcoRI-digested, calf intestinal alkaline phosphatase-treated pGSV3-lux. The recombinant plasmids were transformed into Escherichia coli SM10λpir or S17-1 (30). The transformed E. coli containing the recombinant pGSV3-lux plasmid was conjugated with B. pseudomallei DD503 (24), and transconjugants were selected on LB-gentamicin-polymyxin Bagar. The transconjugants were screened for lux-mediated light production by assaying 100 μl of overnight broth cultures of individual colonies. Cultures which displayed light production were then tested for insertion of pGSV3-lux into the target gene using PCR. B. pseudomallei strains ATL0378, ATL1876, ATL2025, ATL2515, ATL3320, ATL3050, ATS0067, ATS0464, ATS1388, ATS1489, and ATS1530 were insertion mutant derivatives based on genes BPSL0378, BPSL1876, BPSL2025, BPSL2515, BPSL3320, BPSL3050, BPSS0067, BPSS0464, BPSS1388, BPSS1489, and BPSS1530, respectively.
Virulence assessment of mutant strains.
The animal model of acute melioidosis has been previously described (10). Syrian hamsters (females, 6 to 8 weeks old) were intraperitoneally challenged with a 100-μl suspension of a diluted mid-exponential-phase culture of the 11 B. pseudomallei mutant strains containing 100 CFU (10 times higher than LD50 of DD503) of bacteria. The number of dead animals in each group (five per group) was determined after 48 h. LD50 evaluation was performed only for the mutant strains which demonstrated less virulence at this infective dose. The LD50 evaluation was performed with a number of serial dilutions corresponding to 101 to 105 CFU of the mutant strains, and the LD50 values were calculated using the Reed and Muench method (28). All animals used in these studies were cared for and used humanely according to the Guide to the Care and Use of Experimental Animals published by the Canadian Council on Animal Care.
Complementation of ΔBPSL2025 and ΔBPSS0067 mutants.
DNA fragments of full BPSL2025 or BPSS0067 genes including the upstream promoter region were amplified from B. pseudomallei 1026b genomic DNA by PCR using the primers listed in Table 2. Both PCR products were cloned as a BamHI-XbaI fragment into pUCP28T (35) to generate recombinant plasmids, pCPL2025 and pCPS0067. E. coli SM10λpir containing recombinant plasmid pCPL2025 or pCPS0067 was conjugated to B. pseudomallei ATL2025 or ATS0067, respectively, and Tmpr colonies were screened by plasmid DNA isolation to confirm the presence of pCPL2025 and pCPS0067 in the mutant strains.
RESULTS
Comparative gene expression profiles of in vivo- and in vitro-grown B. pseudomallei.
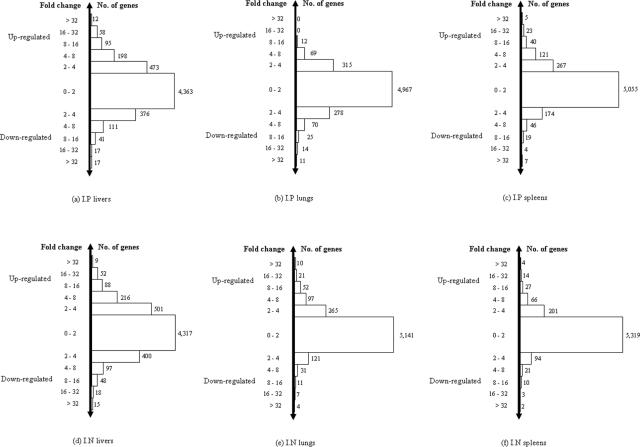
Syrian hamsters were infected with B. pseudomallei 1026b intraperitoneally and intranasally. Gene expression profiles were obtained from a comparison of transcriptional levels of expressed genes observed in each infected organ including liver, lung, and spleen with those from bacterial growth at mid-exponential phase in LB medium. To select differentially expressed genes, we initially grouped genes according to their expression levels as seen in Fig. 1. We then applied a COV value of ≤0.25 to the group of genes which had low expression levels, i.e., levels between twofold and less-than-fourfold different (1 ≤ log2 ratio < 2 or −2 < log2 ratio ≤ −1), and a COV value of ≤0.5 to the groups containing high expression levels, i.e., levels at least fourfold different (2 ≤ log2 ratio ≤ −2). Subsequently, gene lists from those two groups were combined and defined as differentially expressed genes. The SD of the selected genes did not exceed 25% and 50% of the mean log2 ratio from those low- and high-relative-expression groups, respectively. This ensured that the selected genes were chosen from consistently expressed genes, and the minimum difference defined as significant was 1.68-fold (log2 ratio = 0.75). Thus, 429, 83, and 128 genes were differentially expressed in liver, lung, and spleen, respectively, in i.p. infected animals. In i.n. infected animals, 581, 136, and 58 genes were differentially expressed from liver, lung, and spleen, respectively. A total of six organ infection models (i.p. infected liver, i.p. infected lung, i.p. infected spleen, i.n. infected liver, i.n. infected lung, and i.n. infected spleen) were generated from this study; gene lists and details are shown in Tables 1 to 6 posted at ftp://136.159.173.80/. We noted that the highest numbers of differentially expressed genes in animals infected intraperitoneally or intranasally were seen in infected liver.
FIG. 1.
Genes grouped according to expression levels (changes [n-fold]) from six organ infection models (i.p. infected liver, i.p. infected lung, i.p. infected spleen, i.n. infected liver, i.n. infected lung, and i.n. infected spleen). The highest numbers of up- or down-regulated genes were from infected livers (a and d) for both routes of infection.
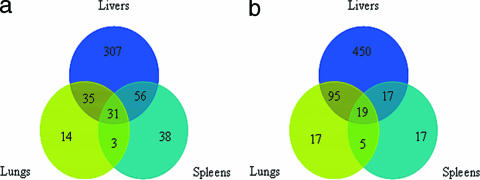
We searched for common genes among those six organ infection models using two different criteria: (i) common genes and routes of infection and (ii) common genes and types of infected organs.
(i) Common genes and routes of infection.
