Abstract
Shigella flexneri is the causative agent of dysentery, and its pathogenesis is mediated by a type III secretion system (T3SS). S. flexneri secretes effector proteins into the eukaryotic cell via the T3SS, and these proteins usurp host cellular functions to the benefit of the bacteria. OspF and OspC1 are known to be secreted by S. flexneri, but their functions are unknown. We transformed S. flexneri with a plasmid that expresses a two-hemagglutinin tag (2HA) in frame with OspF or OspC1 and verified that these proteins are secreted in a T3SS-dependent manner. Immunofluorescence of HeLa cells infected with S. flexneri expressing OspF-2HA or OspC1-2HA revealed that both proteins localize in the nucleus and cytoplasm of host cells. To elucidate the function of these T3SS effectors, we constructed ΔospF and ΔospC1 deletion mutants by allelic exchange. We found that ΔospF and ΔospC1 mutants invade host cells and form plaques in confluent monolayers similar to wild-type S. flexneri. However, in the polymorphonuclear (PMN) cell migration assay, a decrease in neutrophil migration was observed for both mutants in comparison to the migration of wild-type bacteria. Moreover, infection of polarized T84 intestinal cells infected with ΔospF and ΔospC1 mutants resulted in decreased phosphorylation of extracellular signal-regulated kinase 1/2 in comparison to that of T84 cells infected with wild-type S. flexneri. To date, these are the first examples of T3SS effectors implicated in mitogen-activated protein kinase kinase/extracellular signal-regulated kinase pathway activation. Ultimately, OspF and OspC1 are essential for PMN transepithelial migration, a phenotype associated with increased inflammation and bacterial access to the submucosa, which are fundamental aspects of S. flexneri pathogenesis.
Shigella species are responsible for dysentery (shigellosis) in humans, which starts as an acute infection in the large intestine, which is followed by cramps, diarrhea, and fever. The infection is usually self-limiting, but it can also cause damage to the colonic mucosa, intestinal bleeding, and death if untreated. Worldwide, Shigella spp. infections are responsible for approximately 163 million cases of dysentery and 1 million deaths each year (23). The majority of infections occur in third-world countries, where contaminated food and drinking water are common (23); however, even developed nations still have multiple Shigella outbreaks every year (47). Therefore, studying the pathogenesis of these gram-negative bacteria is of utmost importance, particularly in light of emerging antibiotic resistance, the lack of an appropriate vaccine, and the potential for use of Shigella as a bioweapon (23, 33, 42).
Following ingestion, Shigella flexneri eventually reaches the large intestine, the target site for infection, where access to the basolateral membrane is a prerequisite for the invasion of epithelial cells (see reference 48 for a review). M cells in the large intestine phagocytose and subsequently transcytose the bacteria from the lumen to the submucosal side of the epithelial barrier (48). Once S. flexneri cells reach the submucosa, they are engulfed by resident macrophages (48). S. flexneri cells escape from the macrophage phagosome and kill the macrophage quickly, thus providing access to the basolateral side of the epithelial barrier (22, 36, 53, 62). S. flexneri cells invade the colonic epithelial cells at the basolateral membrane and, following invasion, are found inside a membrane-bound vacuole (48). S. flexneri cells escape from this vacuole like they escape from the macrophage phagosome; however, instead of killing the epithelial cells, S. flexneri cells replicate in the cytoplasm and spread cell to cell throughout the colonic epithelium (48).
Another aspect of S. flexneri pathogenesis is the disruption of tight junctions, which leads to the loss of cell-to-cell contact between polarized epithelial cells of the colon (45). One result of this disruption is facilitation of the transepithelial migration of polymorphonuclear leukocytes (PMN) to the luminal side of the epithelial barrier (31, 41, 48). Lipopolysaccharide and unidentified factors activate the mitogen-activated protein kinase kinase/extracellular signal-regulated kinase signaling pathway (MEK/ERK pathway), as measured by the phosphorylation of ERK1/2 (21). In turn, the phosphorylation of ERK1/2 and its localization to the nucleus result in the production of signaling molecules required to recruit PMN into the lumen (21, 59). PMN migration through the colonic epithelium may also provide additional openings to the submucosa for S. flexneri (41, 48). As infection progresses, increased tight junction disruption and PMN recruitment lead to severe inflammation and to the destruction of the epithelial barrier associated with dysentery (21, 31, 41, 45, 48).
S. flexneri possesses a 218-kb plasmid that harbors most of its virulence determinants (2, 49). A 31-kb region of this plasmid, termed the mxi-spa locus, encodes the proteins necessary to assemble a type III secretion system (T3SS) (40, 50). The primary function of the T3SS is to secrete proteins, called effectors, into the host to modify cell function and overcome cell defenses (15, 34). In S. flexneri pathogenesis, the T3SS is required for invasion, vacuolar escape, cell-to-cell spread, and PMN transepithelial migration (5, 17, 31, 48).
The transcription and expression of the S. flexneri T3SS are induced by the VirF/VirB system when the temperature is shifted from 30°C to 37°C (54). The expression of the first set of effector proteins secreted by S. flexneri is activated by VirB, and these proteins are essential for invasion (25, 50). Secretion from the S. flexneri T3SS is dependent on contact with the host cell, specifically an interaction between IpaB and cholesterol (4, 12, 57). However, secretion can also be artificially induced in liquid growth media (1).
A second set of putative S. flexneri effectors are designated OspB to OspG (outer Shigella protein) (2). These proteins are secreted by S. flexneri into the growth medium in a ΔipaB deletion mutant background (2), but most have undefined functions. Some osp genes appear to be the result of gene duplication during evolution, and each gene is at least >70% homologous to the other genes in the same group (2). Sequencing of the virulence plasmid from S. flexneri serotype 5 revealed three ospD genes (ospD1 to ospD3), four ospC genes (ospC1 to ospC4), and two ospE genes (ospE1 and ospE2). Interestingly, osp gene regulation varies, even between homologues. For example, ospD1 and ospD2 are up-regulated solely by VirB, while ospD3 is regulated solely by MxiE, a separate transcription factor active only when S. flexneri is inside the host cell (17, 25, 30). Finally, a third group of osp genes, which includes ospF and ospC1, is regulated by both VirB and MxiE, suggesting that these proteins are required throughout the course of infection (25).
The purpose of this study was to identify the contribution of the ospF and ospC1 genes to S. flexneri pathogenesis. We determined that the OspF and OspC1 proteins are indeed secreted by the T3SS, and we elucidated their localization inside the host cell. Furthermore, we generated ospF and ospC1 deletion mutants to examine the impact of these genes on phenotypes associated with virulence. While ospF and ospC1 deletion mutations did not obstruct invasion or cell-to-cell spreading, these mutations did interfere with postinvasion aspects of S. flexneri pathogenesis. OspC1 was found to be required for the inflammation and swelling characteristics associated with a positive Serény reaction in an animal model. Both OspF and OspC1 were essential for Shigella-induced transepithelial PMN migration and the up-regulation of the MEK/ERK pathway that is required for PMN recruitment.
MATERIALS AND METHODS
Strains and growth conditions.
