Abstract
Streptococcus agalactiae is part of the normal flora of the human gastrointestinal tract and also the leading cause of bacterial infections in human newborns and immunocompromised adults. The colonization and infection of different regions within the human host require a regulatory network in S. agalactiae that senses environmental stimuli and controls the formation of specific virulence factors. In the present study, we characterized an Rgg-like transcriptional regulator, designated RovS (regulator of virulence in Streptococcus agalactiae). Deletion of the rovS gene in the genome of S. agalactiae resulted in strain 6313 ΔrovS, which exhibited an increased attachment to immobilized fibrinogen and a significant increase in adherence to the eukaryotic lung epithelial cell line A549. Quantification of expression levels of known and putative S. agalactiae virulence genes by real-time PCR revealed that RovS influences the expression of fbsA, gbs0230, sodA, rogB, and the cyl operon. The altered gene expression in mutant 6313 ΔrovS was restored by plasmid-mediated expression of rovS, confirming the RovS deficiency as the cause for the observed changes in virulence gene expression in S. agalactiae. DNA electrophoretic mobility shift assays showed that RovS specifically binds to the promoter regions of fbsA, gbs0230, sodA, and the cyl operon, indicating that RovS directly regulates their expression. Deletion and mutation studies in the promoter region of fbsA, encoding the main fibrinogen receptor in S. agalactiae, identified a RovS DNA motif. Similar motifs were also found in the promoter regions of gbs0230, sodA, and the cyl operon, and alignments allowed us to propose a consensus sequence for the DNA-binding site of RovS.
Streptococcus agalactiae is a commensal bacterium that colonizes the gastrointestinal tract of a significant percentage of the human population (1). However, it is also the leading cause of invasive infections in neonates, in which it causes septicemia, meningitis, and pneumonia (50, 52). In addition, it has emerged as an increasingly common cause of invasive diseases in the elderly and in immunocompromised persons.
For the establishment of a successful infection, S. agalactiae needs to adhere to epithelial surfaces, eventually penetrate them, and also protect itself against the immune system. For this purpose, S. agalactiae possesses several surface proteins that interact with proteins of the host extracellular matrix (ECM). The Lmb protein from S. agalactiae mediates the binding of the bacteria to human laminin (41). Beckman et al. and Cheng et al. demonstrated binding of C5a peptidase from S. agalactiae to fibronectin and showed that fibronectin binding of S. agalactiae is important for the invasion of the bacteria into host cells (2, 11). Binding of S. agalactiae to human fibrinogen is brought about by the fibrinogen receptor FbsA, which interacts with fibrinogen by repetitive units and which is widely distributed in different S. agalactiae strains (38). FbsA was also demonstrated to mediate the binding of S. agalactiae to lung epithelial cells (39). In general, surface proteins play a significant role in the virulence of S. agalactiae and their synthesis is clearly regulated during infection.
Recently, the genomic sequences of the serotype III S. agalactiae strain NEM316 (16) and of the serotype V strain 2603 V/R (46) were published. Analysis of the sequence data revealed the presence of several putative virulence genes, including genes for putative bacterial surface proteins and virulence regulators. Although a few regulatory systems from S. agalactiae have been studied on the molecular level (12, 31, 40), the targets and stimuli of most transcriptional regulators are still unknown. Among gram-negative bacteria, virulence regulatory genes have been studied intensively, and it could be shown that many of these are members of several large conserved protein families. Much less is known about virulence regulatory networks in gram-positive bacteria. However, the recent influx of genomic information has revealed large conserved families of regulators that are unique to gram-positive bacteria. One such example is the Rgg protein family of transcriptional regulators. This family includes Rgg of Streptococcus gordonii, which is necessary for extracellular glycosyltransferase G expression (43, 44); GadR of Lactococcus lactis, which is required for glutamate-dependent acid tolerance (37); and MutR, which controls the expression of the mutacin l antibiotic (MutA) of Streptococcus mutans (34). Rgg of Streptococcus pyogenes, also known as RopB, is necessary for the expression of the secreted cysteine protease SpeB (6, 27). Subsequent genome analysis revealed that Rgg from S. pyogenes influences the expression of additional secreted proteins (7, 8). In addition, rgg inactivation changed the expression of known and putative transcriptional regulators, including several two-component regulatory systems, which are important in the transcriptional response to changing environmental conditions (7).
