Abstract
NF-κB is a crucial mediator of macrophage inflammatory responses, but its role in the context of pathogen-induced adaptive immune responses has yet to be elucidated. Here, we demonstrate that classical NF-κB activation delays phagocytosis-induced cell death (PICD) in Raw 264.7 and bone marrow-derived macrophages (BMDMs) upon ingestion of bacteria from the Escherichia coli laboratory strain Top10. By expression of a nondegradable form of IκBα (superrepressor) and pyrrolidine dithiocarbamate treatment, prolonged activation of NF-κB upon bacterial coculture is suppressed, whereas initial induction is only partially inhibited. This activation pattern results in partial inhibition of cellular activation and reduced expression of costimulatory CD86. Notably, suppression of classical NF-κB activation does not influence bacterial uptake rates but is followed by increased production of oxygen radicals and enhanced intracellular killing in Raw macrophages. This is associated with reduced expression of NF-κB-dependent antiapoptotic c-IAP-2 and a loss of the mitochondrial transmembrane potential. Accordingly, NF-κB inhibition in Raw cells and BMDMs causes increased apoptotic rates within 12 h of bacterial ingestion. Interestingly, accelerated eradication of E. coli in NF-κB-inhibited macrophages is associated with reduced antigen-specific T-cell activation in macrophage-lymphocyte cocultures. These data suggest that E. coli inhibits PICD of macrophages via classical, antiapoptotic NF-κB activation and thus facilitates signaling to T cells. Subsequently, a proper adaptive immune response is likely to be generated. Conclusively, therapeutic inhibition of classical NF-κB activation in macrophages may hamper the initiation of adaptive immunity.
The innate immune response represents the first line of defense during infection. Innate immune cells, such as macrophages, are essential for the host to efficiently control and remove invading pathogens. During this process, macrophages orchestrate the innate and adaptive immunity in response to a wide range of bacterial, viral, and fungal infections. In addition, they also play a key role in stimulating the subsequent clonal response of adaptive immunity (17). Thus, e.g., upon engulfment of bacteria, macrophages act as antigen-presenting cells and provide costimulatory signals necessary for full lymphocyte activation (5).
On a molecular basis, signal transduction via the IκB kinase (IKK)/NF-κB pathway is crucial for control and coordination of the innate as well as the adaptive immune response (21). Among others, proinflammatory cytokines and pathogen-associated molecular patterns induce activation of the IKK complex by signaling via tumor necrosis factor (TNF) receptor- or Toll-like receptor-interleukin-1 (IL-1) receptor superfamilies. The best known form of this complex consists of the IKKα and IKKβ catalytic subunits and the regulatory IKKγ subunit, also known as NEMO (NF-κB essential modulator), which recruits upstream signals to the complex (12). Recently, experimental evidence led to the separation of the classical NF-κB activation pathway, depending on IKKβ and IKKγ, from the alternative IKKα pathway (8, 31). During classical NF-κB signaling, activated IKKβ catalyzes the phosphorylation of inhibitory IκBs, which is followed by their polyubiquitination and proteasomal degradation. Liberated NF-κB, previously retained in the cytoplasm via binding to IκBs, translocates to the nucleus and activates transcription of target genes especially involved in innate immune responses (4, 12). The new alternative pathway is strictly dependent on IKKα, which phosphorylates the NF-κB precursor protein p100. Similar to IκBs, p100 becomes polyubiquitinated, and its inhibitory C-terminal end is degraded by the 26S proteasome (8). As a result, p100 is processed to p52 and mainly p52-RelB heterodimers enter the nucleus to activate transcription of genes that play a central role in development and maintenance of secondary lymphoid organs (4).
Despite the important role of macrophages during immune responses, little is known about macrophage effector functions that crucially depend on classical or alternative NF-κB activation. On the basis of evolutionary considerations, the original function of the NF-κB pathway is the initiation of inflammation and innate immune responses via production of inflammatory mediators and recruitment of immune cells (17, 39). As can be seen in Ikkβ−/− and Ikkγ−/− murine embryonic fibroblasts and macrophages, complete depletion of classical NF-κB activation is associated with dramatically reduced production of proinflammatory cytokines upon stimulation (20, 30, 35). Interestingly, gene-targeting experiments showed that regularly inducible classical NF-κB is dispensable for the production and differentiation of granulocytes and monocytes/macrophages in vivo (18, 32). Regarding its functional role, however, it might be assumed that impaired classical NF-κB signaling in macrophages might cause dramatic immune dysfunction in vivo. Indeed, complete or partial inactivation of this pathway results in a marked increase of susceptibility to infections in vivo, once the embryonic lethality associated with these deficiencies is prevented (2, 10, 32). Yet, it is not clear whether classical NF-κB deficiency primarily causes innate or adaptive immune failure in vivo, as it was shown that defects of classical NF-κB activation also result in impaired formation of secondary lymphoid structures (1). Subsequent to the initial inflammatory response, adaptive immunity to pathogens requires secondary signals generated by antigen-presenting cells, such as macrophages, to activate lymphocytes. This mechanism usually involves phagocytosis of pathogens, followed by killing, processing, and presentation of pathogens. The exact role of classical NF-κB in this respect is not fully elucidated.
Notably, phagocytosis by professional phagocytes, such as macrophages, is a prerequisite to combat, e.g., bacterial infections. Ingestion destroys most bacterial pathogens, although it is known that bacteria that are highly virulent, such as Listeria, Shigella, and Salmonella, are able to actively induce cell death of macrophages (9), which makes up a significant part of their pathology. However, it was shown recently that bacteria that exhibit relatively low virulence, such as Escherichia coli, also induce cell death upon phagocytosis (13). The significance of this form of death is not fully understand. A widely accepted point of view suggests that elimination of macrophages is disadvantageous for the host immune response, because they are relatively long-lived cells residing in tissue that regulate inflammation and subsequent immune responses. Thus, modulation of macrophage cell death by bacteria can be regarded as a mechanism of pathogenesis.
In this study, we shed light on the role of NF-κB signaling during primary macrophage effector functions with particular respect to bacterial clearance in the context of costimulatory signaling. We propose that induction of NF-κB-dependent antiapoptotic mechanisms upon ingestion of nonvirulent bacteria, such as E. coli, has important implications for the generation of an adaptive immune response. Therefore, we stably transfected Raw 264.7 macrophages with a mutant form of IκBα (IκBα superrepressor [IκBαSR]). IκBαSR is resistant to inducible degradation due to replacement of serine residues at positions 32 and 36 with alanine, resulting in reduced NF-κB activation (16). Subsequently, we tested the extent of NF-κB involvement in cell activation, cytokine production, and cell death in response to E. coli. Further, we investigated whether NF-κB mediates the process of bacterial clearance by coordinating the uptake of microbial particles or by inducing the formation of bactericidal oxygen species. Finally, the impact of the macrophage NF-κB pathway on costimulatory signaling to T cells upon E. coli ingestion was studied. The main results were verified by means of pyrrolidine dithiocarbamate (PDTC)-mediated inhibition of NF-κB activation in bone marrow-derived macrophages (BMDMs).
MATERIALS AND METHODS
Materials and reagents.
Murine macrophages (Raw 264.7) were cultured in Dulbecco modified Eagle medium (DMEM) (Invitrogen) supplemented with 10% fetal calf serum (PAA Laboratories), 1% Glutamax I (Invitrogen), and 0.02 mg/ml gentamicin (Refobacin; Merck) at 37°C in 5% CO2. For T cells, β-mercaptoethanol was added to a final concentration of 50 μM. Long-term culturing of transfected cells was performed with 200 μg/ml Geneticin (Invitrogen). Stimulation of cell cultures was carried out with 1 μg/ml lipopolysaccharide (LPS) (from E. coli O26:B6; Sigma), 10 ng/ml murine tumor necrosis factor alpha (TNF-α) (Sigma), and 10 U/ml gamma interferon (IFN-γ) (Calbiochem), respectively. To inhibit NF-κB activation in BMDMs, pyrrolidine dithiocarbamate (10 μM) was used. To induce apoptosis, cells were treated with 1 μM staurosporine for 3 h. Polyclonal antibodies against IκBα (C21), p65 (C-20), actin (C-11), cytochrome c oxidase II (K-20), and Sam 68 (C-20) were obtained from Santa Cruz. Additionally, anti-FasL (catalog no. F37720; Transduction), anti-caspase 3 (8G10) (Cell Signaling), and anti-p100/p52 (K-27) (Cell Signaling), and anti-Bax (catalog no. 06-499; Upstate) antibodies were used. E. coli antibody (clone BDI190) was obtained from Biodesign International.