Venn diagrams in Fig. 2 display the numbers of genes which were differentially expressed and shared (common) among the infected organs for each route of infection. There were 31 and 19 common genes from i.p. and i.n. infected animals, respectively. These common genes are listed and detailed in Tables 3 and 4. Interestingly, there were four common genes differentially expressed in all six organ infection models. These included BPSL3050 (trpE, anthranilate synthase component 1), BPSL2902 (putative phosphoglycerate mutase), BPSL2082 (conserved hypothetical protein), and BPSL0317 (conserved hypothetical protein). All four genes were highly up-regulated in all organs.
FIG. 2.
Venn diagrams showing numbers of common genes differentially expressed in each organ and associated with routes of infection, intraperitoneal (a) and intranasal (b). These genes were selected using the COV applied to an individual infected organ (total of two organs).
TABLE 3.
List of 31 common genes of B. pseudomallei differentially expressed in all three organ infection models following i.p. inoculation
| Gene | Annotation | Expression level for organ and preparation no.
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver
|
Lung
|
Spleen
|
|||||||||||
| 1
|
2
|
1
|
2
|
1
|
2
|
||||||||
| Meana | SD | Meana | SD | Meana | SD | Meana | SD | Meana | SD | Meana | SD | ||
| BPSL0004 | hupA, DNA-binding protein HU-alpha | −4.22 | 0.46 | −5.86 | 0.66 | −6.68 | 1.57 | −4.15 | 0.90 | −5.68 | 1.70 | −5.04 | 0.67 |
| BPSL0060 | AraC family transcriptional regulator | 2.49 | 0.65 | 3.36 | 0.36 | 2.62 | 0.48 | 1.07 | 0.24 | 2.13 | 0.24 | 2.81 | 0.84 |
| BPSL0117 | LysR family transcription regulatory protein | 3.84 | 1.05 | 5.50 | 0.77 | 2.89 | 0.89 | 3.17 | 1.06 | 4.85 | 2.22 | 4.23 | 0.92 |
| BPSL0317 | Conserved hypothetical protein | 3.80 | 0.36 | 3.87 | 0.61 | 4.42 | 0.52 | 2.53 | 0.41 | 3.22 | 1.54 | 4.66 | 0.74 |
| BPSL0379 | Putative short-chain dehydrogenase | −4.11 | 1.03 | −3.93 | 0.54 | −3.66 | 0.53 | −2.91 | 0.96 | −2.87 | 0.49 | −2.18 | 0.82 |
| BPSL0430 | Putative thioesterase | 5.14 | 0.54 | 4.41 | 0.33 | 3.08 | 0.53 | 2.25 | 1.11 | 1.93 | 0.38 | 3.78 | 0.48 |
| BPSL0522 | Putative membrane protein | −8.78 | 1.06 | −5.96 | 0.59 | −8.49 | 1.30 | −4.12 | 0.45 | −6.34 | 2.93 | −5.52 | 0.87 |
| BPSL0635 | Conserved hypothetical protein | −5.19 | 1.05 | −4.97 | 0.81 | −3.11 | 0.83 | −5.49 | 1.47 | −3.25 | 0.34 | −2.69 | 0.98 |
| BPSL1058 | Putative transposase | −9.40 | 1.03 | −7.52 | 0.37 | −6.93 | 0.64 | −8.24 | 2.30 | −7.34 | 0.42 | −6.02 | 0.56 |
| BPSL1119 | Putative lipopolysaccharide biosynthesis-related protein | 4.62 | 0.46 | 5.37 | 0.23 | 3.45 | 0.76 | 3.04 | 1.39 | 4.96 | 0.93 | 4.70 | 1.27 |
| BPSL1356 | ftsH, cell division protein | −3.11 | 0.34 | −3.40 | 0.37 | −2.85 | 0.20 | −3.09 | 0.63 | −2.73 | 0.12 | −4.07 | 0.76 |
| BPSL1392 | Hypothetical protein | 3.63 | 1.43 | 5.59 | 0.62 | 3.05 | 0.57 | 1.88 | 0.20 | 2.78 | 1.10 | 5.40 | 1.74 |
| BPSL1592A | Conserved hypothetical protein | −5.57 | 0.25 | −4.79 | 0.29 | −4.43 | 0.75 | −4.77 | 0.99 | −4.00 | 0.45 | −3.58 | 0.37 |
| BPSL1836 | cysA, sulfate transport ATP-binding protein | 4.20 | 0.49 | 4.58 | 0.45 | 3.56 | 0.08 | 1.78 | 0.38 | 3.45 | 0.81 | 4.65 | 0.52 |
| BPSL2040 | Hypothetical protein | 4.32 | 0.78 | 5.31 | 0.96 | 4.41 | 0.77 | 2.91 | 0.75 | 3.60 | 0.92 | 5.82 | 0.90 |
| BPSL2082 | Conserved hypothetical protein | 4.34 | 1.72 | 6.26 | 0.95 | 4.72 | 0.87 | 2.40 | 0.48 | 5.18 | 0.73 | 5.74 | 1.57 |
| BPSL2219 | Putative phosphoserine aminotransferase | −5.40 | 1.15 | −3.40 | 0.28 | −3.28 | 0.70 | −3.76 | 0.26 | −3.89 | 1.30 | −2.36 | 0.48 |
| BPSL2557 | cspA, cold shock-like protein | −6.15 | 2.79 | −8.83 | 0.57 | −7.20 | 1.88 | −5.22 | 1.86 | −5.62 | 1.12 | −5.11 | 0.79 |
| BPSL2881 | Putative membrane protein | −6.99 | 0.26 | −4.74 | 1.10 | −4.39 | 1.15 | −4.86 | 1.52 | −3.44 | 0.83 | −1.83 | 0.24 |
| BPSL2902 | Putative phosphoglycerate mutase | 4.19 | 0.93 | 5.17 | 0.38 | 2.73 | 0.96 | 2.20 | 0.63 | 3.25 | 0.43 | 4.31 | 0.72 |
| BPSL3027 | murD, UDP-N-acetylmuramoylalanine-d-glutamate ligase | 5.03 | 0.63 | 4.81 | 0.38 | 2.48 | 1.12 | 3.27 | 1.62 | 2.97 | 0.55 | 2.59 | 0.38 |
| BPSL3050 | trpE, anthranilate synthase component 1 | 3.94 | 1.93 | 5.42 | 0.33 | 2.72 | 0.81 | 2.24 | 1.02 | 2.49 | 0.70 | 3.55 | 1.26 |