The present study describes a protein, named RovS, with similarity to members of the Rgg-like protein family of transcriptional regulators. By deletion of the rovS gene in the chromosome of S. agalactiae, the importance of RovS for the binding of the bacteria to human fibrinogen and the adherence to eukaryotic cells was addressed. Using real-time PCR, the expression of known and putative virulence genes was compared between the rovS mutant and its parental strain. Finally, to further characterize the RovS-dependent regulation, the binding of RovS to the promoter region of fbsA, encoding the main fibrinogen receptor in S. agalactiae, and other virulence genes was analyzed by electrophoretic mobility shift assays (EMSAs). We were able to identify the RovS binding motif in the fbsA promoter region, and we therefore propose a consensus sequence for the RovS DNA-binding site.
MATERIALS AND METHODS
Bacterial strains, eukaryotic cells, and growth conditions.
S. agalactiae 6313 is a serotype III clinical isolate obtained from an infected neonate (48). Escherichia coli DH5α (18) was used for cloning purposes, and E. coli BL21(DE3) (14) served as host for the production of a RovS fusion protein. S. agalactiae was cultivated at 37°C in Todd-Hewitt yeast broth (THY) containing 1% yeast extract. S. agalactiae clones, carrying the plasmid pG+host6 (Appligene) or pAT19 (47), were selected with erythromycin (5 μg/ml). E. coli was grown at 37°C in Luria broth (LB), and clones carrying the plasmid pG+host6, pAT19, or pET28a were selected in the presence of ampicillin (100 μg/ml), erythromycin (300 μg/ml), or kanamycin (50 μg/ml). Growth experiments with S. agalactiae were performed in THY and were monitored by determining the optical density of the culture at 600 nm (OD600). Cell line A549 (ATCC CCL-185) was obtained from the American Type Culture Collection. A549 is a human lung carcinoma cell line which has many characteristics of type II alveolar pneumocytes. A549 cells were propagated in RPMI tissue culture medium (Gibco BRL) supplemented with 10% fetal calf serum. Tissue cultures were incubated in a humid atmosphere at 37°C with 5% CO2.
Antibodies and human proteins.
Peroxidase-labeled goat anti-mouse immunoglobulin G Fab fragments and monoclonal anti-His-tag antibodies were purchased from Dianova (Hamburg, Germany) and Roche Diagnostics (Penzberg, Germany), respectively. Human fibrinogen, collagen I, collagen IV, and laminin were obtained from Sigma-Aldrich (Steinheim, Germany).
General DNA techniques.
Chromosomal S. agalactiae DNA was isolated according to the method of Pospiech and Neumann (30). Conventional techniques for DNA manipulation, such as restriction enzyme digests, PCR, ligation, transformation by electroporation, and Southern blotting, were performed as described by Sambrook et al. (35). Oligonucleotides were purchased from MWG Biotech GmbH (Ebersberg, Germany).
Construction of a rovS deletion mutant.
The rovS gene was deleted in the chromosome of S. agalactiae 6313 according to a previously described procedure (36). In brief, two DNA fragments flanking the rovS gene were amplified by PCR from the genome of S. agalactiae 6313 with the primers rovS_del1 (5′ GAGCGGGGTACCGTTGAGTGAATGAGTTGAC) and rovS_del2 (5′ CCCATCCACTAAACTTAAACATTCTGACTCTCCTCTCTC) and with the primers rovS_del3 (5′ TGTTTAAGTTTAGTGGATGGGCTTGGTGATACTTCTTCAGG) and rovS_del4 (5′ CCGCGGATCCAAAGAAGATACTTCCCTCG). cDNA sequences in the primers rovS_del2 and rovS_del3 are marked in italics, and the KpnI and BamHI restriction sites in the primers rovS_del1 and rovS_del4 are underlined. The rovS-flanking PCR products were mixed in equal amounts with each other and subjected to a crossover PCR with the primers rovS_del1 and rovS_del4, resulting in one PCR product that carried the two rovS-flanking regions. The crossover PCR product and the thermosensitive vector pG+host6 were digested with KpnI and BamHI, ligated, and transformed into E. coli DH5α. The resulting plasmid, pG+ΔrovS, was electroporated into S. agalactiae 6313, and the resultant strains were grown at 37°C in the presence of erythromycin selection and subsequently passaged at 30°C in the absence of erythromycin selection as described previously (36). Successful deletion of the gene rovS was confirmed by Southern blotting with PvuII-digested chromosomal DNA of the S. agalactiae parental strains and of the putative deletion mutant. A digoxigenin-labeled probe for the detection of deletions in rovS was obtained by PCR with the primers rovS_SB1 (5′ GCTTGACCAGTTACATACC) and rovS_SB2 (5′ ATGCAAAGGCATCACA GC). Southern blot analysis confirmed the successful deletion of the rovS gene in the chromosome of the S. agalactiae ΔrovS strain (data not shown).