BMDMs.
Bone marrow from BALB/c mice was flushed from femurs in DMEM (Invitrogen) supplemented with 10% fetal calf serum (PAA Laboratories), 1% Glutamax I (Invitrogen), and 0.02 mg/ml Refobacin (Merck) and cultured in DMEM conditioned with L929 medium (30%). After 24 h, floating cells were recultured, and BMDMs were used for experiments after 1 week.
DNA constructs and transfection.
IκBα-superrepressor plasmid (SR) was kindly provided by R. Zwacka (IZKF, University of Ulm, Germany). The pcDNA3 construct contains the IκBα sequence mutated at Ser32/Ser36 to form a phosphorylation-resistant (A32/A36) IκBα superrepressor (IκBαSR). Transfection was performed using Fugene 6 (Roche) according to the manufacturer's instructions. Cells were seeded in six-well plates and cultured to 60% confluence.
Proliferation assay.
For proliferation analysis of empty-vector-transfected (Mock) and SR cells, a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was performed according to the manufacturer's instructions (Roche). Briefly, 5 × 104 cells/well were incubated in triplicate in a 96-well plate in a final volume of 100 μl for the indicated time periods at 37°C. Thereafter, 25 μl of MTT solution (5 mg/ml in phosphate-buffered saline [PBS]) was added to each well. After 2 h of incubation at 37°C, 100 μl of the extraction buffer (20% sodium dodecyl sulfate, 50% dimethylformamide) was added, incubation was continued overnight at 37°C, and the optical density was measured at 570 nm using a microplate reader. Alternatively, cell proliferation was determined by counting cells at distinct time points after 0.1% trypan blue treatment.
Nuclear and cytosolic extracts, mitochondrial purification, immunoblotting, electrophoretic mobility shift assay (EMSA), and supershift analysis.
For nuclear extract preparation, cells (2 × 107) were washed two times in NP-40 buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 30 mM sucrose, 0.5% NP-40, pH 7.0), centrifuged (1,500 × g) for 7 min, and then washed twice in CaCl2 buffer (10 M Tris-HCl, 10 M NaCl, 3 mM MgCl2, 30 M sucrose, 0.1 mM CaCl2, pH 7.0). Nuclei were resuspended in lysing buffer (50 mM Tris-HCl, pH 7.6, 250 mM NaCl, 3 mM EDTA, 3 mM EGTA, 1% Triton X-100, 0.5% NP-40, 10% glycerol) and lysed on ice for 30 min, followed by centrifugation at 14,000 rpm for 30 min. The supernatant (nuclear extract) was collected and stored at −80°C. For total cell extracts, cells were directly lysed with lysing buffer. Except for cell death analysis, floating cells were always eliminated by washing steps. All steps were carried out at 4°C. All buffers were supplemented with 1 mM β-glycerolphosphate, 2 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, 10 μM leupeptin, 2 mM p-nitrophenylphosphate, and 0.1 mM orthovanadate. Mitochondria have been enriched as previously described (38). Briefly, PBS-washed cells were resuspended in homogenization buffer and disrupted by 200 strokes in an L-Dounce glass homogenizer. After removal of unbroken cells, membranes, and nuclei by centrifugation (1,500 × g, 10 min at 4°C), mitochondria were pelleted from the supernatant by further centrifugation at 12,000 × g for 10 min at 4°C. Subsequently, the supernatant was centrifuged at 50,000 × g for 60 min at 4°C to obtain the cytosol fraction.
Immunoblotting was performed using Bio-Rad Protean II Minigels with 10 μg protein extracts per lane. Proteins were blotted on nitrocellulose, and membranes were blocked with 7.5 or 10% nonfat dried milk in PBS and 0.3% Tween 20. For band shift assays, nuclear extracts (10 μg) were incubated in a 10-μl reaction mixture for 30 min with 0.1 μg/μl poly(dI-dC) (Pharmacia) and 20,000 cpm [α-32P]dATP-labeled oligonucleotide in 1 mM DTT, 10 mM HEPES, pH 7.6, 50 mM KCl, 6 mM MgCl2, 1.2 mM CaCl2, 1 mM DTT, and 5% glycerin. Complexes were separated in native 4% polyacrylamide gels for 3 h. Gels were dried and exposed to X-ray films. The following oligonucleotides were annealed to form double-stranded gel shift oligonucleotides: NF-κB (HIVκB site), sense (5′GGATCCTCAACAGAGGGGACTTTCCGAGGCCA3′) and reverse (5′GGATCCTGGCCTCGGAAAGTCCCCTCTGTTGA3′); NF-1, sense (5′TTTTGGATTGAAGCCAATATGATAA3′) and reverse (5′TTATCATATTGGCTTCAATCCA3′); AP-1, sense (5′CGCTTGATGACTCAGCCGGAA3′) and reverse (5′TTCCGGCTGAGTCAGCAA 3′). Cold NF-κB (HIVκB site) gel shift oligonucleotide and Ets-1/PEA3 (Santa Cruz) was used for specific and nonspecific competitor analysis, respectively. For supershift analysis, nuclear extracts (10 μg) were preincubated (30 min) with 5 μl p65 (C-20) and 1 μl p50 (sc-114X) from Santa Cruz.
Flow cytometry and measurement of ROS and antioxidative capacity.
Upon priming with 10 U/ml IFN-γ and stimulation with 1 μg/ml LPS for 1 h, detached Raw 264.7 cells (1 × 105/reaction mixture) were preincubated with mouse Fc block (Pharmingen) for 15 min on ice and washed with fluorescence-activated cell sorting buffer (PBS, 0.1% bovine serum albumin, 0.1% NaN3). Cells were then incubated with fluorescein isothiocyanate- or phycoerythrin-labeled antibodies for 1 h on ice and subsequently analyzed by means of flow cytometry. Anti-mouse CD40 (3/23), CD69 (H1.2F3), CD70 (FR70), CD80 (16-10A1), CD86 (GL1), and isotype control antibodies were obtained from BD Pharmingen. Membrane integrity was routinely tested by exclusion of propidium iodide, and all preparations were >95% propidium iodide negative. For the detection of reactive oxygen species (ROS) production upon stimulation, macrophages (1 × 105/well in 96-well plates) were preincubated with 15 μM CM-H2DCFDA [5- (and 6-)chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester] (Molecular Probes) in Hanks balanced salt solution (HBSS) (Invitrogen) for 10 min, allowing the dye to enter the cells. This dye freely permeates the cell membrane and becomes fluorescent upon cleavage by ROS. Cells were washed two times with PBS before stimulation with 5 × 105 E. coli per ml to induce ROS. Plates were analyzed at 485-nm excitation and 535-nm emission on a GeniosPlus (Tecan) microplate reader. Similarly, endogenous antioxidative capacity was determined via incubation of CM-H2DCFDA-preincubated cells with 100 μM H2O2 for 20 min.
Phagocytosis, confocal microscopy, and cell death assays.