| BPSL3085 | Conserved hypothetical protein | −3.46 | 0.29 | −2.87 | 0.16 | −2.56 | 0.60 | −3.62 | 1.12 | −2.60 | 0.67 | −2.07 | 0.58 |
| BPSL3253 | Putative oxidoreductase | 4.83 | 1.18 | 4.82 | 0.58 | 3.05 | 0.92 | 2.90 | 1.05 | 3.56 | 1.18 | 2.15 | 0.52 |
| BPSS0515 | Hypothetical protein | −6.63 | 0.6 | −5.21 | 0.90 | −5.35 | 2.04 | −6.03 | 1.52 | −4.40 | 1.62 | −3.02 | 0.17 |
| BPSS0965 | Putative oxalate decarboxylase | 3.81 | 0.48 | 5.93 | 0.20 | 3.47 | 0.21 | 2.25 | 1.04 | 3.93 | 0.15 | 3.96 | 1.35 |
| BPSS1199 | Hypothetical protein | −6.11 | 1.73 | −7.50 | 0.63 | −6.96 | 1.95 | −4.64 | 2.01 | −6.18 | 1.83 | −4.96 | 0.1 |
| BPSS1388 | Putative membrane protein | 5.56 | 1.53 | 7.86 | 1.05 | 5.40 | 1.33 | 2.34 | 0.65 | 4.59 | 0.44 | 5.14 | 1.36 |
| BPSS1489 | Putative exported protein | 5.94 | 0.53 | 7.45 | 0.37 | 3.97 | 0.40 | 2.68 | 1.34 | 3.64 | 0.13 | 4.53 | 1.17 |
| BPSS1933 | Putative membrane protein | −6.43 | 0.58 | −4.67 | 0.54 | −4.05 | 0.64 | −4.63 | 2.07 | −4.51 | 0.38 | −3.82 | 0.36 |
| BPSS2229 | Conserved hypothetical protein | 2.90 | 1.08 | 3.45 | 0.27 | 2.40 | 1.04 | 2.22 | 0.43 | 2.02 | 0.44 | 3.25 | 1.11 |
Mean value is mean log2 ratio (in vivo/in vitro) calculated from two replicates.
TABLE 4.
List of 19 common genes of B. pseudomallei differentially expressed in all three organ infection models following i.n. inoculation
| Gene | Annotation | Expression level for organ and preparation no.
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Liver
|
Lung
|
Spleen
|
|||||||||||
| 1
|
2
|
1
|
2
|
1
|
2
|
||||||||
| Meana | SD | Meana | SD | Meana | SD | Meana | SD | Meana | SD | Meana | SD | ||
| BPSL0308 | Putative exported protein | 3.58 | 0.10 | 3.71 | 0.23 | 3.20 | 0.37 | 2.41 | 0.56 | 3.09 | 1.26 | 2.88 | 1.27 |
| BPSL0317 | Conserved hypothetical protein | 3.37 | 0.64 | 3.95 | 0.43 | 2.56 | 0.51 | 4.51 | 0.57 | 3.16 | 1.54 | 3.38 | 1.29 |
| BPSL0500 | Putative chitobiase | 2.70 | 0.65 | 3.52 | 0.56 | 3.01 | 0.95 | 2.04 | 0.76 | 2.76 | 1.11 | 2.70 | 1.12 |
| BPSL0548 | Hypothetical protein | 4.43 | 0.38 | 4.46 | 1.23 | 2.44 | 0.86 | 3.98 | 1.17 | 3.49 | 0.35 | 3.43 | 0.50 |
| BPSL0959 | cysD, putative sulfate adenylyltransferase subunit 2 | 4.76 | 0.52 | 5.97 | 0.81 | 3.98 | 0.66 | 3.58 | 1.47 | 3.14 | 1.21 | 3.42 | 0.78 |
| BPSL2082 | Conserved hypothetical protein | 4.29 | 0.38 | 5.32 | 0.69 | 5.79 | 0.64 | 4.64 | 1.27 | 3.19 | 0.92 | 3.12 | 1.55 |
| BPSL2696 | Putative kinase | 5.30 | 0.47 | 6.78 | 0.66 | 5.15 | 0.62 | 6.62 | 2.33 | 4.57 | 1.51 | 4.80 | 1.62 |
| BPSL2756 | Putative exported protein | 2.74 | 0.20 | 3.17 | 0.47 | 3.30 | 0.40 | 2.91 | 0.47 | 2.65 | 1.14 | 2.67 | 1.26 |
| BPSL2786 | Putative acetyltransferase protein | 4.07 | 0.59 | 5.49 | 0.19 | 3.86 | 0.56 | 2.89 | 0.44 | 2.39 | 0.58 | 2.63 | 0.59 |
| BPSL2859 | Conserved hypothetical protein | 2.78 | 0.38 | 3.76 | 0.59 | 2.13 | 0.49 | 4.16 | 1.08 | 2.90 | 0.93 | 3.07 | 0.66 |
| BPSL2902 | Putative phosphoglycerate mutase | 3.73 | 0.75 | 4.73 | 0.27 | 2.61 | 0.64 | 3.12 | 1.17 | 4.88 | 2.06 | 5.04 | 1.65 |
| BPSL3050 | trpE, anthranilate synthase component 1 | 3.29 | 0.46 | 4.57 | 0.35 | 2.22 | 0.67 | 2.42 | 0.81 | 2.20 | 0.47 | 2.29 | 0.75 |
| BPSS0163 | asnO gene, asparagine synthetase (glutamine hydrolyzing) | 4.40 | 0.36 | 4.89 | 0.78 | 2.43 | 0.23 | 2.84 | 0.96 | 2.50 | 0.72 | 2.45 | 0.76 |
| BPSS0204 | tsr gene, methyl-accepting chemotaxis protein I | 3.53 | 0.86 | 4.73 | 0.27 | 3.47 | 0.82 | 2.74 | 1.06 | 3.65 | 1.29 | 3.79 | 0.84 |
| BPSS0464 | potI gene, putrescine transport system permease protein | 4.69 | 0.51 | 5.21 | 1.09 | 3.58 | 1.32 | 3.41 | 1.10 | 5.19 | 0.91 | 4.58 | 0.66 |
| BPSS0622 | Enoyl-CoA hydratase/isomerase family | 3.56 | 0.43 | 3.55 | 0.43 | 3.19 | 0.71 | 4.12 | 0.80 | 3.58 | 0.73 | 3.66 | 0.84 |
| BPSS0834 | Hypothetical protein | 3.83 | 0.65 | 4.63 | 0.37 | 3.05 | 0.93 | 4.40 | 0.77 | 2.86 | 0.76 | 3.06 | 0.64 |
| BPSS0909 | OmpA family protein | 3.79 | 0.43 | 5.06 | 0.73 | 4.94 | 0.42 | 6.02 | 1.40 | 2.47 | 1.12 | 2.65 | 0.81 |
| BPSS0925 | Short-chain dehydrogenase | 3.03 | 0.28 | 3.70 | 0.26 | 4.25 | 0.83 | 4.75 | 1.13 | 3.36 | 1.25 | 3.67 | 1.18 |
Mean value is mean log2 ratio (in vivo/in vitro) calculated from two replicates.