Plasmid-mediated expression of rovS in S. agalactiae.
The rovS structural gene, including its own promoter, was amplified from chromosomal S. agalactiae 6313 DNA by PCR using the primers rovSK1 (5′ GAGCGGGGTACCGTTGAGTGAATGAGTTGAC) and rovSK2 (5′ CCGCGGATCCAAAGAAGATACTTCCCTCG 3′). The KpnI and BamHI sites used for cloning are underlined. The PCR product was cut with KpnI and BamHI and ligated into the KpnI/BamHI-digested E. coli/Streptococcus shuttle vector pAT19 (47). The resultant plasmid was termed pATrovS. The plasmids pAT19 and pATrovS were transformed by electroporation into the corresponding S. agalactiae strains with subsequent erythromycin selection.
Construction of the fbsA-lacZ transcriptional fusion plasmid pTfbsA.
Plasmid pTCV-lac (33) was used for expression analysis of the fbsA gene in S. agalactiae. Plasmid pTfbsA was constructed as described previously (36).
Construction of the plasmid for synthesis of RovS fusion protein.
The vector pET28a (Novagen) was used for the synthesis of a hexahistidyl-tagged RovS fusion protein. The rovS gene was amplified from chromosomal DNA of S. agalactiae 6313 by PCR with the primers rovS_N1 (5′ CATGCCATGGAAAAAGAATTAGGAAAAACACTAAGAAG) and rovS_C1 (5′ CCGCTCGAGGCATTCTTTATTATTGCCAAGTACTTTTTG). The NcoI and XhoI restriction sites used for cloning are underlined. After digestion of the rovS-derived PCR products and of plasmid pET28a with NcoI and XhoI, the rovS derivate was ligated into pET28a and transformed into E. coli BL21(DE3).
The RovS fusion protein was synthesized in recombinant E. coli BL21(DE3) by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) after the culture had reached an OD600 of about 1.0. The cells were harvested, washed once with His-tag buffer B (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0), and then disrupted using a French press cell. Purification of the fusion protein was performed according to the instructions of QIAGEN (Hilden, Germany) using Ni2+ affinity chromatography. The RovS fusion protein was finally dialyzed against 1× binding buffer (10 mM Tris-HCl [pH 7.8], 5% glycerol, 10 mM NaCl, 5 mM dithiothreitol, 1 mM MgCl2, 1 mM EDTA).
Western blot analysis.
For Western blot experiments, proteins were size separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted onto nitrocellulose. The membrane was subsequently blocked overnight with 1% (wt/vol) blocking reagent (Roche Diagnostics) in phosphate-buffered saline (PBS). The membrane was subsequently incubated for 1 h with mouse anti-His-tag antibodies (1:250 in PBS). Afterwards, the membrane was washed three times with PBS. The membrane was subsequently incubated for 1 h with peroxidase-labeled anti-mouse immunoglobulin G Fab fragments (1:10,000). To remove unbound antibodies, the membrane was washed three times with PBS containing 0.05% (wt/vol) Tween 20 and twice with PBS. Finally, peroxidase-conjugated antibodies were detected by chemiluminescence, using the ECL kit from Amersham Biosciences (Braunschweig, Germany).
Size-exclusion chromatography.
A total of 250 μl of protein solution at a concentration of 0.3 mg/ml in running buffer (50 mM phosphate buffer, 150 mM NaCl, pH 7.2) was loaded onto a Superdex 200 HR 10/30 (Amersham Biosciences) gel filtration column connected to an AKTA fast protein liquid chromatography system (Amersham Biosciences). Protein separation at a flow rate of 1 ml/min was monitored by detecting the absorbance (A280 and A219) of the eluant, which was collected in fractions of 1 ml. The column was calibrated using high-molecular-weight and low-molecular-weight gel filtration calibration kits (Amersham Biosciences). The molecular weight calculation of RovS protein was determined from plots of the logarithm of molecular weights of standards versus retention volume. To clearly identify RovS in single fractions, the RovS protein was detected in Western blot analysis with mouse anti-His-tag antibodies.