Cells were plated at 1 × 106 in six-well plates and preincubated overnight, and then the medium was exchanged with HBSS (Invitrogen) containing 5% mouse serum. Subsequently, cells were either untreated (control cells) or primed for 30 min (IFN-γ [10 U/ml]) at 37°C and subsequently cocultured with 4 × 106 living Escherichia coli (Top10) containing an expression vector with green fluorescent protein (GFP) under the control of a cytomegalovirus promoter (kindly provided by A. Borowski, Bonn University). At the indicated time points, cells were washed extensively with HBSS to remove extracellular bacteria, and bacterial uptake was analyzed by flow cytometry. To discriminate between bacterial adhesion and phagocytosis, a separate set of experiments was performed. Following coculture with macrophages, extracellular bacteria were additionally labeled with an antibody that specifically recognizes the core region of E. coli endotoxin and also interacts with protein-bound endotoxin (E. coli outer membrane proteins; fimbrial subunits) (clone BDI190) (catalog no. B47171G; Biodesign International). Detection was performed with tetramethyl rhodamine isocyanate-labeled anti- immunoglobulin G1 secondary antibody. Extracellular adherent bacteria were identified via confocal microscopy. For analysis of microsphere phagocytosis, carboxylated fluorescein isothiocyanate-labeled microspheres (Molecular Probes) were vigorously resuspended by vortexing and sonication and preincubated with 1% bovine serum albumin in PBS. Microspheres (1 × 107) were added per 1 × 106 cells in a six-well plate and incubated at 37°C for 2 h. Upon extensive washing in PBS to remove extracellular microspheres, phagocytosis was analyzed by means of flow cytometry and determination of numbers of beads per cell. To detect mitochondrial transmembrane potential (ψm), treated cells were stained with 200 nM tetramethylrhodamine (TMRM) (Molecular Probes) in PBS (10 min, 37°C), washed, and microscopically analyzed. The rate of cell death was determined by detection of propidium iodide intercalation in isolated nuclei by the method of Nicoletti et al. (27). Apoptotic cell population accumulates as a sub-G1 peak. Additionally, determination of dead cells, characterized by membrane disintegration, was performed by trypan blue exclusion. After staining with trypan blue, 300 cells were examined, and dead cells were recorded as a percentage of cell death.
Bactericidal activity.
To determine bactericidal activity, a classical CFU assay was used with minor modifications (26). Briefly, cells were placed in 24-well culture plates (1 × 105/well) and preincubated overnight. Subsequently, medium was exchanged with antibiotic-free fresh medium, and after 1 h, cells were infected with 1 × 106 CFU of live E. coli (Top10). Control wells contained only bacteria. After 2 h of culture in a 5% CO2 incubator, plates were placed at −80°C for 30 min, and cells were lysed by thawing. Prior control experiments showed that this did not affect bacterial viability. The lysates were serially diluted, plated on LB agar plates, and incubated overnight at 37°C, and the number of bacterial colonies was counted. Bactericidal activity was expressed as the percentage of bacterial death = (CFU from control wells [without cells] − CFU from experimental wells)/(CFU from control wells [without cells]) × 100.
Animals, antigen, and cell coculture.
Female BALB/c mice aged 8 to 10 weeks were purchased from Charles River Breeding Laboratories. All experimental protocols were approved by the local ethics committee. Mice were sensitized intraperitoneally at two sites at days 0 and 10 with 50 μg of ovalbumin (OVA) (Sigma) adsorbed to 20 μl of aluminum hydroxide gel (Pierce) in a final volume of 200 μl. Commencing day 20, single-cell suspensions of splenocytes were prepared by crushing lymphoid organs between two microscope slides in cold PBS. Cells were passed through a 40-μm cell strainer. Erythrocytes were lysed using NH4Cl. Cell viability was verified by trypan blue exclusion. T cells were magnetically separated according to the manufacturer's instructions (Miltenyi Biotec). For macrophage-T-cell coculture, Raw 264.7 cells (Mock and SR) or BMDMs treated with PDTC (10 μM) or not treated with PDTC were plated for 48 h in 12-well plates (1 × 105/well) with 20 U/ml IFN-γ. OVA (10 mg/ml) was added for the final 16 h. Eight hours before isolated T cells were added, macrophages were incubated with 5 × 105 E. coli/well or 1 μg/ml LPS for 1 h. Thereafter, extracellular E. coli bacteria were eliminated by washing with fresh medium (containing 10 mg/ml OVA and 20 U/ml IFN-γ) and further incubated. Then, dead cells and PDTC were removed by three washing steps, and the remaining macrophages were counted and again plated at 2.5 × 105/well in 12-well plates. Isolated, antigen-primed T cells (5 × 105/well) were added and centrifuged onto the macrophages to initiate contact. Activation of T cells was analyzed by flow cytometric detection of CD69 (H1.2F3; BD Pharmingen) expression after 6 h of coculture.
Statistical analysis.
The data were analyzed using Student's t test and Mann-Whitney U test. A P value of <0.05 was regarded as statistically significant. All data were expressed as means ± standard errors of the means (SEMs).
RESULTS
Expression of an IκBα superrepressor in Raw 264.7 macrophages and PDTC treatment of BMDMs results in reduced NF-κB DNA-binding activity upon LPS stimulation and E. coli coculture.
In order to biochemically verify blocked NF-κB signaling, the DNA-binding activity of NF-κB in stably transfected Raw 264.7 SR clones was compared to that in empty-vector-transfected control cells (Mock) upon stimulation with E. coli (Fig. 1A and B) as well as stimulation with TNF-α and LPS (data not shown). The screened SR cell clones clearly reveal reduced NF-κB activation compared to Mock cells (Fig. 1B). Several stable SR clones were tested and showed similar kinetics of NF-κB DNA-binding activity. However, the kinetic patterns regularly show moderately depressed NF-κB activation during the first 30 min, followed by complete absence thereafter (Fig. 1A and D). The initial activation pattern results from degradation of endogenous IκBα, which parallels NF-κB activation. Under baseline conditions, endogenous IκBα and IκBαSR are expressed at levels similar to those observed for SR clone 3 (Fig. 1C). Upon stimulation with E. coli, endogenous IκBα becomes degraded, whereas IκBαSR protein remains stable (Fig. 1C). Thus, IκBαSR expression does not affect stimulus-induced degradation of the endogenous IκBα protein, but it scavenged newly synthesized or exported NF-κB after initial stimulation. To exclude the possibility that IκBα SR does affect other stress pathways, we examined the transcriptional activator AP-1. Only minor reductions of AP-1 activity in Mock and SR cells suggest that IκBαSR does not block other stress pathways that culminate therein (Fig. 1D). Moreover, this effect is specific as demonstrated by the results of competition assays (Fig. 1E) and selectively involves classical NF-κB signaling as demonstrated by p65 and p50 supershifts (Fig. 1F) and induction of nuclear p52 upon stimulation (Fig. 1G). Thus, the NF-κB inhibiting effect of IκBαSR in this system is restricted to the classical NF-κB pathway. Notably, a moderate difference of initial NF-κB induction is detectable upon comparison of clone SR 3 with SR 4 and 19 (Fig. 1A). Thus, these cell lines were analyzed separately, but the results were summarized in this work. Accordingly, significant disparate results of distinct clones are addressed whenever differences were detected. In order to verify the main results of the Raw 264.7 cell line with primary cells, BMDMs were established and analyzed upon suppression of NF-κB activation by PDTC. Similarly, NF-κB activity is inhibited mainly at late time points (Fig. 1H). To rule out effects on macrophage functions that depend on cell growth differences, proliferation of Mock and SR cell lines was determined by an MTT test and by counting cells under baseline cell culture conditions at distinct time points upon 0.1% trypan blue treatment. Under baseline conditions, no proliferation defect was detected in SR cells compared to empty-vector-transfected macrophages (Fig. 1I). Accordingly, Raw 264.7 cell lines stably transfected with an IκBα superrepressor and BMDMs treated with PDTC represent appropriate in vitro systems to investigate essential molecular and cellular effects in macrophages that particularly depend on late NF-κB signaling.
FIG. 1.