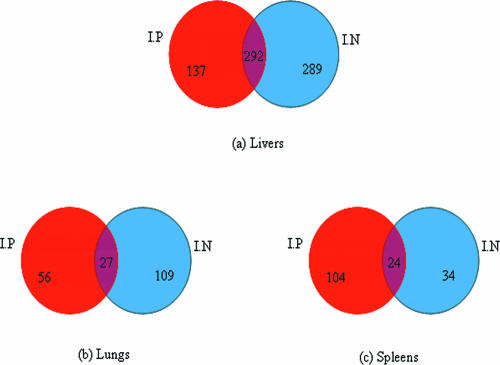
(ii) Common genes and types of infected organs.
There were 292, 27, and 24 genes differentially expressed and shared (common) between i.p. and i.n. infected animals from the liver, lung, and spleen, respectively (Fig. 3). The Student t test was used to evaluate whether the relative transcriptional levels (mean log2 ratio) of the common genes were significantly different. The analysis revealed that the transcriptional levels of genes differentially expressed and shared between the i.p. and i.n. infection models were not significantly different (P > 0.05). We noted that the proportion of the shared genes among i.p. and i.n. infections was significantly low as seen in the lungs and spleens. This would suggest different gene expression profiles between i.p. and i.n. infections especially in the lungs and spleens. However, the common genes found between two different routes of the infection would demonstrate a similar status of bacterial response during the infection. A list of the common genes between i.p. and i.n. in each group of the organ types is found in Tables 7 to 9 posted at ftp://136.159.173.80/.
FIG. 3.
Venn diagrams showing numbers of common genes differentially expressed and associated with infected organ types, livers (a), lungs (b), and spleens (c), for i.p. and i.n. routes of infection. Transcriptional levels of the common genes between i.p. and i.n. routes of each type of the organs were not significantly different.
Bacterial growth and adaptation to the host environment during infection. (i) Determination of bacterial growth status during infection.
Microarray data revealed significant differences in gene expression between B. pseudomallei grown in vivo and that grown in vitro. One major difference was a reduced expression of genes known to be involved in bacterial growth and multiplication. These included genes coding for ribosomal proteins (BPSL3206, BPSL2911, etc.) and a cell division protein (ftsH, BPSL1356). This was clearly seen in data obtained from most organ infection models except the i.n. lungs and i.n. spleens where the ribosomal protein and cell division protein genes were not differentially expressed. The overall gene expression profile for ribosomal protein genes is shown in Fig. 1 posted at ftp://136.159.173.80/. Interestingly, rpsA (BPSL2515, 30S ribosomal protein S1), a housekeeping gene, was highly up-regulated, especially in infected liver. In addition, we also found that an equal amount of total bacterial RNA purified from all tested organs contained less rRNA than that isolated from broth culture. This observation was confirmed by quantitative RT-PCR of the 16S and 23S rRNA genes from both sources of the total RNA samples. The results clearly showed a higher quantity of PCR products of those two target genes from the total RNA isolated from the broth culture relative to infected tissues (see Fig. 5 posted at ftp://136.159.173.80/). Thus, we used purified bacterial mRNA instead of total RNA for microarray analyses in our studies.
(ii) Biochemical pathways.
Gene expression profiles revealed that a number of genes for various metabolic enzymes were differentially expressed between in vivo- and in vitro-grown B. pseudomallei. We used information available for B. pseudomallei biochemical pathways from GenomeNet at The Bioinformatics Centre, Kyoto University, Japan (http://www.genome.jp), together with genome annotation information (13) as a guide to interpret our microarray data. We observed that most genes coding for proteins involved in carbohydrate metabolism including several biochemical pathways such as glycolysis, citrate cycle (tricarboxylic acid [TCA] cycle), pentose phosphate pathway, etc., were not differentially expressed, except for a few genes which were highly up- or down-regulated. The highly up-regulated genes were those coding for proteins known or predicted to be involved in (i) an intermediate step of the glycolytic pathway (or known as the first substrate-level intermediate such as a phosphoglycerate mutase [BPSL2902]), (ii) inositol catabolism (BPSL1996 and BPSL1997), and (iii) glucosamine catabolism such as the glmS3 gene (BPSS2009, glucosamine-fructose-6-phosphate aminotransferase). In contrast, the highly down-regulated genes were genes coding for proteins associated with the TCA cycle such as BPSL0373 (putative isocitrate dehydrogenase), BPSL2469 (fumC, fumarate hydratase), BPSL0780 (sucD, succinyl coenzyme A [CoA] ligase), BPSS1916 (phbB, acetoacetyl-CoA reductase), etc. These down-regulated genes were mostly found in the infected livers from both i.p. and i.n. infected animals.
Moreover, genes coding for proteins involved in catabolism and synthesis of particular amino acids were highly differentially expressed, while most genes were not differentially expressed. The highly up-regulated genes were genes BPSL3050 (trpE, anthranilate synthase component 1), BPSS0163 (asnO, asparagine synthase), BPSL2260 (putative tryptophanyl-tRNA synthase), and BPSS2009 (glmS3, glucosamine-fructose-6-phosphate aminotransferase), etc. The down-regulated genes were genes coding for proteins associated with serine catabolism/synthesis including BPSS0547 (glyA, serine hydroxymethyl-transferase) and BPSL2219 (putative phosphoserine aminotransferase). These differentially expressed genes were mostly found in infected livers and i.n. infected lungs.