RNA preparation and LightCycler real-time PCR.
S. agalactiae 6313 was grown to mid-log phase in 50 ml of THY. Total RNA was prepared from the bacterial culture with the RNeasy Midi extraction kit (QIAGEN) and treated for 1 h with 150 U of RNase-free DNase (Promega). RNA samples were checked for DNA contamination by PCR without prior reverse transcription (RT). As no amplicons were obtained, DNA contamination during RNA preparation could be excluded. Quantitative real-time PCR experiments were performed after RT of RNA with random hexanucleotides and the RevertAid first-strand cDNA synthesis kit (MBI Fermentas) according to the instructions of the manufacturer. Expression analysis of the genes fbsA, gbs0230, sodA, and rogB and the cyl operon was performed with the primers listed in Table 1 in a LightCycler as described previously (17). The quantity of cDNA for the investigated genes was normalized to the quantity of gyrA cDNA in each sample. Each experiment was performed at least four times with two independent RNA preparations.
TABLE 1.
Primers used for quantitative real-time PCR using a LightCycler
| Gene tested | Primer (5′ to 3′)
|
|
|---|---|---|
| Forward | Reverse | |
| cylE | GAGGAGACAGAAATTAGGAC | TCTTGTCCAAAGGTTGGC |
| fbsA | CCGCGGATCCGTAGGTCAACTTATAGG | CCGCGGATCCATTATACTTAATTTTCATTGCG |
| gbs0230 | AAGAGACGTAGCTGGGTTAG | TCCTCGACTATCCCCTTTAG |
| gyrA | GACGTTCAGGTATTCAC | TCAAACTGAGGTACGACG |
| cfb | TGAGGCTATTACTAGCGTGG | AAGTCGACAGCATCACACG |
| lmb | ATGGAAGTCACACAAGGC | ATAGCAGCAACTGAGCCG |
| hsh | GTGCTTTGCCATGGTTGGCAAGCAGTTAACAG | ACACTTTCTGAGGCTGACG |
| rogB2 | TACGATCTGTCTGCTCTAG | CAGGATAGAATGTTGAAGG |
| cpsA | GGTGATAGTCAAGCTATGG | TCTATCGTTATCGCCTCC |
| hylA | CCTATTATCCAACGTACCG | GAACCTGTAACTGATAACGG |
| scpB | AACAGTAGCAGATGACGC | AGCTAGTGCAGCATTACC |
| lytR | GATGATGAACCAGTTGCACG | TGCCATTGTTGTAGTGAGCC |
| rogB | CCATCGATGCAGTTGCACAAGATAGTC | GCAGCGGATCCTTTGAGAGAGAGTTTACTG |
| sodA | CATCATGATAAGCACCATGC | TGGAGTATCTTGATTGGCCAG |
Primer extension analyses.
S. agalactiae 6313 RNA was isolated by using the RNeasy kit and QIAshredder (QIAGEN GmbH) according to the manufacturer's instructions. Primer extension analyses were carried out as described elsewhere (26) with the IRD800-labeled primers fbsA-PEX1 (5′ GCCAAAGCTTTCCTGATTTCC) and fbsA-PEX2 (5′ TTTCCTGATTTCCAAGTTC). The signals were analyzed with an automatic sequencer (LICOR 4000L; Licor, Inc., Lincoln, Nebr.) at 1,500 V and 50°C using a 6% (wt/vol) polyacrylamide gel.
DNA electrophoretic mobility shift assay.