All bacterial strains used in this study are listed in Table 1. Escherichia coli and Salmonella enterica strains used for molecular biology applications were routinely cultured in Luria-Bertani broth (LB). S. flexneri was routinely cultured in tryptic soy broth. All bacteria grown overnight were incubated at 37°C with aeration. Unless indicated otherwise, antibiotics were used in growth media at the following concentrations: kanamycin, 50 μg/ml; chloramphenicol, 25 μg/ml; spectinomycin, 100 μg/ml; streptomycin, 25 μg/ml; and ampicillin, 200 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype and/or description | Reference or source |
|---|---|---|
| S. flexneri strains | ||
| 2457T | Wild-type Shigella flexneri 2a | 7 |
| BS103 | Virulence plasmid-cured derivative of 2457T | 30 |
| BS652 | 2457T/Δspa47 (spa47::aadA), Specr | Lab stock |
| BS766 | 2457T transformed with pKM208 Ampr | This study |
| BS771 | 2457T/ΔospF (ospF::cat), Cmr | This study |
| BS772 | 2457T/ΔospC1 (ospC1::cat), Cmr | This study |
| BS784 | 2457T transformed with pDZ2, Cmr | This study |
| BS785 | BS103 transformed with pDZ2, Cmr | This study |
| BS792 | 2457T transformed with pDZ3, Cmr | This study |
| BS793 | BS103 transformed with pDZ3, Cmr | This study |
| BS794 | BS652 transformed with pDZ3, Cmr | This study |
| BS801 | 2457T transformed with pDZ7, Cmr | This study |
| BS814 | BS771 with cat eliminated using FLP recombinase | This study |
| BS815 | BS814 transformed with pDZ2, Cmr | This study |
| BS816 | BS772 with cat eliminated using FLP recombinase | This study |
| BS817 | BS817 transformed with pDZ3, Cmr | This study |
| E. coli strains | ||
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 (ΔlacZYA-argF)U169 deoR φ80dlacZΔM15 | 10 |
| BL21/pLysS | F−ompT hsdSB(rB− mB−) dcm gal λ(DE3) pLysS(Cmr) tonA | Novagen |
| S. enterica 1344 | Wild-type S. enterica serovar Typhimurium 1344 hisG46 | 13 |
| Plasmids | ||
| pKD3 | bla cat oriR6K | 3 |
| pKM208 | Temperature-sensitive red-, gam-, and lacI-expressing plasmid driven by PTac promoter, Ampr | 35 |
| pACB C-2HA | PipB2-2HA fusion, pACYC184 backbone, Cmr | 20 |
| pDZ1 | Cloning intermediate | This study |
| pDZ2 | OspF-2HA expression driven by POspB, Cmr | This study |
| pDZ3 | OspC1-2HA fusion | This study |
| pDZ7 | IpaH9.8-2HA fusion | This study |
| pGEX 6P-1 | Protein expression vector results in GST fusion, Ampr | Amersham |
| pGEX::OspF | OspF cloned into pGEX 6P-1 with BamHI/XhoI, Ampr | This study |
| pGEX::OspC1 | OspC1 cloned into pGEX 6P-1 with BamHI/XhoI, Ampr | This study |
| pBAD24 | Arabinose-inducible vector, pBR322ori, Ampr | 8 |
| pBAD24::OspF | OspF cloned into NcoI/BamHI sites of pBAD24 | This study |
| pBAD24::OspC1 | OspC1 cloned into NcoI/BamHI sites of pBAD24 | This study |
| pDsRed2-C1 | Red fluorescent protein vector, Kanr | Clontech |
| pEGFP-C1 | Green fluorescent protein vector, Kanr | Clontech |
| pRFP::OspF | ospF N-terminal RFP fusion | This study |
| pEGFP::OspC1 | ospC1 N-terminal GFP fusion | This study |
| pGFP-actin | GFP-actin fusion | 58 |
| pEGFP-tub | GFP-tubulin fusion | Clontech |
Tissue culture.
HeLa cells and L2 mouse fibroblast cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The J774.1 murine macrophage cell line was maintained in DMEM containing 10% FBS supplemented with 2 mM glutamine and essential amino acids. The human epithelial colon cancer-derived cell line T84 (passages 46 to 66) was maintained in DMEM/F-12 supplemented with 15 mM HEPES (pH 7.5) and 10% FBS. To obtain polarized monolayers, T84 cells were grown on 0.33- or 4.7-cm2 collagen-coated permeable polycarbonate filters (Costar) that had pore sizes of 5.0 and 3.0 μm, respectively, and they were utilized after they reached a confluent and differentiated state, as previously described (5, 21, 31). All tissue culture media were acquired from Invitrogen, and all cell lines were maintained in the presence of 5% CO2 at 37°C.
Plasmid construction.
All plasmids and primers used in this study are described in Tables 1 and 2. S. flexneri genes were amplified by PCR using Vent polymerase (New England Biolabs), cloned into pGEM-T (Promega), and sequenced. ospF was amplified with BamHI and Acc65I sites engineered at the 5′ end of its forward primer and a BglII site engineered at the 3′ end of the reverse primer. ospC1 and ipaH9.8 were cloned into pGEM-T with Acc65I and BglII restriction enzyme sites engineered into the amplifying primers. To make the pBAD constructs, both ospF and ospC1 were excised from pGEM-T by NcoI digestion and BglII digestion and cloned into pBAD24 cut with the same enzymes.
TABLE 2.
Primers used in this study
| Purpose | Forward primer
|
Reverse primer
|
||
|---|---|---|---|---|
| Designation | Sequencea | Designation | Sequencea | |
| Amplify the cassette for deletion of ospF | OspFMutF | TATATCTATTTTATAGAGGACGTTTTCTATGCCCATAAAAAAGCCCTGTTGTGTAGGCTGGAGCTGCTTC | OspFMutR | GCTACAAGGTGGTGTAGCTGGCATCTTCTCTACTCTATCATCAAACGATACATATGAATATCCTCCTTAG |
| Amplify the cassette for deletion of ospC1 | OspC1MutF | ATTAAAACTGTTTTCATATAAGGTTCATTTTATGAATATATCAGAAACATGTGTAGGCTGGAGCTGCT | OspC1MutR | CTTTTGCTAAACGATATTCAATTTTGATTAAATATATTTATTGTCAGATGCATATGAATATCCTCCTTAG |
| Clone POspB into pDZ1 | OspBProF | AATAGGGGATCCTACCTGACGCTT | OspBProR | CTACATGGATCCGACTCCATATATTG |
| Clone ospF into pGEM-T | OspFF | GGATCCGGTACCATGCCCATAAAAAAGCCCTG | OspFR | AGATCTCTCTATCATCAAACGATAAAATGG |
| Clone ospC1 into pGEM-T | OspC1For | GGTACCATGAATATATCAGAAACACTG | OspC1Rev | AGATCTAATATATTTATTGTCAGATGTTG |
| Clone ipaH9.8 into pGEM-T | IpaH9.8F | GGTACCATGTTACCGATAAATAATAAC | IpaH9.8R | AGATCTTGAATGGTGCAGTTGTGAGCCG |
| Clone spvC into pGEM-T | SpvCF | GGTACCATGCCCATAAATAGGCCTAATC | SpvCR | AGATCTCTCTGTCATCAAACGATAAAACG |
Underlining identifies an engineered Acc65I, BglII, or EcoRI restriction site.
To make the tagged fusion plasmids, ospF was liberated from pGEM-T using the SapI and BglII restriction enzymes and was cloned into pACB C2-HA that was cut with SalI (blunted with T4 DNA polymerase) and BglII. The resulting vector was designated pDZ1. The ospB promoter, at positions −300 to −1 upstream of ospB, was amplified with primers with BamHI ends and cloned into the BamHI site engineered into pDZ1. The resulting plasmid was designated pDZ2. ospC1 and ipaH9.8 were subcloned into pDZ2 digested with Acc65I and BglII.
For cloning into pDsRed2-C1 or EGFP-C1, ospF or ospC1 was excised from pGEM-T using the Acc65I and BglII restriction enzymes. The fragments were ligated into the Acc65I and BamHI sites found in the multicloning site of both red fluorescent protein (RFP) and green fluorescent protein (GFP) vectors. The spvC gene from Salmonella enterica serovar Typhimurium 1344 was amplified by PCR with Acc65I and BglII restriction sites engineered at the 5′ and 3′ ends as well and was cloned into pGEM-T. spvC was digested with these restriction enzymes and cloned into pDsRed2-C1 in the same way that ospF was cloned.
Mutant construction.
The ΔospF and ΔospCI deletions in S. flexneri were generated using a modification of the method of Datsenko and Wanner (3), where the lambda red recombination genes, red and gam, were driven by the PTAC promoter on the pKM208 plasmid (35). Briefly, PCR was used to generate a chloramphenicol resistance cassette gene (cat) with sequences at the 5′ and 3′ ends identical to sequences 20 bp internal to and 30 bp upstream of the gene being deleted (Table 2). BS766 (S. flexneri carrying pKM208) was grown overnight at 30°C, subcultured in LB without NaCl, and grown at 30°C. When late log phase (optical density at 600 nm [OD600], 0.9 to 1.0) was reached, isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was added to the medium to induce expression of the lambda red recombination genes, and the bacteria were shifted to 37°C for 30 min. After heat shock at 42°C and electroporation (35), bacteria were recovered in SOC medium and plated on tryptic soy broth plates containing Congo red (CR) and chloramphenicol at a concentration of 5 μg/ml or 10 μg/ml. Cmr colonies on these plates were purified and screened by PCR using three different primer sets to identify the deletion mutants, and positive recombinants were verified by sequencing. To remove cat from BS771 and BS772, these strains were transformed with pCP20 and incubated at 42°C (3) to generate BS814 and BS816. We verified that all mutations that were created did not have an effect on S. flexneri growth.
Congo red secretion assay.
Bacterial cultures following overnight incubation were subcultured and grown at 37°C. Once late log phase (OD600, 0.7 to 0.8) was reached, bacterial samples were normalized to the same OD600, and CR (0.7 μg/ml) was added. After 1 h, whole-cell lysates and supernatants were prepared. Whole-cell lysates were generated by centrifugation of 1 ml of bacterial culture, followed by one wash with ice-cold 1× phosphate-buffered saline (PBS). Cells were centrifuged again and resuspended in equal amounts of Tris-glycine gel sample buffer (24). CR supernatants were generated by centrifugation of 20 ml of S. flexneri grown in liquid medium. The supernatants were removed, centrifuged again, and passed through a 0.45-μm-pore-size filter (Millipore). T3SS secreted proteins were precipitated by addition of trichloroacetic acid (final concentration, 10%), and this was followed by acetone washing and resuspension of the pellet in 50 μl gel sample buffer.