In addition to the metabolic genes described above, we also observed that most genes coding for proteins commonly involved in energy metabolism such as ATP-synthase (BPSL3401, BPSS1953, etc.) and NADH-dehydrogenase enzymes (BPSL1223, BPSL1224, etc.) were down-regulated. However, oxidative phosphorylation genes coding for enzymes involved in metabolism of alternative energy sources such as the genes cyoA (BPSS1897) and cyoB (BPSS1896), coding for ubiquinol oxidase; BPSS0555, coding for ferredoxin oxidoreductase; and BPSL2528 (fdsG), coding for NAD-dependent formate dehydrogenase, were highly induced. This provided evidence for the use of alternative pathways for energy during slower growth in infected organs. Figures 2 to 4 posted at ftp://136.159.173.80/ display an overview of metabolic genes and their expression profiles in all six organ infection models.
Expression profile of known virulence genes.
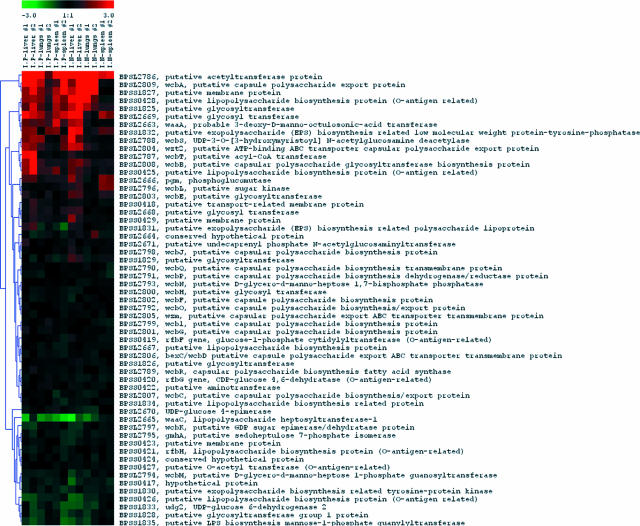
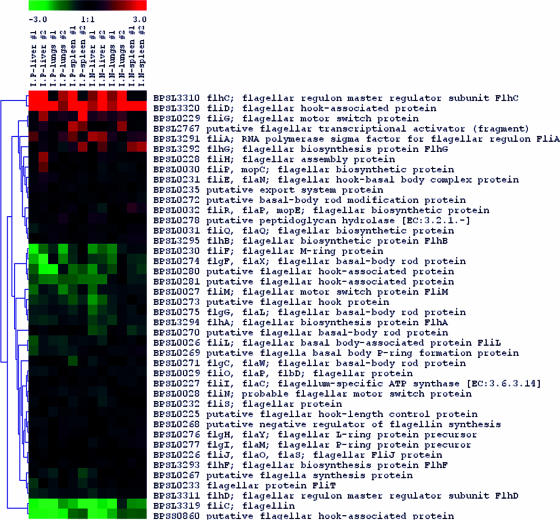
We observed several groups of genes coding for proteins which have been well characterized and known to be involved in bacterial pathogenesis and virulence in B. pseudomallei. These included genes for capsular biosynthesis, type III secretion system (TTSS), and flagellar biosynthesis and secreted hydrolytic enzymes such as phospholipases and proteases. We found that a putative acetyltransferase gene (BPSL2786) involved in capsular biosynthesis was highly up-regulated in most organ infection models except in one out of two replicates of the i.p. infected lungs (Fig. 4). In addition, at least two genes in TTSS such as BPSS1526 (putative invasion protein) and BPSS1530 (putative H-NS-like regulatory protein) were up-regulated in infected livers. Furthermore, BPSS1489, a possible additional gene in TTSS3 (25), was also highly up-regulated in most tested organs. We also observed that two genes involved in flagellar biosynthesis, including fliD (BPSL3320, flagellar hook-associated protein) and flhC (BPSL3310, flagellar regulon master regulator subunit FlhC), were highly up-regulated, while the fliC gene (BPSL3319, flagellin) was strongly down-regulated (Fig. 5). We also found that genes coding for phospholipase enzymes such as genes BPSS0067 (nonhemolytic phospholipase C) and BPSS1876 (putative phospholipase) were strongly up-regulated, especially in infected liver.
FIG. 4.
Gene expression profile in vivo versus in vitro for capsular biosynthesis genes. A particular gene coding for acetyltransferase enzyme activity (BPSL2786) was highly up-regulated, while most genes for the capsular biosynthesis were not differentially expressed. LPS, lipopolysaccharide.
FIG. 5.
Gene expression profile in vivo versus in vitro of flagellar biosynthesis genes. The fliC gene coding for flagellin was significantly down-regulated, while fliD, encoding a flagellar hook-associated protein, was highly induced. Most flagellar biosynthesis genes were not differentially expressed.
Virulence assessment of B. pseudomallei mutant strains.
To validate the microarray data, at least 11 highly up-regulated genes from this study were characterized by insertional mutagenesis and the mutants were evaluated for virulence in the hamster model of acute melioidosis. The mutant strains ATL0378, ATL1876, ATL2025, ATL2515, ATL3050, ATL3320, ATS0067, ATS0464, ATS1388, ATS1489, and ATS1530 were made from the mutation in genes BPSL0378, BPSL1876, BPSL2025, BPSL2515, BPSL3050, BPSL3320, BPSS0067, BPSS0464, BPSS1388, BPSS1489, and BPSS1530, respectively. These genes have not previously been characterized in any study before and were chosen based on their various putative functions such as genes for bacterial growth and metabolism (i.e., BPSL2515, BPSL3050, and BPSS0464), regulatory systems (i.e., BPSL0378 and BPSS1530), and putative virulence molecules (i.e., BPSL1876, BPSL3320, and BPSS0067) and genes with unknown functions (i.e., BPSL2025, BPSS1388, and BPSS1489). Our data showed that only the mutants ATL2025 and ATS0067 had significant LD50 values higher than that of the parent strain B. pseudomallei DD503, which were determined to be 1.4 × 104 and 4.5 × 104 CFU, respectively. In addition, two complemented mutants, ATCPL2025 and ATCPS0067, were also evaluated for virulence in the hamster model. We found that both complemented mutants were able to restore the virulence in hamsters to levels equivalent to that of the wild-type strain. Details of the insertional mutants and their virulence assessment data are shown in Table 5.