The binding of His-tagged RovS protein to the fbsA, gbs0230, sodA, rogB, and cyl promoter regions was tested by DNA EMSA using the DNA fragments or double-stranded oligonucleotides indicated and shown in Results. The DNA fragments were generated by PCR using the primers fbsA-BS1 (5′AACTATTGCTCCCCTGC), fbsA-BS2 (5′ TTTCCTGATTTCCAAGTTC), gbs0230-BS1 (5′ GTAAAGGCACAGGTAGAT), gbs0230-BS2 (5′ CACCCCTCTCAAATCGTGAT), sod-BS1 (5′GTGGCTCAAGCGCATCATAT), sod-BS2 (5′ GTCTTACTTGGCAGGATAG), rogB-BS1 (5′AACCAGTTGAGACATG), rogB-BS2 (5′ TTTTACAACTCCTATTGTGC), cyl-BS1 (5′ TATGAGAGTGCGGGTTCTTG), and cyl-BS2 (5′ GGAGGACAGGGTAATGTTCA). All DNA fragments were purified by the Nucleospin extract kit (Macherey Nagel). For generating native and mutated double-stranded oligonucleotides, two complementary primers were mixed and heated to 95°C for 10 min in TE buffer (10 mM Tris [pH 7.8], 1 mM EDTA) and allowed to anneal by slow cooling at room temperature. In the binding assay, 100 ng of the fragments or oligonucleotides was incubated with various amounts of RovS (0 to 1.2 μg) in 20 μl of 10 mM Tris-HCl reaction buffer, pH 7.6, containing 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 10% (wt/vol) glycerol, and 1 μg of poly(dI-dC) for 20 min at room temperature. Subsequently the mixture was separated on a 2% agarose gel in 1× TAE buffer (200 mM Tris-HCl [pH 7.5], 100 mM acetate, 5 mM EDTA) at 70 V and 80 mA and stained with ethidium bromide.
Binding of fluorescein isothiocyanate-labeled group B streptococci to immobilized human proteins.
Terasaki microtiter plates were coated with human fibrinogen, collagen I, collagen IV, and laminin, and the binding of fluorescein isothiocyanate-labeled S. agalactiae to the immobilized proteins was measured as described previously (17). Results were measured in triplicate, and each assay was repeated at least four times.
Adherence and internalization assays.
Adherence of S. agalactiae to and internalization into A549 epithelial cells were assayed as described previously (17). Briefly, A549 cells were transferred to 24-well tissue culture plates at approximately 4 × 105 cells per well and cultivated overnight in RPMI tissue culture medium supplemented with 10% fetal calf serum. After replacement of the medium with 1 ml of fresh medium, the cells were infected with S. agalactiae at a multiplicity of infection of 10:1 and incubated at 37°C for 2 h. The infected cells were subsequently washed three times with PBS. The number of cell-adherent bacteria was determined by lysis of the eukaryotic cells with distilled water and subsequent determination of CFU by plating appropriate dilutions of the lysates on THY agar. Intracellular bacteria were determined after a further incubation of the infected cells for 2 h with RPMI medium containing penicillin G (10 U) and streptomycin (0.01 mg) to kill extracellular bacteria. After three washes with phosphate-buffered saline, the epithelial cells were lysed with distilled water and the amount of intracellular bacteria was quantitated by plating serial dilutions of the lysate onto THY agar plates. All samples were tested in triplicate, and experiments were repeated at least three times.
Determination of hemolytic activity.
For the preparation of hemolysin extracts, bacteria were grown in 50 ml of THY medium containing 2% soluble starch and subsequently pelleted by centrifugation, washed, and resuspended in 500 μl of PBS containing 0.2% glucose, 1% dextran 500 (Serva, Heidelberg, Germany), and 3% Tween 80. After 30 min of incubation at 37°C in 5% CO2 the suspension was centrifuged and the supernatant containing the hemolysin was removed. For determination of hemolytic activity different volumes of hemolysin extracts were diluted in 500 μl PBS. Fresh ovine erythrocytes were washed and adjusted to a 1% suspension in PBS. The diluted hemolysin extract and the erythrocyte suspension were mixed 1:2 in a volume of 1 ml and incubated at 37°C. After 10 min the probes were centrifuged for 1 min and the extinction of each supernatant was measured at 540 nm. To determine the hemolytic activity of each strain the quotient OD540/OD600 was calculated. All samples were tested in triplicate, and experiments were repeated at least three times.
Determination of β-galactosidase activity.
Recombinant S. agalactiae strains harboring the plasmid pTCV-lac or pTfbsA were grown aerobically in THY broth at 37°C with shaking (100 rpm). Strains were pretreated with a 0.5% toluene-4.5% ethanol mixture as described previously (33), and β-galactosidase activity was measured as described previously (36).
Statistical analysis.
Differences between the parental, the rovS mutant, and the complemented strain in binding to human fibrinogen, in the adherence to A549 lung epithelial cells, and in the expression of different virulence genes were assessed using the two-tailed Student t test. P values of ≤0.05 were considered statistically significant.