OspF antibody production.
Glutathione S-transferase (GST)-OspF was generated by cloning the ospF gene from pGEM-T into the BamHI/XhoI sites of the pGEX 6P-1 vector (Amersham) and transforming the preparation into BL21/pLysS. Subsequently, GST-OspF was expressed by addition of IPTG (1 mM) and was purified from bacterial lysates using glutathione beads (Amersham). OspF was cleaved from GST using PreScission protease (Amersham), purified, and dialyzed against 1× PBS. The molecular mass of this purified OspF was ∼27.5 kDa, as determined by Coomassie blue staining (data not shown), and 200 μg/ml was injected into New Zealand White rabbits every other week for a total of 6 weeks. Bleeding of rabbits and serum purification were carried out 8 and 10 weeks after the initial injection (Spring Valley Antibodies).
Virulence assays.
S. flexneri invasion assays were carried out as previously described (9), and CFU were counted and compared to the amount of input bacteria to calculate the invasion efficiency. Plaque assays were performed as previously described (38). Briefly, L2 fibroblast monolayers were grown to confluence and infected with S. flexneri strains whose concentrations were standardized to an OD600 of 0.30. Two dilutions of bacteria were used, and the assay was performed at least twice in triplicate. Plaques were enumerated after 3 days. The Serény test was used to assess invasion and the in vivo inflammatory response in guinea pigs as previously described (52). Briefly, 2.5 × 108 CFU of wild-type S. flexneri, BS771, or BS772 was used to infect one guinea pig eye. At least three guinea pigs were used to evaluate each strain used for each experiment, and symptoms were monitored for 4 days. Experiments were repeated at least twice.
To assess macrophage killing, ∼5 × 104 J774.1 macrophages were seeded and grown on acid-washed (0.1 N HCl) 12-mm coverslips overnight in 24-well tissue culture plates. Bacteria were subcultured, grown to late log phase, and added to wells at a multiplicity of infection (MOI) of ∼200. After 1 h, macrophage death was evaluated using a Live/Dead kit (Invitrogen), and the numbers of dead cells (orange, ethidium) and live cells (green, fluorescein) per 100 total visible cells were determined. This assay was done a minimum of two times in triplicate.
The PMN migration assay was performed as previously described (31). Shigella cells were added to the basolateral side of 0.33-cm2 transwells at an MOI of ∼100 for 90 min. Cells were washed and then incubated in fresh media containing gentamicin (50 μg/ml) for 90 min. PMN were added to the basolateral compartment, and transmigration to the apical compartment was quantified by assaying for the PMN azurophilic granule marker myeloperoxidase. For inducible complementation of deletion mutations, arabinose was added at a final concentration of 0.6% to tissue culture media. Unpaired Student t tests were used to analyze statistical significance in comparisons between the mutant strains and 2457T.
ERK1/2 phosphorylation was detected as previously described (21). Briefly, polarized T84 monolayers on 4.7-cm2 transwells were infected and harvested with lysis buffer (1% Triton X-100, 100 mM NaCl, 10 mM HEPES, 2 mM EDTA, 4 mM Na3VO4, 40 mM NaF, 200 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail [Roche]). Samples were centrifuged, and the supernatant representing the cytosol was saved and stored at −80°C until it was used.
Polyacrylamide gel electrophoresis and immunoblot analysis of proteins.
For protein analysis, samples were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on Tris-glycine gels (24). For immunoblotting, proteins were transferred to pure nitrocellulose membranes, and hemagglutinin (HA)-tagged proteins were detected by addition of mouse anti-HA monoclonal antibody HA.11 (Covance). ERK1/2 samples were immunoblotted using mouse monoclonal antibody against phosphorylated ERK1/2 and goat polyclonal antibody against ERK1/2 (Santa Cruz). Bands were visualized using sheep anti-mouse and sheep anti-goat secondary antibodies conjugated to horseradish peroxidase (Amersham). Rabbit anti-OspF antibody was detected using donkey anti-rabbit polyclonal antibody conjugated to horseradish peroxidase (Amersham). All primary and secondary antibodies were used at a 1:1,000 dilution. All blots were developed using Visualizer (Upstate), and images were captured with a charge-coupled device camera from the LAS-3000 CH imaging system (Fuji) or on film.
Immunofluorescence and transfection analysis.
A total of ∼5.0 × 104 HeLa cells were seeded and grown on acid-treated, 12-mm coverslips overnight in 24-well tissue culture plates. S. flexneri was subcultured and grown at 37°C until late log phase was reached. Bacterial cell numbers were normalized to an OD600 of 0.72, washed, and resuspended in 1 ml of DMEM. Bacteria were added to cells at an MOI of ∼200 bacteria/HeLa cell. The 24-well plates were centrifuged at 37°C for 10 min at 3,000 × g and incubated at 37°C in the presence of 5% CO2 for 30 min. Then cells were washed twice with PBS, followed by the addition of DMEM with gentamicin (50 μg/ml). After 90 min, cells were washed three times with PBS, and DMEM with gentamicin was added again. Cells were washed once with PBS and fixed by addition of 3% paraformaldehyde for 10 min at 37°C for each time.
After fixation, infected HeLa cells were analyzed by immunofluorescence. Briefly, cells were washed twice with PBS and then blocked and permeabilized with 10% natural goat serum (NGS) and 0.1% saponin, respectively, for 30 min at room temperature. Cells were washed once with PBS and incubated in a PBS solution containing mouse anti-HA monoclonal antibody HA.11 (Covance), 10% NGS, and 0.1% saponin for 1 h. After three PBS washes, cells were incubated with goat anti-mouse antibody conjugated to Alexa Fluor 488 (Invitrogen), 10% NGS, and 0.1% saponin for 1 h. The primary antibody was used at a 1:1,000 dilution, and the secondary antibody was used at a 1:800 dilution. Cells were washed three times with PBS and once with sterile distilled H2O before they were mounted on slides using Anti-Fade reagent (Invitrogen).
For transfection experiments, HeLa cells were seeded as described above in 24-well plates. Then 1.0 μg of RFP-OspF, GFP-OspC1, pDs-Red2, pEGFP-C1, GFP-actin, or pEGFP-tub DNA was complexed with GeneJammer reagent and incubated with cells according to the manufacturer's specifications (Stratagene). At 24 h posttransfection, cells were washed twice with PBS and fixed by addition of 3% paraformaldehyde for 10 min at 37°C. HeLa cells were occasionally stained with 4′,6′-diamidino-2-phenylindole (DAPI) (0.5 μg/ml) for 20 min or with anti-beta-tubulin antibody conjugated to fluorescein isothiocyanate (FITC) (Sigma) at a 1:50 dilution for 1 h. All cell images were acquired with an Olympus 1X81 fluorescent microscope using a SensiCam charge-coupled device camera (Cooke).
RESULTS
Secretion of OspF and OspC1 is dependent on the S. flexneri T3SS.
While it was previously shown by N-terminal protein sequencing that OspF and at least one OspC protein are secreted by S. flexneri (2), we wanted to determine if OspF and OspC1 secretion is T3SS dependent. Therefore, we constructed a plasmid in which the ospF and ospC1 genes were cloned in frame with a two-hemagglutinin (2HA) tag. Transcription of ospF and ospC1 was driven by the ospB promoter (POspB), which was cloned upstream of both genes. We chose POspB to drive expression because this promoter is activated to the highest degree inside eukaryotic cells compared to the activation of other MxiE-regulated genes (17).
The OspF-2HA plasmid (pDZ2) and the OspC1-2HA plasmid (pDZ3) were transformed into 2457T to produce BS784 and BS792, respectively, and were transformed into BS103 (a strain with a virulence plasmid-cured background) to produce BS785 and BS793, respectively (Table 1). The pDZ3 plasmid was also transformed into a Δspa47 mutant (BS652) to generate BS794 in order to evaluate the direct role of the T3SS in OspC1 secretion. Spa47 is the ATPase of the S. flexneri T3SS and is required for a functional T3SS (16). In order to evaluate OspF secretion in a Δspa47 mutant background, a rabbit polyclonal antibody against recombinant OspF was generated. However, cross-reactivity with other OspC proteins (OspC2 to OspC4) prevented the use of anti-OspC1 antibody in a similar experiment.