TABLE 5.
Virulence assessments of B. pseudomallei mutants in the hamster model of acute melioidosis
| Strain | Description/gene for mutation | LD50 (CFU) |
|---|---|---|
| DD503 | Parent strain | <10 |
| ATL0378 | ΔBPSL0378 (MarR family regulatory protein) | <10 |
| ATL1876 | ΔBPSL1876 (putative phospholipase) | <10 |
| ATL2025 | ΔBPSL2025 (putative two-component regulatory system) | 1.4 × 104 |
| ATL2515 | ΔBPSL2515 (rpsA; 30S ribosomal protein S1) | <10 |
| ATL3050 | ΔBPSL3050 (trpE; anthranilate synthase component 1) | <10 |
| ATL3320 | ΔBPSL3320 (fliD, flagellar hook-associated protein) | <10 |
| ATS0067 | ΔBPSS0067 (putative phospholipase C precursor) | 4.5 × 104 |
| ATS0464 | ΔBPSS0464 (potI, putrescine transport system permease protein) | <10 |
| ATS1388 | ΔBPSS1388 (putative membrane protein) | <10 |
| ATS1489 | ΔBPSS1489 (putative exported protein) | <10 |
| ATS1530 | ΔBPSS1530 (bprA, putative H-NS-like regulatory protein) | <10 |
| ATCPL2052 | Complemented mutant of ATL2052 | <10 |
| ATCPS0067 | Complemented mutant of ATS0067 | <10 |
DISCUSSION
The current studies detail the results from the analysis of gene expression in B. pseudomallei in an animal model of acute melioidosis using whole-genome microarrays and comparing different routes of infection. Recently, the relationship between routes of infection and virulence of B. pseudomallei has been investigated in BALB/c and C57BL/6 mouse models of systemic melioidosis (2). This study demonstrated that the virulence of B. pseudomallei is dependent upon the route of infection. LD50 values for animals infected with B. pseudomallei inoculated subcutaneously, intranasally, and orally were higher than those for animals inoculated intravenously or intraperitoneally. In our present studies, we used the hamster infection model of systemic acute melioidosis, as hamsters are exquisitely sensitive to B. pseudomallei and do not display individual susceptibility variation (10).
Our studies have conclusively demonstrated that B. pseudomallei genes are differentially expressed in vivo compared to in vitro growth. A number of genes were up- or down-regulated in infected organs, especially in the liver. Gene expression profiles and additional evidence demonstrated that bacterial growth in infected organs was slower than growth at mid-exponential phase in LB medium. This was demonstrated by reduced expression of numerous housekeeping genes coding for bacterial growth and multiplication proteins including genes for ribosomal proteins and cell division proteins. Interestingly, the total bacterial RNA from infected tissues contained lower amounts of rRNA than did that from organisms grown in LB. Thus, it could be suggested that the lower growth rate of B. pseudomallei in infected organs characterizes acute melioidosis in the hamster model. Further, the identification of the differentially expressed genes in these organs provides an understanding of bacterial response to the host environment during the infection. A similar observation was found in a recent in vivo gene expression study of B. mallei, a closely related organism, using a mouse model of infection which demonstrated that a number of housekeeping genes and metabolic genes responsible for bacterial growth and multiplication were relatively reduced in expression (17).
Four genes were commonly found to be highly induced in vivo. These included gene trpE (anthranilate synthase component 1), a gene for a putative phosphoglycerate mutase, and two genes coding for conserved hypothetical proteins. The trpE gene codes for an enzyme anthranilate synthase known to be involved in tryptophan amino acid biosynthesis. It has been reported that mutagenesis of trpE and aroA, an essential gene in Bordetella bronchiseptica, produces an aromatic amino acid auxotrophic mutant, and this mutant has reduced abilities to invade and survive within the macrophage cells in vitro and in vivo. This mutant was also tested for its ability to serve as a potential live-attenuated vaccine against this pathogen (22). Another up-regulated gene, a putative phosphoglycerate mutase, is known to be involved in the isomerization of the first substrate-level intermediate, 3-phosphoglycerate, to produce 2-phosphoglycerate for ATP. This would suggest that the organism is able to access this high-energy molecule during infection. Functional information for trpE, BPSL2902, and the two conserved hypothetical protein genes in B. pseudomallei is presently unavailable. In addition, insertion mutagenesis of the trpE gene in B. pseudomallei did not reduce virulence in the hamster model of acute melioidosis.
Our studies have demonstrated that B. pseudomallei uses novel pathways to generate and maintain cellular energy in vivo. The down-regulation of genes coding for enzymes known to be involved in the glycolytic pathway and the TCA cycle could contribute to the lower growth rate in vivo. We have demonstrated that genes coding for enzymes involved in the catabolism of d-glucosamine, an amino sugar, and inositol are highly up-regulated. Both of these substrates could serve as alternative carbon sources for carbohydrate metabolism. In addition, the induced expression of specific amino acid catabolism and biosynthesis genes such as asnO (asparagine synthase) and BPSL2260 (putative tryptophanyl-tRNA synthase) suggests that the organism uses novel metabolic pathways for amino acid metabolism. In addition to the alternative pathways utilized for carbohydrate and amino acid metabolism during in vivo growth, genes coding for enzymes known or predicted to be involved in alternative energy metabolism such as ubiquinol oxidases were also induced. Further, genes coding for enzymes required for primary energy metabolism such as ATP-synthase and NADH-dehydrogenase were down-regulated. We made a similar observation for bacteria grown under low-iron conditions (33). In low-iron conditions which are postulated to imitate the host environment, siderophore biosynthesis genes including a hydroxamate siderophore and pyochelin as well as heme-hemin receptor transport protein genes are highly induced (33). In the present studies, the pyochelin biosynthesis gene (BPSS0588) was induced in infected livers (see Table 7 posted at ftp://136.159.173.80/). Multiple in vitro studies designed to mimic the host environment have identified the expression of genes necessary for bacterial growth and survival under these conditions. These in vitro studies have involved an examination of growth under oxidative stress and the identification of the expression of the dpsA gene as necessary for survival (21) and stress due to the presence of reactive nitrogen intermediates and the identification of the expression of the ahpC-katG genes as important for survival (20). In addition, macrophage killing assays have been used in several studies to mimic the environment of host defense mechanisms, and genes responsible for the bacterial protective mechanisms have been identified. In particular, expression of the type III secretion bsa genes has been shown to be involved in the induction of the caspase-1-dependent macrophage death (32) and shown to play an essential role in modulating the intracellular behavior of B. pseudomallei (31). Interestingly, these well-characterized genes were not found in any organ infection models in our present studies. This is likely due to the equal expression of these under in vivo and in vitro conditions. This same observation was noted with other genes known to be involved in virulence such as genes for capsular biosynthesis, other genes in the type III secretion system, and flagellar biosynthesis genes. The products of these genes have been demonstrated to be virulence determinants in various animal models. It is likely that the transcriptional levels of these genes are equal during in vivo and in vitro growth, as it is well known that capsule, flagellum, and other virulence determinants are constitutively produced in vitro.