Characterization of Raw 264.7 Mock and SR macrophages. Several stable transfected clones of SR macrophages were independently established by transfection of normal Raw 264.7 macrophages with IκBαSR pcDNA3, which introduces a superrepressor mutant of IκBα, and neo. Mock cells are mock-transfected control macrophages that express neo alone. (A) NF-κB nuclear DNA-binding activity of Mock cells and three stable SR clones upon stimulation with E. coli Top10 (1.5 × 107 cells/ml culture volume) for 4 h. Raw 264.7 cells and bacteria were incubated at normal cell culture conditions, and nuclear extraction was performed at the indicated time points (e.g., 0 to 60 min [60′]). NF-κB activity was measured by an EMSA. Clones 3 and 19 were selected for further experiments. (B) Relative NF-κB activity. Calculation was performed via phosphorimaging relative to NF-1 activity. C, control. (C) Western blot analysis of IκBα and IκBαSR expression. Mock and SR cells were cocultured with E. coli Top10 (described above) for the indicated time points. (D) Comparison of NF-κB with AP-1 DNA-binding activity in Mock and SR cells. Nuclear extracts were performed upon incubation with E. coli Top10 for the indicated time points. (E) Competitor analysis of NF-κB DNA-binding activity. Cells were stimulated with E. coli for 30 min. For EMSA, 10 μg (Mock, BMDM) and 20 μg (SR 3, SR 19) of nuclear extracts were incubated with either 32P-labeled oligonucleotides alone (⊘) or together with nonspecific inhibitor (ns) or with specific, cold oligonucleotides (s). (F) NF-κB supershift analysis of Raw 264.7 macrophages. Cells were stimulated with E. coli for 1 h, and nuclear extracts were prepared at the indicated time points. Supershifted bands are indicated. (G) Nuclear p52. Western blot of nuclear extracts 12 h after E. coli stimulation. (H) Inhibition of NF-κB activation in BMDMs by PDTC. Normal BMDMs and BMDMs treated with PDTC (10 μM) were stimulated with E. coli as described above. Nuclear extracts were prepared at the indicated time points, and EMSAs were performed. (I) Raw 264.7 macrophage proliferation. Proliferation kinetics of cells were determined under normal basal cell culture conditions either by means of an MTT test (left) or by counting living cells upon trypan blue exclusion (right). These data are representative of three independent experiments.
Suppression of NF-κB activation moderately affects cellular activation but impairs the ability of macrophages to express costimulatory molecules.
To investigate the impact of NF-κB on macrophage cell functions, the expression of diverse macrophage-specific differentiation and activation markers was examined by flow cytometry. No differences were found between Mock and SR macrophages with respect to expression of major histocompatibility complex class II, F4/80, and CD11b with and without IFN-γ priming, followed by LPS stimulation (data not shown). Next we tested the extent of cellular activation via the expression of conventional macrophage activation markers, such as CD70 (CD27L) and CD69 (25), as well as by the determination of cytokine production. As shown in Fig. 2A and B, stimulation with LPS and IFN-γ activates SR and Mock cells regarding CD69/CD70 expression to a similar extent. In addition, the production of proinflammatory cytokines, such as TNF-α (data not shown) and IL-1α (Fig. 2C), is decreased but remains inducible in SR macrophages. However, macrophages that interact with pathogens become activated. Due to some suppressed SR cell activation, it can be assumed that there are also differences in subsequently provided signals for the adaptive immune system to sufficiently support specific immune responses. To evaluate the role of NF-κB signaling in this context, the capability of Mock and SR macrophages to interact with cells of the adaptive immune system was indirectly studied in treated with IFN-γ and LPS via analysis of the expression of CD40 and the costimulatory molecules CD80 (B7-1) and CD86 (B7-2), as well as by analysis of the expression of proinflammatory IL-12 and anti-inflammatory IL-10. Baseline and inducible CD40 expression is decreased (Fig. 2D and E), whereas baseline expression of CD80 and CD86 (Fig. 2D) is similar in Mock and SR cells. Upon stimulation with IFN-γ and LPS, CD86 is significantly reduced and CD80 (Fig. 2D and E) is partially reduced in SR macrophages compared to Mock cells. The production of the anti-inflammatory IL-10 shows no difference between SR and Mock cells over a period of 24 h, whereas IL-12 is suppressed (Fig. 2C). These data suggest that cellular activation of Raw 264.7 macrophages only partially depends on prolonged NF-κB activation. Moreover, the suppression of molecules associated with costimulatory functions in SR macrophages supports the idea that NF-κB signaling in these cells essentially contributes to the transfer of, e.g., bacterial information between the innate and adaptive immune system in the course of defense mechanisms.
FIG. 2.
Cellular characterization of Mock and SR macrophages. (A) Activation characteristics. Activation markers of Mock and SR macrophages were analyzed by means of flow cytometry under basal conditions and upon priming with 10 U/ml IFN-γ and stimulation with 10 μg/ml LPS for 1 h. (B) Quantitative summary of activation marker analysis (mean fluorescence intensity [MFI] = percentage of activated cells × mean fluorescence). C, control. (C) Production of cytokines. Upon stimulation with 10 U/ml IFN-γ and 10 μg/ml LPS for up to 24 h, levels of IL-1α, IL-12, and IL-10 were determined by means of enzyme-linked immunosorbent assays at the indicated time points. The values for Mock and SR cells were significantly different (P < 0.05) (*). (D) Costimulatory signaling. Expression of costimulatory molecules was analyzed by flow cytometry upon priming with 10 U/ml IFN-γ and stimulation with 10 μg/ml LPS. (E) Quantitative summary of costimulatory molecule analysis. Values represent means ± SEMs (error bars). *, P < 0.05. These data are representative of three independent experiments.
Cellular uptake of E. coli is not affected upon inhibition of NF-κB induction.
Phagocytosis is a key mechanism of the innate immune system to fight invading pathogens and, subsequently, to present foreign antigens as signals for the initiation of an adaptive immune response. To rule out the possibility that defective bacterial phagocytosis accounts for impaired cellular activation, Mock and SR macrophages were primed with 10 U IFN-γ and incubated with living E. coli bacteria expressing GFP. Bacterial uptake was determined at different time points by flow cytometry. Engulfment of E. coli did not show any difference between Mock and SR macrophages for up to 120 min (Fig. 3A and B). To exclude potential differences of adherence and uptake, extracellularly bound bacteria were counterstained with an anti-E. coli antibody, and immunofluorescence analysis was performed. Analysis by means of confocal microscopy revealed similar bacterial adherence and uptake by Mock and SR macrophages (Fig. 3C). Also, phagocytic capacity did not reveal any differences between Mock and SR macrophages, as in both cases, equal numbers of fluorescent microspheres per cell were taken up (data not shown). Similarly, no difference of phagocytic activity could be detected between BMDMs with and without NF-κB inhibition upon incubation with fluorescent E. coli (Fig. 3D).
FIG. 3.
Phagocytosis of Raw 264.7 macrophages and BMDMs upon coculture with E. coli. (A) Uptake of bacteria by Raw 264.7 macrophages. Cells were incubated under normal culture conditions with GFP-expressing E. coli for the indicated time points and washed, and uptake was assessed as GFP fluorescence of Raw 264.7 cells by flow cytometry. (B) Quantitation of bacterial uptake derived from three independent experiments. Values represent means ± SEMs (error bars). (C) Qualitative analysis of bacterial uptake. Raw 264.7 cells and E. coli were cultured as described above on coverslips. At the indicated time points (30 and 60 min [30′and 60′]), incubation was stopped by placing the cells on ice, and the cells were washed, counterstained with anti-E. coli antibody, and analyzed using confocal microscopy. Adherent bacteria (white arrows) and internalized bacteria (white arrowheads) are indicated. (D) Uptake of bacteria by BMDMs. Cells were incubated under BMDM-specific culture conditions with GFP-expressing E. coli for the indicated times and washed, and uptake was assessed as GFP fluorescence of BMDMs by flow cytometry. Histograms show data from three independent experiments.