Nucleotide sequence accession number.
The genomic DNA sequence of S. agalactiae NEM316 can be accessed in the EMBL database under accession number AL732656. In the SwissProt TREMBL database, the RovS polypeptide described in this report possesses the accession number Q8E447.
RESULTS
Analysis of the rovS gene, encoding a transcriptional regulator.
The annotated genome sequence of the S. agalactiae strains NEM316 and 2603V/R revealed the presence of at least 107 putative transcriptional regulatory genes, representing about 5% of the total genome (16, 46). Proteins encoded by the genes gbs0230, gbs1555, and gbs2117 exhibit significant similarity to the Rgg family of transcriptional regulators (16), which control in S. pyogenes the expression of several known or putative virulence genes (7, 9). Among the three Rgg-like proteins in S. agalactiae, Gbs1555 reveals the highest identity to Rgg from S. pyogenes. This prompted us to characterize the importance of Gbs1555 for the virulence of S. agalactiae. As subsequent functional analysis revealed that gbs1555 encodes a regulator of virulence in Streptococcus agalactiae, the gbs1555 gene was termed rovS. In silico analysis of the rovS gene showed that it has a size of 849 bp and encodes a protein of 282 amino acids. The RovS protein has a predicted molecular mass of 33,128 Da and carries a helix-turn-helix motif of 54 amino acids at its N terminus. In database analysis, the RovS protein revealed 28% identity to the transcriptional regulator RggD of Streptococcus pneumoniae (21), 25% identity to the regulator Rgg of S. gordonii (43, 44), and 21% identity to the regulator RopB of S. pyogenes (28). Gene gbs1554 upstream of rovS encodes a protein with similarity to formamidopyrimidine-DNA glycosylase, a DNA repair enzyme that excises oxidized purines from damaged DNA. Gene gbs1556 downstream of rovS encodes a protein with no similarity to polypeptides of other organisms.
Deletion of rovS increases the binding of S. agalactiae to human fibrinogen.
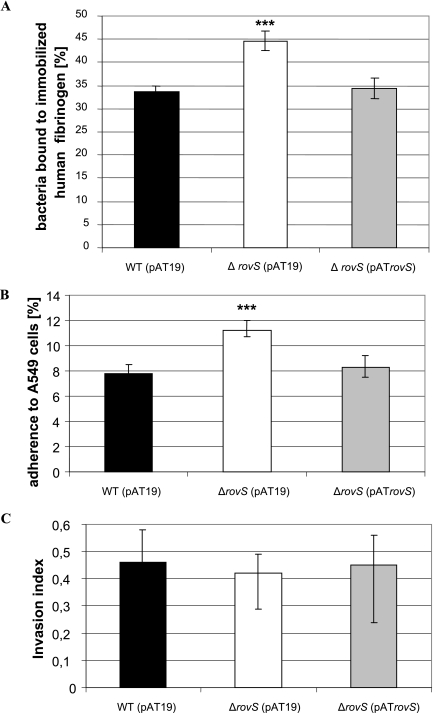
To analyze the importance of rovS for the pathogenicity of S. agalactiae, the rovS gene was deleted in the genome of S. agalactiae 6313, resulting in a ΔrovS mutant. Southern blot analysis confirmed the successful disruption of the rovS gene in the chromosome of the S. agalactiae ΔrovS strain (data not shown). No difference in the growth rate, final optical cell density, chain length, or clumping between the S. agalactiae ΔrovS mutant and the wild-type strain 6313 was observed (data not shown). For complementation studies, the rovS gene was cloned into the vector pAT19, resulting in plasmid pATrovS. The vector pAT19 was subsequently transformed into the S. agalactiae strain 6313 and the ΔrovS strain, and plasmid pATrovS was introduced into the S. agalactiae ΔrovS strain. To analyze the importance of rovS for the binding of S. agalactiae to host proteins, the abilities of the S. agalactiae 6313(pAT19), ΔrovS(pAT19), and ΔrovS(pATrovS) strains to bind to different proteins of the extracellular matrix were quantitated. The three S. agalactiae strains revealed no binding to immobilized collagen I, collagen IV, and laminin (data not shown) but exhibited significant binding to immobilized fibrinogen (Fig. 1A). Compared to the wild-type strain 6313(pAT19), the S. agalactiae ΔrovS(pAT19) strain showed a 35% increased binding to immobilized fibrinogen (Fig. 1A). However, plasmid-mediated expression of rovS in the ΔrovS(pATrovS) strain restored its binding to human fibrinogen to a wild-type level (Fig. 1A). These data indicate that the interaction of S. agalactiae with human fibrinogen is downregulated by RovS.