Flagella have been shown to be a virulence determinant in the BALB/c mouse model; mutation in the fliC gene caused virulence attenuation during intranasal and intraperitoneal infection in this animal model (7). fliC mutants remain highly virulent in the Syrian hamster and diabetic rat models (9). In the current studies, the fliC gene was strongly down-regulated, while fliD, a gene coding for a flagellar hook-associated protein, was highly induced (Fig. 5). An insertion mutation in the fliD gene did not lead to attenuation in the hamster infection model. These results confirm our earlier results indicating that flagella do not play a role in virulence in the hamster model of acute melioidosis. This would appear to be a deficiency of the hamster model, as the important role of flagella in different animal models of melioidosis is generally accepted, and a recent study has demonstrated the potential of using plasmid-encoded flagellin as a melioidosis vaccine (5).
Mutagenesis of a total of 11 up-regulated genes allowed us to identify two genes which were involved in virulence in the hamster model of acute melioidosis. These genes were BPSL2025 (putative two-component regulatory system) and BPSS0067 (putative phospholipase C precursor). A different two-component regulatory system known as irlRS has been characterized previously in B. pseudomallei, and its mutant was deficient in the ability to invade a human type II pneumocyte cell line, but it is not a virulence determinant in the infant diabetic rat and Syrian hamster models of acute B. pseudomallei infection (14). In addition, a different phospholipase C gene encoding a phosphatidylcholine-hydrolyzing phospholipase C (PC-PLC) has also been characterized previously in B. pseudomallei, and its product was recognized by human immunoglobulin M antibodies (18). However, there has been no report of this gene's involvement in bacterial virulence. Since our studies have shown a potential role of a putative two-component regulatory system (BPSL2025) and a putative phospholipase C (BPSS0067), a full understanding of these two genes in B. pseudomallei pathogenesis warrants further investigation. Our studies demonstrate the value of using gene expression profiles during bacterial infection in vivo combined with mutagenesis to identify potential virulence genes in B. pseudomallei.
In summary, our studies have identified differentially expressed genes which include potential virulence genes, and they have also provided a better understanding of bacterial metabolism in response to the host environment during acute melioidosis. The observation relative to the induction of specific genes for alternative biochemical pathways and a number of unknown genes most certainly warrants further experimentation.
Acknowledgments
This work was funded by the Canadian Institutes of Health research grant MOP-36343 to D.E.W., a Canada Research Chair in Microbiology. The printing of the B. pseudomallei whole-genome microarray slides was supported by a Los Alamos Research and Development grant to J.D.
We thank Mayi Arcellana-Panlillo and Xiuling Wang at the Southern Alberta Microarray Facilities, University of Calgary, for technical assistance, and we also thank Paige Pardington and Robert Cary at Los Alamos National Laboratory for printing the B. pseudomallei whole-genome microarray slides.
Editor: J. T. Barbieri
REFERENCES
- 1.Allworth, A. M. 2005. Tsunami lung: a necrotising pneumonia in survivors of the Asian tsunami. Med. J. Aust. 182:364. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, J. L., and N. Ketheesan. 2005. Route of infection in melioidosis. Emerg. Infect. Dis. 11:638-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett, P. J., D. DeShazer, and D. E. Woods. 1997. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol. Infect. 118:137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaowagul, W., N. J. White, D. A. Dance, Y. Wattanagoon, P. Naigowit, T. M. Davis, S. Looareesuwan, and N. Pitakwatchara. 1989. Melioidosis, a major cause of community-acquired septicemia in northeastern Thailand. J. Infect. Dis. 159:890-899. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y. S., Y. S. Hsiao, H. H. Lin, C. M. Yen, S. C. Chen, and Y. L. Chen. 2006. Immunogenicity and anti-Burkholderia pseudomallei activity in Balb/c mice immunized with plasmid DNA encoding flagellin. Vaccine 24:750-758. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, A. C., J. N. Hanna, R. Norton, S. L. Hills, J. Davis, V. L. Krause, G. Dowse, T. J. Inglis, and B. J. Currie. 2003. Melioidosis in Northern Australia, 2001-02. Commun. Dis. Intell. 27:272-277. [DOI] [PubMed] [Google Scholar]
- 7.Chua, K. L., Y. Y. Chan, and Y. H. Gan. 2003. Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 71:1622-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dance, D. A. 2000. Melioidosis as an emerging global problem. Acta Trop. 74:115-119. [DOI] [PubMed] [Google Scholar]
- 9.DeShazer, D., P. J. Brett, R. Carlyon, and D. E. Woods. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeShazer, D., and D. E. Woods. 1999. Animal models of melioidosis, p. 199-203. In O. Zak and M. Sande (ed.), Handbook of animal models of infection. Academic Press, London, United Kingdom.