Suppression of NF-κB activation is associated with increased bacterial killing and ROS production.
Effective combat of microbial invaders involves not only efficient phagocytosis but also the elimination of pathogens. We hence examined bactericidal activity in empty-vector-transfected and IκBαSR-transfected macrophages. Interestingly, as shown in Fig. 4A, bactericidal activity in Mock macrophages was significantly lower than that from NF-κB-inhibited macrophages. Consequently, as intracellular bacterial killing strongly depends on the production of oxygen radicals, the formation of ROS was determined upon coculture with bacteria. Thus, in line with the results of bacterial killing, ROS production by SR macrophages upon challenge with E. coli is increased compared to cells with normal NF-κB activation (Fig. 4B). These results are rather striking, as in murine macrophages, e.g., regulation of inducible nitric oxide synthetase (iNOS) expression is governed predominantly by the transcription factor NF-κB (28). Surprisingly, induction of iNOS is moderately increased in SR cells upon E. coli stimulation compared to Mock cells (Fig. 4C), which suggests slightly improved production of oxidizing substances, such as NO, following NF-κB inhibition. However, several enzymes and molecules with antioxidative properties, such as Mn superoxide dismutase or members of the glutathione system, are under the control of NF-κB (28). This indicates some antioxidative function that is linked to the NF-κB pathway. To shed more light on the role of NF-κB signaling in this context, the antioxidative capacity of Mock and SR macrophages was analyzed during oxidative stress. As expected, increased scavenging of H2O2-induced ROS in Mock macrophages compared to SR cells demonstrates their increased endogenous antioxidative capacity (Fig. 4D). In conclusion, accelerated killing of E. coli in macrophages deficient of prolonged NF-κB activation is most likely due to reduced levels of ROS-scavenging molecules.
FIG. 4.
Bacterial killing. (A) Cells and bacteria were cultured as described in the text. At the indicated time points, cells were washed, lysed, diluted by serial dilutions, and incubated on agar. After 24 h, bacterial colonies were counted. Values were calculated as described in Materials and Methods and represent means ± SEMs (error bars). The values for Mock and SR cells were significantly different (P < 0.05) (*). (B) ROS induction by E. coli bacteria in Raw 264.7 cells. Cells were preincubated with 15 μM CM-H2DCFDA. Thereafter, macrophages were exposed to bacteria, and cells were analyzed by means of a fluorescence microplate reader at the indicated time points. Values are means ± SEMs (error bars) of three replicate samples in three independent experiments. RFU, relative fluorescence units. (C) Immunoblot analysis of iNOS expression upon bacterial challenge for 1 h of Mock and SR macrophages. The changes in induction relative to actin was calculated by means of densitometry. These data are representative of two independent experiments. (D) Oxidative capacity of Raw 264.7 cells. Cells were preincubated with 15 μM CM-H2DCFDA. Thereafter, macrophages were incubated with 100 μM H2O2 for 20 min, and cells were analyzed by means of a fluorescence microplate reader at the indicated time points. Values are means ± SEMs (error bars) of three replicate samples in three independent experiments. *, P < 0.05.
E. coli prevents PICD of macrophages via induction of NF-κB-dependent antiapoptotic proteins.
NF-κB activation is considered an essential mechanism to prevent programmed cell death during cellular stress (21). In order to address this mechanism in our SR macrophages with moderately decreased initial but suppressed prolonged NF-κB activation, the percentage of hypodense sub-G1 (apoptotic) nuclei after propidium iodide staining (27) was quantified. For a positive control, staurosporine-treated Mock cells are shown (Fig. 5A). LPS alone was not found to significantly induce apoptosis in Mock or SR Raw 264.7 macrophages or in BMDMs treated with PDTC or not treated with PDTC over a period of 48 h (data not shown). Interestingly, TNF-α stimulation, which induces apoptosis in cells completely devoid of NF-κB (3, 32), does not increase cell death rates in SR cells (data not shown). In contrast, apoptotic rates of SR cells are significantly increased upon incubation with E. coli (Fig. 5A and B), suggesting an essential antiapoptotic role of NF-κB upon bacterial ingestion. These data could be confirmed by the trypan blue exclusion test, which also demonstrates increased numbers of dead SR cells upon E. coli treatment (Fig. 5C). Moreover, NF-κB inhibition of BMDMs via PDTC also leads to increased death rates upon E. coli engulfment compared to untreated BMDMs (Fig. 5C). It is conceivable that this mechanism reflects a more general property of NF-κB activation to prevent cell death during stress. Since prolonged NF-κB activation kinetics and its inhibition after 1 h of E. coli coculture may be different from long-term E. coli stimulation (Fig. 1A), NF-κB DNA-binding activity was determined in Mock and SR macrophages following 1 h of E. coli coculture. Thus, upon bacterial phagocytosis, prolonged activation of NF-κB is suppressed in SR cells compared to Mock macrophages (Fig. 5D). To gain more insight on the form of cell death, we analyzed the expression of certain apoptosis-associated proteins. Expression of proapoptotic NF-κB target genes Bax and FasL in total cell lysates did not show any difference in both cell lines upon phagocytosis of E. coli (Fig. 5E). Antiapoptotic effects mediated by NF-κB involve the transcription of caspase inhibitors, such as proteins of the IAP (inhibitor of apoptosis) family or antiapoptotic Bcl-2 family members (21, 29). Immunoblot analysis revealed increased expression of Bcl-2 and even induction of c-IAP-2 (cellular inhibitor of apoptosis protein 2) upon phagocytosis of E. coli in total lysates of Mock cells but suppressed expression levels in SR macrophages (Fig. 5E). However, levels of bax and bcl-2 expression in mitochondrial and cytosol extracts are more stable, suggesting other mechanisms leading to increased SR death (Fig. 5F). It is known that a loss of the mitochondrial transmembrane potential precedes apoptosis (34). Therefore, mitochondrial homeostasis was analyzed 6 h after 1 h of coculture of Mock and SR macrophages with E. coli. TMRM staining and confocal microscopic analysis revealed a marked decrease of ψm in Mock cells compared to SR cells (Fig. 5G). These data suggest that phagocytosis of E. coli by macrophages induces an NF-κB-dependent antiapoptotic program in both ways, by stabilizing ψm and by blocking effector caspases.
FIG. 5.
Phagocytosis-induced cell death of macrophages upon coculture with E. coli. (A) Induction of PICD. Raw 264.7 cells and bacteria were cultured under normal culture conditions for 1 h, washed, and again cultured under normal conditions. Twelve hours later, apoptotic cell death was determined by measuring propidium iodide intercalation of isolated nuclei by the method of Nicoletti et al. (27). The percentages are the relative frequencies of sub-G1 nuclei. Staurosporin-induced apoptosis (Sts, 1 μM) was used as positive control. (B) Quantitative assessment of PICD. Values represent means ± SEMs (error bars) of the percentage of sub-G1 cells stained by the method of Nicoletti et al. (27) from four independent experiments. *, P < 0.05. (C) Trypan blue exclusion test. Raw 264.7 macrophages and BMDMs treated with PDTC or not treated with PDTC were challenged with E. coli as described in Materials and Methods. At 0 h (control) and 12 h, the relative frequency of trypan blue-positive dead cells was determined by counting. Values are means ± SEMs (error bars) of three replicate samples in three independent experiments. Values that were significantly different (P < 0.05) from the values for Mock cells are indicated (*). (D) Prolonged NF-κB activation upon bacterial challenge. Raw 264.7 cells were incubated at normal cell culture conditions. E. coli was administered for 1 h, cells were washed, and nuclear extracts were performed at the indicated time points. NF-κB activity was determined by EMSA. C, control. (E) E. coli modulates expression of NF-κB-dependent antiapoptotic proteins and processing of procaspase 3. Raw 264.7 cells and bacteria were incubated as described above, total protein extraction was performed at the indicated time points, and total cell lysates were immunoblotted with antibodies as indicated. (F) Stable mitochondrial and cytosolic bax and bcl-2 levels. Raw 264.7 cells were incubated with E. coli as described above, and mitochondrial and cytosolic extracts were prepared and subjected to immunoblotting. CytC Ox, cytochrome c oxidase. (G) Uptake of E. coli induces a decrease in Δψm in SR cells. Mock and SR Raw 264.7 cells (2 × 105) were seeded into 12-well plates on coverslips. The following day, some wells were exposed to E. coli for 1 h and washed thereafter. Subsequently, plates were incubated under normal culture conditions. After 6 h, cells were incubated with TMRM and photographed under a fluorescence microscope. In the top two panels, control (C) Mock and SR cells show regular staining of Δψm. In the lower left panel, E. coli phagocytosis has no clear effect on Δψm of Mock macrophages. The lower right panel shows diminished Δψm upon bacterial uptake in SR cells (arrows). Data are representative of at least three independent experiments.