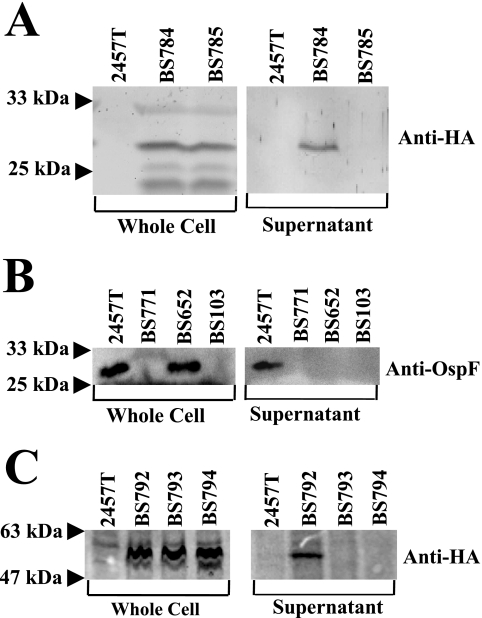
2457T, BS784 (OspF-2HA in 2457T), BS103, and BS785 (OspF-2HA in BS103) were grown separately in medium containing CR to induce the T3SS. After 1 h of exposure to CR, OspF-2HA (∼29 kDa) was found in the whole-cell and supernatant fractions of BS784; in contrast, no signal was detected in the supernatant of BS785 (Fig. 1A). These results verify that OspF secretion requires the virulence plasmid and that the 2HA tag does not inhibit the secretion of this protein.
FIG. 1.
T3SS-dependent secretion of OspF and OspC1. Congo red was added to a final concentration of 7 μg/ml to activate the secretion of T3SS effectors. After 1 h, two sets of samples were saved: whole cell fractions and supernatant. OspF samples were run on a 12% SDS-PAGE gel, and OspC1 samples were run on a 10% SDS-PAGE gel. (A) Samples were immunoblotted with anti-HA antibody to visualize OspF-2HA (∼29-kDa band). (B) Samples were immunoblotted with anti-OspF antibody (∼27.5-kDa band). (C) Samples were immunoblotted with anti-HA antibody to visualize OspC1-2HA (∼54-kDa band).
To verify our 2HA results and to confirm the involvement of the T3SS, 2457T, BS652 (Δspa47), and BS103 were grown in the presence of CR and analyzed using anti-OspF antibody. BS771, a ΔospF deletion mutant, was used as a negative control for the antibody. OspF was detected in only the supernatant of the 2457T sample (Fig. 1B), and no OspF was detected in the supernatant of BS652 (Δspa47), which confirmed that the T3SS is required for secretion of OspF.
Analysis of OspC1-2HA yielded results similar to the OspF secretion profile. OspC1-2HA (BS792) was secreted only when the T3SS was functional (Fig. 1C), and addition of the 2HA tag to the C terminus also had no effect on OspC1 secretion. A BS103 (virulence plasmid-cured) background (BS793) or a deletion in spa47 (BS794) abrogated the secretion of OspC1-2HA (Fig. 1C), which implicated OspC1 as a T3SS effector as well. It should be noted that the total protein secreted was analyzed by Coomassie blue staining, and no discernible difference was observed between the overall secretion profiles of 2457T and the ΔospF and ΔospC1 mutants, which indicates that mutations in ospF and ospC1 do not appear to affect the secretion of other T3SS effectors (data not shown).
Virulence phenotypes of ΔospF and ΔospC1 mutants. (i) Invasion assay.
To gain insight into the function of the S. flexneri effectors, OspF and OspC1, we used the lambda red recombination system to generate allelic exchange mutants, where the target gene was deleted and replaced with a Cmr cassette. The resulting mutants, BS771 (ΔospF) and BS772 (ΔospC1), were then characterized to determine their S. flexneri virulence phenotypes. First, we compared the mutant strains to wild-type parent strain 2457T with a standard invasion assay using HeLa cells. In a typical experiment, 2457T showed an invasion efficiency of 0.32% ± 0.04%. In the same assay, BS771 had an invasion efficiency of 0.37% ± 0.06%, and BS772 had an invasion efficiency of 0.33% ± 0.05%. We also performed an invasion assay using polarized T84 cells, and no difference in invasion efficiency was observed between BS771, BS772, and 2457T (data not shown). Therefore, deletion of either ospF or ospC1 did not alter the invasion of epithelial cells by S. flexneri in either a nonpolarized or polarized tissue culture model.
(ii) Plaque assay.
In order to assess the ability of the mutant strains to invade, replicate, and spread cell to cell, BS771 and BS772 were examined with the plaque assay (38). Again, no significant differences were observed between the mutants and 2457T when the numbers of plaques or plaque sizes were compared. MxiE regulates the transcription of ospF and ospC1, and a ΔmxiE mutant was previously shown to produce fewer plaques and plaques having reduced sizes (17). Therefore, the loss of OspF or OspC1 does not contribute to this ΔmxiE phenotype.
(iii) Macrophage killing.
S. flexneri rapidly causes cell death in infected macrophages (36, 62). As a model system, J774.1 murine macrophage cells in tissue culture are killed by S. flexneri after 1 h of infection (36, 62). We incubated J774.1 macrophages for 1 h with BS771, BS772, and 2457T and measured cell death using fluorescent reporters. Both BS771 and BS772 retained the ability to kill J774.1 macrophages similar to wild-type bacteria (data not shown).
(iv) Serény test.
The Serény test (52) was used to analyze the effects of the ΔospF and ΔospC1 mutations on S. flexneri infection in vivo. Guinea pig eyes were exposed to BS771, BS772, and 2457T at a dose of 2.5 × 108 CFU. After 4 days, no significant differences between the BS771- and 2457T-infected guinea pigs were observed, as both groups developed conjunctivitis and a strong inflammatory response (data not shown). However, in all three guinea pigs infected with the BS772 strain the production of symptoms of conjunctivitis was delayed and there was less swelling and inflammation compared to the guinea pigs infected with 2457T. In fact, one of the BS772-infected guinea pigs cleared the bacterial infection after 2 days (data not shown). These results demonstrate that OspC1 contributes to S. flexneri virulence when this animal model of infection is used.
(v) PMN transepithelial migration.
Not only does PMN transepithelial migration provide a mechanism for S. flexneri access to the basolateral side of the epithelial barrier, but neutrophil recruitment is also responsible for the inflammation associated with disease as modeled in the Serény test (41, 48). The PMN transepithelial migration assay has been used previously to demonstrate that a functional T3SS of S. flexneri is required for neutrophil migration (21, 31). In light of the reduced inflammation with the ΔospC1 strain in the Serény test and the fact that OspF and OspC1 are T3SS effectors, we wanted to assess the ability of the ΔospF and ΔospC1 strains to induce PMN transepithelial migration in a polarized tissue culture assay.
BS771 and BS772 were used to infect a monolayer of polarized T84 cells and compared to 2457T. BS103 served as a negative control. After 180 min of infection, neutrophils were added to the basolateral side of the transwell, and PMN transepithelial migration was evaluated. We found that when both deletion mutant strains were normalized to 2457T, there was a statistically significant decrease (P ≤ 0.0001) in the amount of neutrophils that migrated from the basolateral compartment to the apical compartment of the polarized monolayer (Fig. 2). We constructed plasmids with ospF or ospC1 cloned into the pBAD24 vector (Table 1) and transformed these plasmids into BS771 and BS772 to complement the deletion mutations. The PMN migration assay was repeated in the presence of arabinose to induce expression of the cloned ospF and ospC1 genes, and the complementing strains (BS802 and BS803, respectively) restored PMN migration to levels that were equal to or greater than the wild-type levels. We also complemented ospF and ospC1 deletion mutants (BS814 and BS816) with pDZ2 and pDZ3 (2HA-tagged proteins) and obtained similar results (data not shown), suggesting that the tagged versions of these proteins are still functional. These results suggest that OspF and OspC1 are essential for the signaling events that stimulate PMN transepithelial migration.
FIG. 2.
Transepithelial PMN migration induced by wild-type S. flexneri. T84 polarized monolayers were infected with 2457T, BS771 (ΔospF), BS772 (ΔospC1), BS802(pBAD-OspF), BS803(pBAD-OspC1), and BS103, and then PMN migration was evaluated. HBSS, uninfected monolayers (buffer only). All strains were normalized to wild-type strain 2457T. Experiments were performed three times in triplicate. The data are means ± standard deviations (error bars) of triplicate samples and represent one of the three experiments performed in which similar results were obtained. Samples were compared using an unpaired t test, and statistically significant differences are indicated by an asterisk.
OspF and OspC1 contribute to Shigella-induced ERK1/2 phosphorylation.