- 11.DeShazer, D., and D. E. Woods. 1999. Pathogenesis of melioidosis: use of Tn5-OT182 to study the molecular basis of Burkholderia pseudomallei virulence. J. Infect. Dis. Antimicrob. Agents 16:91-96. [Google Scholar]
- 12.Gan, Y. H., K. L. Chua, H. H. Chua, B. Liu, C. S. Hii, H. L. Chong, and P. Tan. 2002. Characterization of Burkholderia pseudomallei infection and identification of novel virulence factors using a Caenorhabditis elegans host system. Mol. Microbiol. 44:1185-1197. [DOI] [PubMed] [Google Scholar]
- 13.Holden, M. T., R. W. Titball, S. J. Peacock, A. M. Cerdeno-Tarraga, T. Atkins, L. C. Crossman, T. Pitt, C. Churcher, K. Mungall, S. D. Bentley, M. Sebaihia, N. R. Thomson, N. Bason, I. R. Beacham, K. Brooks, K. A. Brown, N. F. Brown, G. L. Challis, I. Cherevach, T. Chillingworth, A. Cronin, B. Crossett, P. Davis., D. DeShazer, T. Feltwell, A. Fraser, Z. Hance, H. Hauser, S. Holroyd, K. Jagels, K. E. Keith, M. Maddison, S. Moule, C. Price, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, M. Simmonds, S. Songsivilai, K. Stevens, S. Tumapa, M. Vesaratchavest, S. Whitehead, C. Yeats, B. G. Barrell, P. C. Oyston, and J. Parkhill, J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. USA 101:14240-14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, A. L., D. DeShazer, and D. E. Woods. 1997. Identification and characterization of a two-component regulatory system involved in invasion of eukaryotic cells and heavy-metal resistance in Burkholderia pseudomallei. Infect. Immun. 65:4972-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, A. L., T. J. Beveridge, and D. E. Woods. 1996. Intracellular survival of Burkholderia pseudomallei. Infect. Immun. 64:782-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kespichayawattana, W., P. Intachote, P. Utaisincharoen, and S. Sirisinha. 2004. Virulent Burkholderia pseudomallei is more efficient than avirulent Burkholderia thailandensis in invasion of and adherence to cultured human epithelial cells. Microb. Pathog. 36:287-292. [DOI] [PubMed] [Google Scholar]
- 17.Kim, H. S., M. A. Schell, Y. Yu, R. L. Ulrich, S. H. Sarria, W. C. Nierman, and D. DeShazer. 2005. Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics 6:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korbsrisate, S., N. Suwanasai, A. Leelaporn, T. Ezaki, Y. Kawamura, and S. Sarasombath. 1999. Cloning and characterization of a nonhemolytic phospholipase C gene from Burkholderia pseudomallei. J. Clin. Microbiol. 37:3742-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leaky, A. K., G. C. Ulett, and R. G. Hirst. 1998. BALB/c and C57Bl/6 mice infected with virulent Burkholderia pseudomallei provide contrasting animal models for the acute and chronic forms of human melioidosis. Microb. Pathog. 24:269-275. [DOI] [PubMed] [Google Scholar]
- 20.Loprasert, S., R. Sallabhan, W. Whangsuk, and S. Mongkolsuk. 2003. Compensatory increase in ahpC gene expression and its role in protecting Burkholderia pseudomallei against reactive nitrogen intermediates. Arch. Microbiol. 180:498-502. [DOI] [PubMed] [Google Scholar]
- 21.Loprasert, S., W. Whangsuk, R. Sallabhan, and S. Mongkolsuk. 2004. DpsA protects the human pathogen Burkholderia pseudomallei against organic hydroperoxide. Arch. Microbiol. 182:96-101. [DOI] [PubMed] [Google Scholar]
- 22.McArthur, J. D., N. P. West, J. N. Cole, H. Jungnitz, C. A. Guzman, J. Chin, P. R. Lehrbach, S. P. Djordjevic, and M. J. Walker. 2003. An aromatic amino acid auxotrophic mutant of Bordetella bronchiseptica is attenuated and immunogenic in a mouse model of infection. FEMS Microbiol. Lett. 211:7-16. [DOI] [PubMed] [Google Scholar]
- 23.Miller, W. R., L. Pannell, L. Cravitz, W. A. Tanner, and T. Rosebury. 1948. Studies on certain biological characteristics of Malleomyces mallei and Malleomyces pseudomallei. II. Virulence and infectivity for animals. J. Bacteriol. 55:127-135. [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, R. A., D. DeShazer, S. Reckseidler, A. Weissmann, and D. E. Woods. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore, R. A., S. Reckseidler-Zenteno, H. Kim, W. C. Nierman, Y. Yu, A. Tuanyok, J. Warawa, D. DeShazer, and D. E. Woods. 2004. The contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 72:4172-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ngauy, V., Y. Lemeshev, L. Sadkowski, and G. Crawford. 2005. Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II. J. Clin. Microbiol. 43:970-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Quinn, A. L., E. M. Wiegand, and J. A. Jeddeloh. 2001. Burkholderia pseudomallei kills the nematode Caenorhabditis elegans using an endotoxin-mediated paralysis. Cell. Microbiol. 3:381-393. [DOI] [PubMed] [Google Scholar]
- 28.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 29.Rolim, D. B. 2005. Melioidosis, northeastern Brazil. Emerg. Infect. Dis. 11:1458-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 31.Stevens, M. P., M. W. Wood, L. A. Taylor, P. Monaghan, P. Hawes, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649-659. [DOI] [PubMed] [Google Scholar]
- 32.Sun, G. W., J. Lu, S. Pervaiz, W. P. Cao, and Y. H. Gan. 2005. Caspase-1 dependent macrophage death induced by Burkholderia pseudomallei. Cell. Microbiol. 7:1447-1458. [DOI] [PubMed] [Google Scholar]
- 33.Tuanyok, A., H. S. Kim, W. C. Nierman, Y. Yu, J. Dunbar, R. A. Moore, P. Baker, M. Tom, J. M. L. Ling, and D. E. Woods. 2005. Genome-wide expression analysis of iron regulation in Burkholderia pseudomallei and Burkholderia mallei using DNA microarray. FEMS Microbiol. Lett. 252:327-335. [DOI] [PubMed] [Google Scholar]
- 34.Warawa, J., and D. E. Woods. 2005. Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol. Lett. 242:101-108. [DOI] [PubMed] [Google Scholar]
- 35.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 36.White, N. J. 2003. Melioidosis. Lancet 361:1715-1722. [DOI] [PubMed] [Google Scholar]
- 37.Whitmore, A., and C. S. Krishnaswami. 1912. An account of glander-like disease occurring in Rangoon. J. Hyg. 13:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woods, D. E. 2002. The use of animal infection models to study the pathogenesis of melioidosis and glanders. Trends Microbiol. 10:483-484. [DOI] [PubMed] [Google Scholar]