Increased elimination of E. coli antigens upon phagocytosis by SR macrophages.
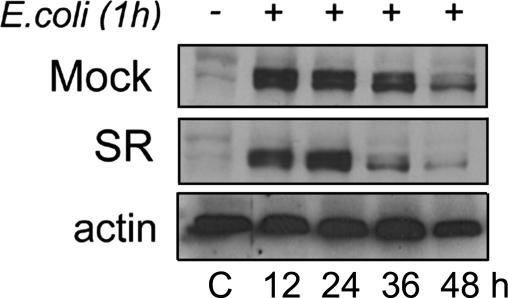
In order to evaluate whether accelerated bacterial killing is followed by increased intracellular elimination of bacterial antigens in SR macrophages, we performed immunoblot analysis of an E. coli-specific antigen (see Materials and Methods). Figure 6 demonstrates a more rapid eradication of this antigen after 36 and 48 h in SR macrophages compared to Mock cells following E. coli phagocytosis.
FIG. 6.
Accelerated elimination of E. coli antigen in SR cells. Immunoblot analysis of intracellular bacterial antigen (see Materials and Methods) in Mock and SR cells upon ingestion of E. coli. Raw 264.7 cells and bacteria were incubated with bacteria as described in the text, total protein extraction was performed at the indicated time points, and cell lysates were immunoblotted with an antibody that specifically recognizes the core region of E. coli endotoxin and also interacts with protein-bound endotoxin (E. coli outer membrane proteins; fimbrial subunits). Data are representative of two independent experiments. C, control.
Decreased activation of lymphocytes upon coculture with E. coli-stimulated SR macrophages.
To further investigate the effect of macrophage NF-κB inhibition on lymphocyte responses, we cocultured Mock, SR macrophages, and BMDMs treated with PDTC or not treated with PDTC (H-2d) with spleen T cells isolated from H-2d restricted BALB/c mice previously immunized with ovalbumin. Initially, macrophages were primed with IFN-γ, loaded with OVA overnight, and cocultured (1 h) with E. coli or LPS 8 h before T cells were added. After 6 h of interaction, T cells were harvested, and their activation was determined via expression of early activation marker CD69 by flow cytometry. As shown in Fig. 7A, coculture of T cells with Mock macrophages, treated with either E. coli or LPS, induces T-cell activation. However, suppression of classical NF-κB induction in E. coli-treated Raw 264.7 macrophages reduces the activation of T cells. Interestingly, LPS-treated SR cells are able to activate T cells to an extent comparable to that by Mock cells (Fig. 7A). Similarly, normal BMDMs activate T cells, but inhibition of NF-κB by PDTC markedly reduces T-cell activation upon E. coli challenge (Fig. 7B). This suggests defective communication of SR macrophages with T cells upon antigenic challenge. Increasing concentrations of IFN-γ or OVA were not able to efficiently compensate for this defect (data not shown), which therefore accounts for a specific function of NF-κB. Most likely, this defect is associated with rapid elimination of E. coli antigens in SR cells (Fig. 6), as LPS-treated SR cells fully activate T cells. In conclusion, these data provide sufficient evidence that the absence of NF-κB activation in macrophages results in marked defects of providing secondary signals to efficiently induce adaptive immune responses.
FIG. 7.
Activation of T cells by macrophages is impaired upon NF-κB inhibition. T cells were isolated from the spleens of OVA-immunized BALB/c mice and cocultured with OVA/IFN-γ-primed and E. coli- or LPS-challenged macrophages. Analysis was performed by means of flow cytometry. (A) Histograms of CD69-positive cells of CD4-gated lymphocytes following coculture of E. coli- or LPS-stimulated Mock and SR cells. The percentages are the relative frequencies of positive cells. These data are representative of three independent experiments. (B) Histograms of CD69-positive cells of CD4-gated lymphocytes following coculture of E. coli- or LPS-stimulated BMDMs treated with PDTC (+ PDTC) or not treated with PDTC (− PDTC). The percentages are the relative frequencies of positive cells. These data are representative of two independent experiments.
DISCUSSION
By stable expression of a gene coding for the IκBα superrepressor in Raw 264.7 macrophages, an in vitro system primarily devoid of prolonged NF-κB activation was established and verified by primary bone marrow-derived macrophages treated with the NF-κB inhibitor PDTC. However, some remaining initial NF-κB induction can be detected in both cell systems, which is central to the results reported here. It is known that during inflammatory responses NF-κB is induced in a biphasic mode in macrophages: first, a transient IκBα degradation-dependent phase of activity and a second, persistent phase of activation (14, 15). Previous studies suggest that a second NF-κB activation phase is caused by newly produced proinflammatory cytokines (14). Thus, the absence of prolonged NF-κB activation in our macrophages might simply be due to reduced proinflammatory cytokine production. However, this mechanism as well as the fact that IκBαSR represses the constitutive activity of NF-κB probably does not play a major role in this system, as SR macrophages are initially activated and TNF-α as well as IL-1α is inducible upon stimulation, though to a smaller extent. More importantly, the biochemical data suggest that suppression of NF-κB activation is derived from the scavenging of exported and newly synthesized NF-κB by the accumulated IκBαSR. In addition, TNF-α stimulation does not increase NF-κB activity in SR macrophages during time points corresponding to the second activation phase. This suggests a definitive and specific block of classical NF-κB activation via IκBαSR as demonstrated by p50 and p65 supershifts and similar levels of nuclear p52 upon stimulation. In this context it has to be noted that NF-κB has a precisely defined role during programmed cell death. Mainly considered a survival transcription factor, prevention of TNF-α-induced programmed cell death is a hallmark of classical NF-κB signaling (3). It occurs in different cell types, such as hepatocytes (2, 3, 11) and lymphocytes (32). Moreover, it has been shown that Ikkβ−/− macrophages, which are completely devoid of NF-κB activation, are very sensitive to LPS-induced apoptosis (19). These survival defects upon suppression of NF-κB activation are less dramatic in the stable SR clones reported herein, most likely due to moderate initial NF-κB activation upon stimulation. Similarly, NF-κB inhibition in BMDMs does not sensitize these cells to undergo LPS-induced cell death, which is due to some initial NF-κB activation under the PDTC concentrations used. Hence, this model serves to primarily investigate cellular mechanisms associated with the second activation peak of the classical, IκBα-dependent NF-κB pathway.