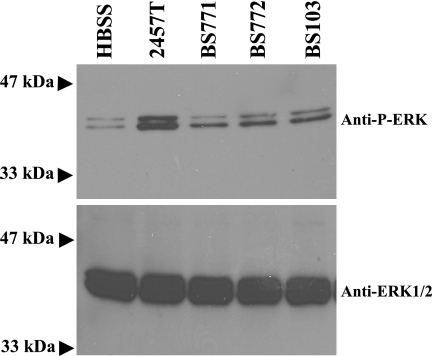
S. flexneri synthesizes factors during the course of infection that initiate a host signaling cascade in which a downstream target, the ERK1/2 protein, becomes phosphorylated on threonine residues (5, 21). Phosphorylated ERK1/2 (P-ERK) has at least 160 substrates in both the cytosol and nucleus that regulate genes or proteins that are associated with proliferation, differentiation, the cytoskeleton, and cell cycle progression (59). Some S. flexneri mutants that show a reduction in the PMN migration assay also have reduced MEK/ERK pathway activation, as measured by the phosphorylation of ERK1/2 (5, 21). Since both the ΔospF and ΔospC1 mutants displayed reduced activity in the PMN migration assay, we predicted that we would also observe a reduction in MEK/ERK signaling in cells infected with these mutants. Therefore, we infected polarized T84 cells with 2457T, BS771, BS772, and BS103 and directly compared the amounts of P-ERK by Western blotting using a monoclonal antibody specific for the phosphorylated form of the ERK1/2 protein. Both BS771- and BS772-infected cells showed a significant reduction in the amount of P-ERK (Fig. 3). Densitometric analysis indicated that there were decreases of 62% and 43% in phosphorylated ERK1/2 for BS771- and BS772-infected cells, respectively, compared to cells infected with wild-type S. flexneri. Cells infected with BS103, the noninvasive mutant, displayed a 45% decrease in phosphorylated ERK1/2 compared to the amount in wild-type infections. These results suggest that the OspF and OspC1 proteins are secreted into the cytoplasm of the host cell and play a role in MEK/ERK pathway activation.
FIG. 3.
ERK phosphorylation in S. flexneri-infected cells. T84 polarized monolayers were infected with 2457T, BS771 (ΔospF), BS772 (ΔospC1), and BS103, and then ERK phosphorylation was evaluated 180 min postinvasion. HBSS, uninfected monolayers (buffer only). Lysates of monolayers were immunoblotted with monoclonal anti-P-ERK antibody (anti-P-ERK), which recognized the phosphorylated ERK1/2 proteins (p44 and p42), or anti-ERK1/2 antibody (anti-ERK1/2), which was a total protein control and demonstrated equal loading. The blot is representative of one experiment that was repeated three times.
OspF and OspC1 T3SS effectors target the nucleus of the host cell.
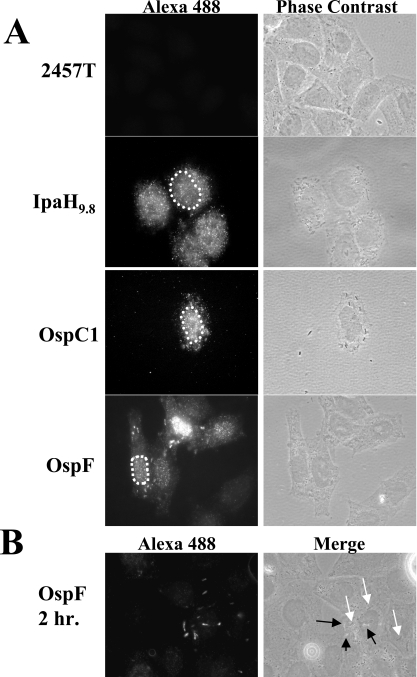
Since the 2HA tag fusions of OspF and OspC1 were secreted via the T3SS, we used BS784 and BS792 to determine the intracellular localization of OspF and OspC1 in HeLa cells infected with these strains. As a control, the gene encoding IpaH9.8 was also cloned into the pDZ2 vector (Table 1), generating a 2HA fusion. IpaH9.8 is an MxiE-regulated T3SS effector of S. flexneri that localizes to the nucleus and cytoplasm of HeLa cells (17, 25, 55). We confirmed that IpaH9.8 was secreted by the T3SS and that the 2HA tag did not disrupt its secretion (data not shown).
Strains that harbored the plasmids with 2HA-tagged effectors were used to infect HeLa cells, and immunofluorescence using anti-HA antibody was used to evaluate protein localization over time after invasion. Untransformed 2457T (no 2HA plasmid) served as a negative control. The first time analyzed was 4 h following invasion because IpaH9.8 is found in the nucleus and cytoplasm by this time (55). At 4 h postinvasion, IpaH9.8-2HA was found in both the cytoplasm and the nucleus (Fig. 4A), as previously shown for native IpaH9.8 (55). OspC1-2HA was found in the nucleus of host cells, similar to IpaH9.8, as well as in the vicinity of invading bacteria (Fig. 4A). Similarly, OspF-2HA also localized to the nucleus at this time (Fig. 4A), suggesting that the nucleus is a common destination for some of the MxiE-regulated effectors of S. flexneri.
FIG. 4.
Localization of OspF-2HA and OspC1-2HA in infected host cells. (A) Strains 2457T, BS801 (IpaH9.8-2HA), BS792 (OspC1-2HA), and BS784 (OspF-2HA) were used to infect semiconfluent monolayers of HeLa cells. After 4 h, cells were fixed and evaluated by immunofluorescence using anti-HA antibody and a secondary goat anti-mouse antibody conjugated to Alexa Fluor 488. The images in the left panels are representative of the immunofluorescence signals observed with each strain, and the images in the right panels are phase-contrast images of the same cells. The nucleus is outlined by a dotted line in some images. (B) The experiment described above was repeated, but preparations were fixed and stained at 2 h. The OspF-2HA signal (black arrows) did not colocalize with bacteria (white arrows). The panel on the left shows the immunofluorescence signal. The panel on the right is a phase-contrast image of the same cells merged with the immunofluorescence signal.
We also observed another signal with OspF-2HA that was localized in the cytoplasm. Subsequently, the OspF-2HA signal was evaluated at 2, 3, 4, and 5 h postinvasion. At 2 h postinvasion, we observed a signal with the anti-HA antibody only in the cytoplasm, and no nuclear localization was observed (Fig. 4B). At 3, 4, and 5 h postinvasion, we observed that the nuclear signal became more intense with greater time, suggesting that there was accumulation of OspF in the nucleus (data not shown). We did not observe any nuclear OspC1-2HA signal before 4 h, suggesting that the majority of the protein was too diffuse in the cytoplasm or that not enough protein was produced by this point to observe a signal (data not shown). However, based on these results we believe that cytoplasmic OspF is targeted to the nucleus, where it accumulates over time.
Ectopically expressed RFP-OspF and GFP-OspC1 display nuclear localization.
To confirm the nuclear localization of OspF and OspC1 using a different tag, we cloned ospF and ospC1 into RFP and GFP mammalian expression vectors to generate N-terminal fusions of fluorescent proteins to OspF and OspC1. Vectors were transiently transfected into a semiconfluent monolayer of HeLa cells, and after 24 h, cells were fixed and analyzed. RFP-OspF was found in the nucleus of HeLa cells, and counterstaining with DAPI confirmed the nuclear localization (Fig. 5). The nuclear localization of the RFP-OspF fusion not only supported the results obtained with Shigella-infected cells but also demonstrated that additional S. flexneri T3SS effectors are not required for OspF localization to the nucleus. The same was true for OspC1 as the GFP-OspC1 signal was found throughout the transfected cell, including the nucleus and cytoplasm (Fig. 5). We also observed that the nuclear signals for both RFP-OspF and GFP-OspC1 increased as the time posttransfection increased (data not shown), suggesting that both proteins accumulate in the nucleus.
FIG. 5.
Ectopic expression of RFP-OspF and GFP-OspC1 in HeLa cells. Semiconfluent monolayers of HeLa cells were transfected with mammalian expression vectors RFP-OspF, GFP-OspC1, and RFP-SpvC. Following transfection, the nuclei were stained with DAPI (left panels), and the GFP or RFP fluorescence of the fusion proteins was evaluated (right panels). In each panel the nucleus is outlined by a dotted line.
SpvC, a protein encoded on the virulence plasmid of S. enterica serovar Typhimurium, exhibits 63% homology with full-length OspF (2, 26). When the C termini (amino acids 100 to 237) of SpvC and OspF were compared, more than 80% homology was detected (BLAST2). We postulated that SpvC has a localization similar to that of OspF inside the host cell because the N-terminal differences could most likely be attributed to secretion signals for the T3SS (34). Therefore, spvC was cloned into the same mammalian expression vector as OspF to generate an RFP-SpvC N-terminal fusion. Surprisingly, RFP-SpvC did not target the nucleus and displayed a punctate pattern in the cytoplasm of transfected HeLa cells (Fig. 5).
RFP-OspF colocalizes with microtubules in HeLa cells.