The precise biological significance of prolonged NF-κB activation is poorly understood. Presumably, the initial phase primarily supports proinflammatory tasks, whereas prolonged NF-κB activation probably serves different, cell type-specific functions (14). Recently, Lawrence et al. (24) reported that the late phase activation of NF-κB is associated with the resolution of inflammation. However, these data were obtained from a noninfectious model, whereas other studies suggest a direct prolonged activation of NF-κB by bacterial virulence factors within the macrophage (15) that may also result in a proinflammatory status (6). The present investigation demonstrates moderate to marked suppression of proinflammatory properties in stimulated SR macrophages. This study is therefore in agreement with reports that describe a proinflammatory role of prolonged classical NF-κB activation (23). Further, our data suggest that prolonged NF-κB activation mainly consists of the classical way, whereas the alternative way is induced independently of classical NF-κB signaling. So far, the contributions of the classical and alternative pathways to prolonged NF-κB activation in innate immune cells have not been fully elucidated. Interestingly, inactivation of the alternative pathway in mice enhances inflammation upon gram-positive bacterial challenge, which is due to suppressed downregulation of classical, apparently prolonged NF-κB activity in macrophages (23). The role of the alternative IKKα-dependent pathway for downregulation of classical induced NF-κB activation is further confirmed by the fact that p52 is accumulated in the nucleus 12 h after E. coli stimulation. As a result, macrophages devoid of alternative NF-κB activation demonstrate aggravated proinflammatory activity and thus exhibit the opposite phenotype of the SR macrophages described here. Thus, classical NF-κB activation in macrophages predominantly fulfills proinflammatory tasks.
Following bacterial ingestion, the innate ability of phagocytes to kill bacteria is crucial for the host defense, because it is immediate, nonspecific, and not dependent on previous pathogen exposure. It is known that this process can cause apoptosis in neutrophils and macrophages, a process called phagocytosis-induced cell death. Regarding neutrophils, this process was shown to be beneficial for the host, as it contributes to the clearance and killing of pathogens and accelerates the resolution of inflammation (9). In contrast to neutrophils, macrophages not only kill but eliminate pathogens. They also provide an important link to initiate adaptive immune responses. Therefore, increased PICD of macrophages might impair the induction of a complete adaptive immune response in which classical NF-κB is likely to play a key role.
In order to study basic cellular and bactericidal mechanisms, a low-virulence laboratory strain of E. coli was used in the present study. Notably, macrophages completely defective of NF-κB activation underwent rapid apoptosis upon coculture with E. coli (22). In principle, our results confirm these data and suggest that macrophage PICD is induced by E. coli but is antagonized by classical NF-κB activation. An antiapoptotic approach is known to be used extensively by distinct microbes, such as Chlamydia pneumoniae, to try to prevent the death of infected cells in order to give the pathogen the opportunity to replicate (36). In our model the prevention of cell death is likely to depend on induction of NF-κB-dependent antiapoptotic proteins, such as c-IAP-2, and is associated with a loss of the mitochondrial membrane potential. As a consequence, the bactericidal function of SR macrophages should be impaired. Strikingly, our data show that suppressed NF-κB activation leads to normal phagocytosis but enhanced bactericidal activity. This is most likely due to increased production of oxygen radicals, which is known to be an important antimicrobial mechanism. There are several possibilities that might contribute to increased ROS production upon classical NF-κB suppression in Raw 264.7 cells. First, moderately increased iNOS induction in the SR cells reported here should produce higher levels of NO. Second, several recent studies indicate that ROS production is critical to PICD (9). In particular, phagocytosis of E. coli by neutrophils is accompanied by production of ROS, which is in turn linked to cell death (37). Thus, aberrant regulation of cell death pathways leads to impaired survival and may at least in part explain increased ROS production by stimulated SR macrophages. Third and most important, the antioxidative capacity of SR cells is reduced as a result of diminished transcription of NF-κB-dependent scavenging proteins, such as Mn superoxide dismutase (7). This releases ROS effects and ultimately leads to increased bactericidal function.
What is the biological importance of NF-κB-associated bacterial resistance? In contrast to neutrophil apoptosis, which is accepted to be beneficial for the host, cell death of macrophages is considered to be disadvantageous for the host regulatory response to infection (9). In the case of E. coli with low virulence, delayed PICD of macrophages via activation of classical NF-κB suggests a positive effect on the induction of adaptive immune responses. Indeed, it was shown here that macrophages deficient of classical NF-κB activation fail to activate T cells. In addition to reduced expression of costimulators, impaired secondary signaling of NF-κB-defective macrophages to T cells in the present study results from rapid intracellular killing and, consequently, accelerated elimination of E. coli antigens. As a consequence, T cells without costimulation are either unresponsive and enter a state of anergy or become apoptotic (33). In this context, it is conceivable that NF-κB-mediated antiapoptosis improves the availability of bacterial antigens. The transfer of bacterial information from macrophages to T cells is essential to mount an adaptive immune response (9). Conclusively, certain antiapoptotic aspects of macrophages that are NF-κB dependent and induced by bacteria are likely to be necessary for achieving an efficient immune response to infection.
Our data support the idea that—rather than simply being a proinflammatory pathway—classical NF-κB-mediated inhibition of macrophage death along with the preservation of bacterial information depicts an important prerequisite to initiate adaptive immune responses. Hence, from an evolutionary point of view, it would appear that the innate, pathogen-removing function of the classical NF-κB pathway in fruit flies is extended by a supportive role in mammalian innate immune cells. Thus, under certain circumstances, new NF-κB-activating treatment strategies might provide a means to more efficiently mount an adaptive immune response in treating infectious diseases.
.
Acknowledgments
This work was supported by a grant of the Deutsche Forschungsgemeinschaft to U.S.
We thank R. Zwacka for the generous gift of the IκBα superrepressor construct; Eberhart Reithmeier for experimental advice; and Bettina Stahl, Sina Heydrich, and Rosemarie Mayer for technical support.
Editor: F. C. Fang
REFERENCES
- 1.Alcamo, E., N. Hacohen, L. C. Schulte, P. D. Rennert, R. O. Hynes, and D. Baltimore. 2002. Requirement for the NF-kappaB family member RelA in the development of secondary lymphoid organs. J. Exp. Med. 195:233-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcamo, E., J. P. Mizgerd, B. H. Horwitz, R. Bronson, A. A. Beg, M. Scott, C. M. Doerschuk, R. O. Hynes, and D. Baltimore. 2001. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-kappa B in leukocyte recruitment. J. Immunol. 167:1592-1600. [DOI] [PubMed] [Google Scholar]
- 3.Beg, A., and D. Baltimore. 1996. An essential role for NF-kappa B in preventing TNF-alpha-induced cell death. Science 274:782-784. [DOI] [PubMed] [Google Scholar]
- 4.Bonizzi, G., and M. Karin. 2004. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25:280-288. [DOI] [PubMed] [Google Scholar]
- 5.Carreno, B. M., and M. Collins. 2002. The B7 family of ligands and its receptors: new pathways for costimulation and inhibition of immune responses. Annu. Rev. Immunol. 20:29-53. [DOI] [PubMed] [Google Scholar]
- 6.Connelly, L., M. Palacios-Callender, C. Ameixa, S. Moncada, and A. J. Hobbs. 2001. Biphasic regulation of NF-kappa B activity underlies the pro- and anti-inflammatory actions of nitric oxide. J. Immunol. 166:3873-3881. [DOI] [PubMed] [Google Scholar]
- 7.Das, K. C., Y. Lewis-Molock, and C. W. White. 1995. Activation of NF-kappa B and elevation of MnSOD gene expression by thiol reducing agents in lung adenocarcinoma (A549) cells. Am. J. Physiol. 269:L588-L602. [DOI] [PubMed] [Google Scholar]
- 8.Dejardin, E., N. M. Droin, M. Delhase, E. Haas, Y. Cao, C. Makris, Z. W. Li, M. Karin, C. F. Ware, and D. R. Green. 2002. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity 17:525-535. [DOI] [PubMed] [Google Scholar]
- 9.Deleo, F. R. 2004. Modulation of phagocyte apoptosis by bacterial pathogens. Apoptosis 9:399-413. [DOI] [PubMed] [Google Scholar]
- 10.Doffinger, R., A. Smahi, C. Bessia, F. Geissmann, J. Feinberg, A. Durandy, C. Bodemer, S. Kenwrick, S. Dupuis-Girod, S. Blanche, P. Wood, S. H. Rabia, D. J. Headon, P. A. Overbeek, F. Le Deist, S. M. Holland, K. Belani, D. S. Kumararatne, A. Fischer, R. Shapiro, M. E. Conley, E. Reimund, H. Kalhoff, M. Abinun, A. Munnich, A. Israel, G. Courtois, and J. L. Casanova. 2001. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat. Genet. 27:277-285. [DOI] [PubMed] [Google Scholar]
- 11.Doi, T. S., M. W. Marino, T. Takahashi, T. Yoshida, T. Sakakura, L. J. Old, and Y. Obata. 1999. Absence of tumor necrosis factor rescues RelA-deficient mice from embryonic lethality. Proc. Natl. Acad. Sci. USA 96:2994-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-kappaB puzzle. Cell 109(Suppl.):S81-S96. [DOI] [PubMed] [Google Scholar]
- 13.Hacker, H., C. Furmann, H. Wagner, and G. Hacker. 2002. Caspase-9/-3 activation and apoptosis are induced in mouse macrophages upon ingestion and digestion of Escherichia coli bacteria. J. Immunol. 169:3172-3179. [DOI] [PubMed] [Google Scholar]
- 14.Han, S. J., H. M. Ko, J. H. Choi, K. H. Seo, H. S. Lee, E. K. Choi, I. W. Choi, H. K. Lee, and S. Y. Im. 2002. Molecular mechanisms for lipopolysaccharide-induced biphasic activation of nuclear factor-kappa B (NF-kappa B). J. Biol. Chem. 277:44715-44721. [DOI] [PubMed] [Google Scholar]
- 15.Hauf, N., W. Goebel, F. Fiedler, Z. Sokolovic, and M. Kuhn. 1997. Listeria monocytogenes infection of P388D1 macrophages results in a biphasic NF-kappaB (RelA/p50) activation induced by lipoteichoic acid and bacterial phospholipases and mediated by IkappaBalpha and IkappaBbeta degradation. Proc. Natl. Acad. Sci. USA 94:9394-9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hettmann, T., J. DiDonato, M. Karin, and J. M. Leiden. 1999. An essential role for nuclear factor kappa B in promoting double positive thymocyte apoptosis. J. Exp. Med. 189:145-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann, J. A., F. C. Kafatos, C. A. Janeway, and R. A. Ezekowitz. 1999. Phylogenetic perspectives in innate immunity. Science 284:1313-1318. [DOI] [PubMed] [Google Scholar]
- 18.Horwitz, B. H., M. L. Scott, S. R. Cherry, R. T. Bronson, and D. Baltimore. 1997. Failure of lymphopoiesis after adoptive transfer of NF-kappa B-deficient fetal liver cells. Immunity 6:765-772. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, L. C., J. M. Park, K. Zhang, J. L. Luo, S. Maeda, R. J. Kaufman, L. Eckmann, D. G. Guiney, and M. Karin. 2004. The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature 428:341-345. [DOI] [PubMed] [Google Scholar]
- 20.Kanters, E., M. Pasparakis, M. J. Gijbels, M. N. Vergouwe, I. Partouns-Hendriks, R. J. Fijneman, B. E. Clausen, I. Forster, M. M. Kockx, K. Rajewsky, G. Kraal, M. H. Hofker, and M. P. de Winther. 2003. Inhibition of NF-kappaB activation in macrophages increases atherosclerosis in LDL receptor-deficient mice. J. Clin. Investig. 112:1176-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karin, M., and A. Lin. 2002. NF-kappaB at the crossroads of life and death. Nat. Immunol. 3:221-227. [DOI] [PubMed] [Google Scholar]
- 22.Kitamura, M. 1999. NF-kappaB-mediated self defense of macrophages faced with bacteria. Eur. J. Immunol. 29:1647-1655. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, T., M. Bebien, G. Y. Liu, V. Nizet, and M. Karin. 2005. IKKalpha limits macrophage NF-kappaB activation and contributes to the resolution of inflammation. Nature 434:1138-1143. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence, T., D. W. Gilroy, P. R. Colville-Nash, and D. A. Willoughby. 2001. Possible new role for NF-kappaB in the resolution of inflammation. Nat. Med. 7:1291-1297. [DOI] [PubMed] [Google Scholar]
- 25.Marzio, R., E. Jirillo, A. Ransijn, J. Mauel, and S. B. Corradin. 1997. Expression and function of the early activation antigen CD69 in murine macrophages. J. Leukoc. Biol. 62:349-355. [DOI] [PubMed] [Google Scholar]
- 26.Matsukawa, A., K. Takeda, S. Kudo, T. Maeda, M. Kagayama, and S. Akira. 2003. Aberrant inflammation and lethality to septic peritonitis in mice lacking STAT3 in macrophages and neutrophils. J. Immunol. 171:6198-6205. [DOI] [PubMed] [Google Scholar]
- 27.Nicoletti, I., G. Migliorati, M. C. Pagliacci, F. Grignani, and C. Riccardi. 1991. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods 139:271-279. [DOI] [PubMed] [Google Scholar]
- 28.Pahl, H. L. 1999. Activators and target genes of rel/NF-kappaB transcription factors. Oncogene 18:6853-6866. [DOI] [PubMed] [Google Scholar]
- 29.Roy, N., Q. L. Deveraux, R. Takahashi, G. S. Salvesen, and J. C. Reed. 1997. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 16:6914-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt-Supprian, M., W. Bloch, G. Courtois, K. Addicks, A. Israel, K. Rajewsky, and M. Pasparakis. 2000. NEMO/IKK gamma-deficient mice model incontinentia pigmenti. Mol. Cell 5:981-992. [DOI] [PubMed] [Google Scholar]
- 31.Senftleben, U., Y. Cao, G. Xiao, F. R. Greten, G. Krahn, G. Bonizzi, Y. Chen, Y. Hu, A. Fong, S. C. Sun, and M. Karin. 2001. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 293:1495-1499. [DOI] [PubMed] [Google Scholar]
- 32.Senftleben, U., Z. W. Li, V. Baud, and M. Karin. 2001. IKKbeta is essential for protecting T cells from TNFalpha-induced apoptosis. Immunity 14:217-230. [DOI] [PubMed] [Google Scholar]
- 33.Sharpe, A. H., and G. J. Freeman. 2002. The B7-CD28 superfamily. Nat. Rev. Immunol. 2:116-126. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu, S., Y. Eguchi, W. Kamiike, Y. Funahashi, A. Mignon, V. Lacronique, H. Matsuda, and Y. Tsujimoto. 1998. Bcl-2 prevents apoptotic mitochondrial dysfunction by regulating proton flux. Proc. Natl. Acad. Sci. USA 95:1455-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka, M., M. E. Fuentes, K. Yamaguchi, M. H. Durnin, S. A. Dalrymple, K. L. Hardy, and D. V. Goeddel. 1999. Embryonic lethality, liver degeneration, and impaired NF-kappa B activation in IKK-beta-deficient mice. Immunity 10:421-429. [DOI] [PubMed] [Google Scholar]
- 36.Wahl, C., F. Oswald, U. Simnacher, S. Weiss, R. Marre, and A. Essig. 2001. Survival of Chlamydia pneumoniae-infected Mono Mac 6 cells is dependent on NF-κB binding activity. Infect. Immun. 69:7039-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watson, R. W., H. P. Redmond, J. H. Wang, C. Condron, and D. Bouchier-Hayes. 1996. Neutrophils undergo apoptosis following ingestion of Escherichia coli. J. Immunol. 156:3986-3992. [PubMed] [Google Scholar]
- 38.Zamora, M., C. Merono, O. Vinas, and T. Mampel. 2004. Recruitment of NF-kappaB into mitochondria is involved in adenine nucleotide translocase 1 (ANT1)-induced apoptosis. J. Biol. Chem. 279:38415-38423. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, G., and S. Ghosh. 2001. Toll-like receptor-mediated NF-kappaB activation: a phylogenetically conserved paradigm in innate immunity. J. Clin. Investig. 107:13-19. [DOI] [PMC free article] [PubMed] [Google Scholar]