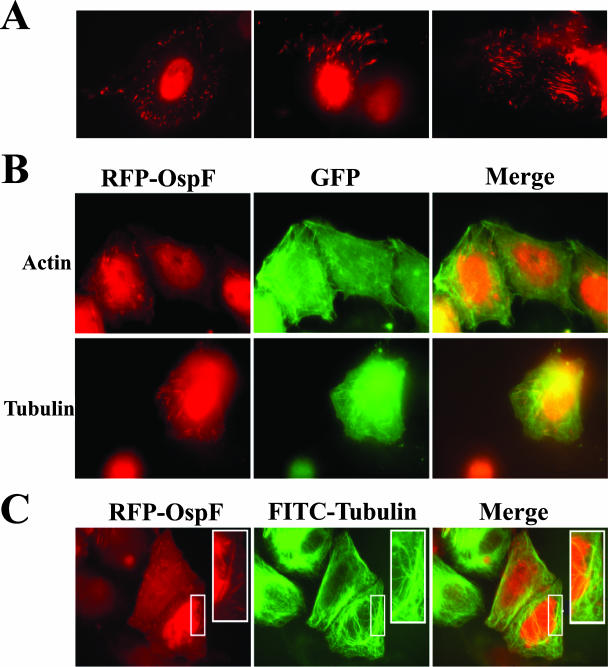
When analyzing the other focal planes of RFP-OspF-transfected cells by microscopy, we noticed signals in the cytoplasm. These signals had different characteristics, occasionally displaying a punctate pattern of localization and sometimes resembling cytoskeletal filaments (Fig. 6A). RFP-OspF signal was also found on these structures before the nuclear localization was observed, suggesting that there was initial targeting in the cytoplasm before nuclear targeting. We wanted to identify these structures because the localization could aid in discovering a host cell mechanism that OspF exploits during S. flexneri infection.
FIG. 6.
RFP-OspF localizes to a target in the cytoplasm. (A) HeLa cells were transfected with RFP-OspF. Each panel shows a different field of cells. (B) HeLa cells were cotransfected with RFP-OspF and GFP-actin or with RFP-OspF and GFP-tubulin. The panels on the left show the RFP-OspF signal (red), the panels in the middle show the GFP signal (green), and the panels on the right show the merged signals. (C) HeLa cells were transfected with RFP-OspF, fixed, and stained with anti-beta-tubulin conjugated to FITC. The panel on the left shows the RFP-OspF signal (red), the panel in the middle shows anti-beta-tubulin FITC signal (green), and the panel on the right shows the merged signals. The insets show magnified areas of interest. The micrographs are representative of the results of experiments repeated at least three times.
Shigella, along with many other gram-negative pathogens, modulates cytoskeletal components (actin and microtubules) during infection (48, 60). Therefore, we cotransfected HeLa cells with RFP-OspF and either GFP-actin or GFP-tubulin plasmids. When signals were merged, RFP-OspF and GFP-actin did not have similar localizations (Fig. 6B); however, the GFP-tubulin signal did colocalize with the filament-like signal of RFP-OspF (Fig. 6B). This result was verified by transfecting HeLa cells with RFP-OspF and after 24 h fixing the cells and staining them with anti-beta-tubulin conjugated to FITC. Again, we observed colocalization of the RFP-OspF signal with the anti-tubulin signal (Fig. 6C).
DISCUSSION
A multitude of T3SS effectors have been identified on the S. flexneri virulence plasmid that contribute to invasion of the host epithelial cell. However, only a few genes have been shown to play a role in the postinvasion aspects of S. flexneri pathogenesis; these genes are icsA, mxiE, ospG, and ipaH (6, 17, 19, 39, 48). Recent studies have shown that a large number of putative S. flexneri T3SS effectors encoded on the virulence plasmid are regulated by MxiE, including many of the Osp proteins (17, 25, 30). It appears that most of the Osp proteins are secreted by the bacteria (2); however, the function of the majority of these proteins is still unknown. It is clear from this study that OspF and OspC1 function after invasion with respect to S. flexneri pathogenesis similar to other MxiE-regulated proteins (6, 19, 25, 39).
It was shown previously that the T3SS is essential for Shigella-induced PMN transepithelial migration (31); however, the effectors involved in this phenomenon were not identified. In this study, we identified two T3SS effectors, OspF and OspC1, that contribute to the PMN transepithelial migration phenotype of Shigella virulence. Moreover, the deficiency in PMN migration was directly correlated to a deficiency in activation of the MEK/ERK signaling pathway in cells infected with ΔospC1 and ΔospF mutants. When we evaluated the ΔospC1 and ΔospF mutants with the Serény test, the ΔospC1 mutant caused reduced inflammation and swelling, and the infection did not advance as quickly as a wild-type S. flexneri infection. In fact, one guinea pig cleared the infection with the ΔospC1 mutant. These observations show that the S. flexneri T3SS effectors OspF and OspC1 are required for essential postinvasion aspects of virulence associated with S. flexneri infection.
Modulation of the MEK/ERK pathway by T3SS effectors is a common characteristic of infection by gram-negative pathogens. Salmonella spp. secrete SptP that interacts with the Raf protein upstream of MEK and ERK1/2, resulting in down-regulation of the MEK/ERK pathway during the early stages of infection (27). Yersinia spp. secrete a protease called YopJ that deubiquinates a number of targets in the cytoplasm and, consequently, leads to down-regulation of the MEK/ERK pathway (61). Finally, a functional T3SS of Bordetella bronchiseptica is required for down-regulation of the MEK/ERK pathway (43). However, these are examples of MEK/ERK down-regulation. The results that we report here suggest that the S. flexneri effectors OspF and OspC1 are required for up-regulation of the MEK/ERK pathway. While enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC) have been shown to activate the MEK/ERK pathway, this has not be attributed to T3SS effectors (51). It was recently shown that S. enterica serovar Typhimurium macrophage infection activates the MEK/ERK pathway in a Salmonella pathogenicity island 2-dependent manner; however, the T3SS effector responsible is unknown (56). Therefore, OspF and OspC1 represent the first T3SS effectors shown to play a role in MEK/ERK pathway activation.
Carboxy-terminal and amino-terminal protein tagging strategies demonstrated that both OspF and OspC1 are T3SS-dependent effectors that localize to the nucleus and cytoplasm of HeLa cells. Nuclear localization is also not a novel phenotype for secreted bacterial proteins. There are two distinct classes of proteins secreted by gram-negative bacteria that localize and function within the nucleus of the host cell. First, some bacteria secrete proteins that target the nucleus to modulate the cell cycle of the eukaryotic cell. These proteins have been classified as cyclomodulins and include proteins such as cytolethal distending toxin (secreted by many gram-negative species) and Cif of EPEC and EHEC (37). While we have not observed any evidence that OspF and OspC1 are cyclomodulins (Zurawski and Maurelli, unpublished), we still cannot conclude that these proteins have an effect on cell division.
A second class of gram-negative secreted proteins that target the nucleus of the host cell have a role in suppressing the inflammatory response of the immune system typically caused by lipopolysaccharide. For example, YopM of Yersinia pestis localizes to the nucleus and reduces the amount of interleukin-15 (IL-15) secreted by host cells (18). Xanthomonas campestris pathovar vesicatoria, a pathogen of tomato and pepper plants, secretes XopD that targets the nucleus and hydrolyzes small ubiquitin-related modifier-conjugated proteins to interfere with the plant defense response (14). Finally, IpaH9.8 from S. flexneri (55) and its homologue SspH1 from Salmonella (11) also localize to the nucleus of mammalian cells. IpaH9.8 and SspH1 both down-regulate the inflammatory response postinvasion (11). Specifically, IpaH9.8 binds to the human UAF35 splicing factor to interfere with the transcription of a number of inflammatory response genes that are up-regulated during Shigella invasion (39). These genes include the IL-8, RANTES, colony-stimulating factor 1, and IL-1β genes. The down-regulation of the innate immune response postinvasion is also the function of another Shigella Osp protein, OspG. OspG interacts with ubiquinated proteins to prevent phospho-IκBα degradation and NF-κB activation induced by tumor necrosis factor alpha stimulation (19). While it is possible that OspF and OspC1 modulate cytokine production given their nuclear localization, we did not see a decrease in IL-8 secretion levels in cells infected with the ΔospF or ΔospC1 mutants (unpublished results).
The ectopic expression of RFP-OspF allowed greater fluorescence intensity to visualize OspF and its dynamic localization inside HeLa cells, particularly its colocalization with microtubules. Cytoskeletal targets are a common target for modulation by bacterial pathogens and for Shigella T3SS effectors. IpaH9.8 requires microtubules for nuclear import, and VirA disrupts microtubules to enhance invasion (55, 60). OspF may use microtubules to mediate its transport to the nucleus, like IpaH9.8. On the other hand, the localization of OspF to microtubules could be the result of an interaction with a host target protein(s). Many eukaryotic proteins involved in organelle transport and cell division interact with microtubules, but the fact most relevant to this study is that MEK and ERK1/2 interact with microtubules (44, 46). Because MEK/ERK pathway components and OspF have a common localization and because Shigella-induced ERK1/2 phosphorylation requires OspF, it is possible that OspF directly interacts with components of the MEK/ERK pathway. We are currently investigating this possibility.
Homologues of ospF are found in at least two other gram-negative bacterial species (2, 26). One of these homologues is spvC, which is found on the pSLT virulence plasmid of S. enterica serovar Typhimurium and resides in an operon that encodes the T3SS effector SpvB (28). SpvC is required for full S. enterica serovar Typhimurium virulence in mice (28), but nothing else is known about its function. Pseudomonas syringae DC3000 also has an OspF homologue, HopAI1, which has recently been shown to be involved in the down-regulation of the innate immune response in the tomato plant, and overexpression of transfected copies of the hopAI1 gene generates disease-like symptoms in plants (26). However, the contribution of HopAI1 is unclear and may be redundant, as a deletion mutation does not result in a measurable difference in the plant's immune response (26). The immune response of a guinea pig infected with the ΔospF mutant of S. flexneri also displayed no discernible difference in the Serény test compared to the immune response of a guinea pig infected with 2457T. Therefore, OspF and HopAI1 may both have redundant functions in their hosts, and a more sensitive assay to measure the effect of mutation on virulence is required. For example, an altered virulence phenotype for the ΔospF mutant was not observed until we used the PMN migration assay. Additional studies are required to characterize similar and different functions of SpvC, HopAI1, and OspF.
OspC1 exhibits about 75% homology with the OspC2, OspC3, and OspC4 proteins of S. flexneri (2). This level of similarity also occurs for the OspC genes from other Shigella spp. However, the contribution of the OspC2, OspC3, and OspC4 proteins to virulence after invasion may be different from the contribution of OspC1 for three reasons. First, ospC2, ospC3, and ospC4 are regulated solely by VirB and not by MxiE (25). Therefore, there should be more OspC1 than its homologues inside the host cells because of the MxiE activation (17, 25, 30). Second, OspC2 to OspC4 are 96% homologous to each other, while OspC1 is more divergent evolutionarily (2). Third, T84 cells infected with a ΔospC2 mutant do not show a reduction in PMN transepithelial migration in the way that the ΔospC1 strain does (unpublished observations). It is conceivable that OspC proteins have similar but redundant functions and that OspC1 has the greatest effect on the host. Evidence that supports redundancy comes from the fact that ospC4 found in S. flexneri serotype 5 M90T has a point mutation that generates a stop codon, perhaps suggesting that S. flexneri is evolving to remove the other OspC effectors (2).
In conclusion, in this study we found for the first time that OspF and OspC1 are T3SS effectors secreted by S. flexneri and that these effectors are required for Shigella-induced MEK/ERK pathway activation and PMN transepithelial migration. PMN transepithelial migration is an important aspect not only of Shigella virulence but also of Salmonella, EPEC, and EHEC virulence (32, 51). The results of this study also underscore the fact that T3SS effectors can mediate virulence phenotypes shared by enteric pathogens. Therefore, future studies of molecular mechanisms by which OspF and OspC1 induce host cell signaling not only should improve our understanding of Shigella virulence but also may highlight a paradigm that is shared by other gram-negative, pathogenic bacteria.
Acknowledgments
We express our gratitude to the Olivia Steele-Mortimer laboratory (National Institutes of Allergy and Infectious Diseases, Rocky Mountain Laboratories, Montana), especially Leigh Knodler, for the gift of the pACB C-2HA vector. We acknowledge the Giam laboratory (Uniformed Services University of the Health Sciences, Bethesda, MD) for the gift of the GFP and RFP vectors and the Sanger laboratory (SUNY Upstate Medical University, Syracuse, NY) for the gift of the GFP-actin and GFP-tubulin vectors. We also thank Nancy Adams and Reinaldo Fernandez for technical assistance and D. Scott Merrell for critical reading of the manuscript.
This work was supported by National Institutes of Allergy and Infectious Diseases grant AI24656. B.A.M. was supported by National Institutes of Health grants DK56754 and DK33506. K.L.M. was supported by a T32 training grant sponsored by Harvard Medical School and the Division of Nuclear Medicine at Massachusetts General Hospital.
Editor: J. B. Bliska
REFERENCES
- 1.Bahrani, F. K., P. J. Sansonetti, and C. Parsot. 1997. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect. Immun. 65:4005-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. D'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enninga, J., J. Mounier, P. Sansonetti, and G. Tran Van Nhieu. 2005. Secretion of type III effectors into host cells in real time. Nat. Methods 2:959-965. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez, I. M., M. Silva, R. Schuch, W. A. Walker, A. M. Siber, A. T. Maurelli, and B. A. McCormick. 2001. Cadaverine prevents the escape of Shigella flexneri from the phagolysosome: a connection between bacterial dissemination and neutrophil transepithelial signaling. J. Infect. Dis. 184:743-753. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Prada, C. M., D. L. Hoover, B. D. Tall, A. B. Hartman, J. Kopelowitz, and M. M. Venkatesan. 2000. Shigella flexneri IpaH(7.8) facilitates escape of virulent bacteria from the endocytic vacuoles of mouse and human macrophages. Infect. Immun. 68:3608-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Formal, S. B., G. J. Dammin, E. H. Labrec, and H. Schneider. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J. Bacteriol. 75:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hale, T. L., and S. B. Formal. 1981. Protein synthesis in HeLa or Henle 407 cells infected with Shigella dysenteriae 1, Shigella flexneri 2a, or Salmonella typhimurium W118. Infect. Immun. 32:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 11.Haraga, A., and S. I. Miller. 2003. A Salmonella enterica serovar Typhimurium translocated leucine-rich repeat effector protein inhibits NF-kappa B-dependent gene expression. Infect. Immun. 71:4052-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayward, R. D., R. J. Cain, E. J. McGhie, N. Phillips, M. J. Garner, and V. Koronakis. 2005. Cholesterol binding by the bacterial type III translocon is essential for virulence effector delivery into mammalian cells. Mol. Microbiol. 56:590-603. [DOI] [PubMed] [Google Scholar]
- 13.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 14.Hotson, A., R. Chosed, H. Shu, K. Orth, and M. B. Mudgett. 2003. Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50:377-389. [DOI] [PubMed] [Google Scholar]
- 15.Hueck, C. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouihri, N., M. P. Sory, A. L. Page, P. Gounon, C. Parsot, and A. Allaoui. 2003. MxiK and MxiN interact with the Spa47 ATPase and are required for transit of the needle components MxiH and MxiI, but not of Ipa proteins, through the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 49:755-767. [DOI] [PubMed] [Google Scholar]
- 17.Kane, C. D., R. Schuch, W. A. Day, Jr., and A. T. Maurelli. 2002. MxiE regulates intracellular expression of factors secreted by the Shigella flexneri 2a type III secretion system. J. Bacteriol. 184:4409-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerschen, E. J., D. A. Cohen, A. M. Kaplan, and S. C. Straley. 2004. The plague virulence protein YopM targets the innate immune response by causing a global depletion of NK cells. Infect. Immun. 72:4589-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, D. W., G. Lenzen, A. L. Page, P. Legrain, P. J. Sansonetti, and C. Parsot. 2005. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA 102:14046-14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knodler, L. A., J. Celli, W. D. Hardt, B. A. Vallance, C. Yip, and B. B. Finlay. 2002. Salmonella effectors within a single pathogenicity island are differentially expressed and translocated by separate type III secretion systems. Mol. Microbiol. 43:1089-1103. [DOI] [PubMed] [Google Scholar]
- 21.Kohler, H., S. P. Rodrigues, and B. A. McCormick. 2002. Shigella flexneri interactions with the basolateral membrane domain of polarized model intestinal epithelium: role of lipopolysaccharide in cell invasion and in activation of the mitogen-activated protein kinase ERK. Infect. Immun. 70:1150-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koterski, J. F., M. Nahvi, M. M. Venkatesan, and B. Haimovich. 2005. Virulent Shigella flexneri causes damage to mitochondria and triggers necrosis in infected human monocyte-derived macrophages. Infect. Immun. 73:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotloff, K. L., J. P. Winickoff, B. Ivanoff, J. D. Clemens, D. L. Swerdlow, P. J. Sansonetti, G. K. Adak, and M. M. Levine. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. W. H. O. 77:651-666. [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Le Gall, T., M. Mavris, M. C. Martino, M. L. Bernardini, E. Denamur, and C. Parsot. 2005. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology 151:951-962. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., H. Lin, W. Zhang, Y. Zou, J. Zhang, X. Tang, and J. M. Zhou. 2005. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA 102:12990-12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, S. L., T. X. Le, and D. S. Cowen. 2003. SptP, a Salmonella typhimurium type III-secreted protein, inhibits the mitogen-activated protein kinase pathway by inhibiting Raf activation. Cell. Microbiol. 5:267-275. [DOI] [PubMed] [Google Scholar]
- 28.Matsui, H., C. M. Bacot, W. A. Garlington, T. J. Doyle, S. Roberts, and P. A. Gulig. 2001. Virulence plasmid-borne spvB and spvC genes can replace the 90-kilobase plasmid in conferring virulence to Salmonella enterica serovar Typhimurium in subcutaneously inoculated mice. J. Bacteriol. 183:4652-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurelli, A. T., B. Blackmon, and R. Curtiss III. 1984. Loss of pigmentation in Shigella flexneri 2a is correlated with loss of virulence and virulence-associated plasmid. Infect. Immun. 43:397-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mavris, M., A. L. Page, R. Tournebize, B. Demers, P. Sansonetti, and C. Parsot. 2002. Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol. Microbiol. 43:1543-1553. [DOI] [PubMed] [Google Scholar]
- 31.McCormick, B. A., A. M. Siber, and A. T. Maurelli. 1998. Requirement of the Shigella flexneri virulence plasmid in the ability to induce trafficking of neutrophils across polarized monolayers of the intestinal epithelium. Infect. Immun. 66:4237-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCormick, B. A., S. I. Miller, D. Carnes, and J. L. Madara. 1995. Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infect. Immun. 63:2302-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moran, G. J. 2002. Threats in bioterrorism. II. CDC category B and C agents. Emerg. Med. Clin. N. Am. 20:311-330. [DOI] [PubMed] [Google Scholar]
- 34.Mota, L. J., I. Sorg, and G. R. Cornelis. 2005. Type III secretion: the bacteria eukaryotic cell express. FEMS Microbiol. Lett. 252:1-10. [DOI] [PubMed] [Google Scholar]
- 35.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nonaka, T., T. Kuwabara, H. Mimuro, A. Kuwae, and S. Imajoh-Ohmi. 2003. Shigella-induced necrosis and apoptosis of U937 cells and J774 macrophages. Microbiology 149:2513-2527. [DOI] [PubMed] [Google Scholar]
- 37.Nougayrede, J. P., F. Taieb, J. De Rycke, and E. Oswald. 2005. Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 13:103-110. [DOI] [PubMed] [Google Scholar]
- 38.Oaks, E. V., T. L. Hale, and S. B. Formal. 1985. Plaque formation by virulent Shigella flexneri. Infect. Immun. 48:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okuda, J., T. Toyotome, N. Kataoka, M. Ohno, H. Abe, Y. Shimura, A. Seyedarabi, R. Pickersgill, and C. Sasakawa. 2005. Shigella effector IpaH9.8 binds to a splicing factor U2AF(35) to modulate host immune responses. Biochem. Biophys. Res. Commun. 333:531-539. [DOI] [PubMed] [Google Scholar]
- 40.Parsot, C., R. Menard, P. Gounon, and P. J. Sansonetti. 1995. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol. Microbiol. 16:291-300. [DOI] [PubMed] [Google Scholar]
- 41.Perdomo, J. J., P. Gounon, and P. J. Sansonetti. 1994. Polymorphonuclear leukocyte transmigration promotes invasion of colonic epithelial monolayers by Shigella flexneri. J. Clin. Investig. 93:633-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pickering, L. K. 2004. Antimicrobial resistance among enteric pathogens. Semin. Pediatr. Infect. Dis. 15:71-77. [DOI] [PubMed] [Google Scholar]
- 43.Reissinger, A., J. A. Skinner, and M. H. Yuk. 2005. Downregulation of mitogen-activated protein kinases by the Bordetella bronchiseptica type III secretion system leads to attenuated nonclassical macrophage activation. Infect. Immun. 73:308-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reszka, A. A., R. Seger, C. D. Diltz, E. G. Krebs, and E. H. Fischer. 1995. Association of mitogen-activated protein kinase with the microtubule cytoskeleton. Proc. Natl. Acad. Sci. USA 92:8881-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakaguchi, T., H. Kohler, X. Gu, B. A. McCormick, and H. C. Reinecker. 2002. Shigella flexneri regulates tight junction-associated proteins in human intestinal epithelial cells. Cell. Microbiol. 4:367-381. [DOI] [PubMed] [Google Scholar]
- 46.Samarakoon, R., and P. J. Higgins. 2002. MEK/ERK pathway mediates cell-shape-dependent plasminogen activator inhibitor type 1 gene expression upon drug-induced disruption of the microfilament and microtubule networks. J. Cell Sci. 115:3093-3103. [DOI] [PubMed] [Google Scholar]
- 47.Sandler, R. S., J. E. Everhart, M. Donowitz, E. Adams, K. Cronin, C. Goodman, E. Gemmen, S. Shah, A. Avdic, and R. Rubin. 2002. The burden of selected digestive diseases in the United States. Gastroenterology 122:1500-1511. [DOI] [PubMed] [Google Scholar]
- 48.Sansonetti, P. J. 2001. Microbes and microbial toxins: paradigms for microbial-mucosal interactions. III. Shigellosis: from symptoms to molecular pathogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 280:G319-G323. [DOI] [PubMed] [Google Scholar]
- 49.Sansonetti, P. J., D. J. Kopecko, and S. B. Formal. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasakawa, C., K. Komatsu, T. Tobe, T. Suzuki, and M. Yoshikawa. 1993. Eight genes in region 5 that form an operon are essential for invasion of epithelial cells by Shigella flexneri 2a. J. Bacteriol. 175:2334-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savkovic, S. D., A. Koutsouris, and G. Hecht. 1996. Attachment of a noninvasive enteric pathogen, enteropathogenic Escherichia coli, to cultured human intestinal epithelial monolayers induces transmigration of neutrophils. Infect. Immun. 64:4480-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serény, B. 1957. Experimental keratoconjunctivitis shigellosa. Acta Microbiol. Acad. Sci. Hung. 4:367-376. [PubMed] [Google Scholar]
- 53.Suzuki, T., K. Nakanishi, H. Tsutsui, H. Iwai, S. Akira, N. Inohara, M. Chamaillard, G. Nunez, and C. Sasakawa. 2005. A novel caspase-1/Toll-like receptor 4-independent pathway of cell death induced by cytosolic Shigella in infected macrophages. J. Biol. Chem. 280:14042-14050. [DOI] [PubMed] [Google Scholar]
- 54.Tobe, T., S. Nagai, N. Okada, B. Adler, M. Yoshikawa, and C. Sasakawa. 1991. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5:887-893. [DOI] [PubMed] [Google Scholar]
- 55.Toyotome, T., T. Suzuki, A. Kuwae, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, T. Toyofuku, M. Hori, and C. Sasakawa. 2001. Shigella protein IpaH(9.8) is secreted from bacteria within mammalian cells and transported to the nucleus. J. Biol. Chem. 276:32071-32079. [DOI] [PubMed] [Google Scholar]
- 56.Uchiya, K., and T. Nikai. 2005. Salmonella pathogenicity island 2-dependent expression of suppressor of cytokine signaling 3 in macrophages. Infect. Immun. 73:5587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watarai, M., T. Tobe, M. Yoshikawa, and C. Sasakawa. 1995. Contact of Shigella with host cells triggers release of Ipa invasins and is an essential function of invasiveness. EMBO J. 14:2461-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Westphal, M., A. Jungbluth, M. Heidecker, B. Muhlbauer, C. Heizer, J. M. Schwartz, G. Marriott, and G. Gerisch. 1997. Microfilament dynamics during cell movement and chemotaxis monitored using a GFP-actin fusion protein. Curr. Biol. 7:176-183. [DOI] [PubMed] [Google Scholar]
- 59.Yoon, S., and R. Seger. 2006. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors 24:21-44. [DOI] [PubMed] [Google Scholar]
- 60.Yoshida, S., E. Katayama, A. Kuwae, H. Mimuro, T. Suzuki, and C. Sasakawa. 2002. Shigella deliver an effector protein to trigger host microtubule destabilization, which promotes Rac1 activity and efficient bacterial internalization. EMBO J. 21:2923-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou, H., D. M. Monack, N. Kayagaki, I. Wertz, J. Yin, B. Wolf, and V. M. Dixit. 2005. Yersinia virulence factor YopJ acts as a deubiquitinase to inhibit NF-kappa B activation. J. Exp. Med. 202:1327-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zychlinsky, A., M. C. Prévost, and P. J. Sansonetti. 1992. Shigella flexneri induces apoptosis in infected macrophages. Nature 358:167-169. [DOI] [PubMed] [Google Scholar]