FIG. 1.
(A) Attachment of S. agalactiae to immobilized fibrinogen. The ordinate represents the mean binding of the total bacteria of the S. agalactiae wild-type (WT) 6313(pAT19), ΔrovS(pAT19), or ΔrovS(pATrovS) strain to immobilized fibrinogen. Each assay was performed at least four times in triplicate. ***, P < 0.001. (B and C) Adherence to (B) and invasion into (C) the lung epithelial cell line A549 by the S. agalactiae 6313(pAT19), ΔrovS(pAT19), and ΔrovS(pATrovS) strains. Bacterial adherence and invasion were calculated as follows: adherence = number of adherent bacteria/total number of bacteria in the assay × 100; invasion index = number of adherent bacteria/number of internalized bacteria in the assay × 100. Each experiment was performed at least four times in triplicate. ***, P < 0.001.
The rovS gene is important for the adhesion of S. agalactiae to eukaryotic cells.
In S. agalactiae the major fibrinogen-binding protein FbsA plays an important role in bacterial adhesion to the lung epithelial cell line A549 (39). As rovS was shown in the present study to control fibrinogen binding in S. agalactiae, the S. agalactiae 6313(pAT19), ΔrovS(pAT19), and ΔrovS(pATrovS) strains were tested for their adherence to and invasion into the human epithelial cell line A549. Adherent and invasive bacteria were quantitated by plate viability counts. As depicted in Fig. 1B, the S. agalactiae wild-type strain 6313(pAT19) revealed significant adherence to A549 cells. However, the adherence of the mutant rovS(pAT19) strain to A549 cells was increased by 44% compared to the wild-type strain. Plasmid-mediated expression of rovS downregulated host cell adherence of the S. agalactiae ΔrovS(pATrovS) strain to wild-type level. These data suggest that the rovS gene is involved in the attachment of S. agalactiae to epithelial cells.
To compare the invasion of the different S. agalactiae strains into A549 cells, the number of invasive bacteria of each strain was related to the number of adherent bacteria, resulting in the calculation of the invasion index (15). As depicted in Fig. 1C, the S. agalactiae 6313(pAT19), ΔrovS(pAT19), and ΔrovS(pATrovS) strains revealed similar invasion indices, indicating that rovS is not involved in the invasion of epithelial cells by S. agalactiae.
Expression profiling of virulence genes in the rovS mutant and its wild-type strain.
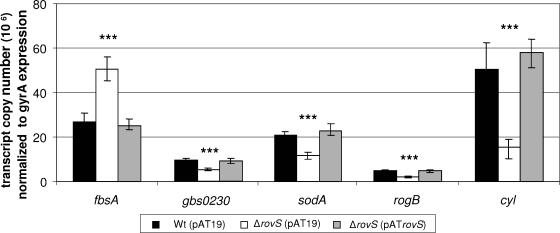
The previous results suggested a significant role of rovS in the pathogenicity of S. agalactiae. As the Rgg protein controls in S. pyogenes the transcription of a variety of virulence genes (7), the expression profile of known and putative S. agalactiae virulence genes was analyzed by quantitative RT-PCR in the S. agalactiae ΔrovS strain, the S. agalactiae ΔrovS(pATrovS) strain, and their parental strain 6313. Expression profiling was performed with the genes cfb, lmb, sodA, cpsA, hylB, scpB, lytR, gyrA, rogB, gbs0230, cylE, and fbsA, encoding CAMP factor, the laminin-binding protein Lmb, superoxide dismutase, an activator of capsule gene expression, hyaluronate lyase, C5a peptidase, an autolysin response regulator, gyrase subunit A, RogB regulator protein, a transcription regulator similar to Rgg, a protein required for the production of hemolysin, and the fibrinogen receptor FbsA, respectively. Likewise the expression of the rovS-flanking genes gbs1554 and gbs1556 was analyzed. Equal amounts of total RNA from exponential-phase cultures (OD600 = 0.3) of the S. agalactiae ΔrovS and 6313 strains were reverse transcribed and used to quantify the transcript levels of the above-mentioned genes by real-time PCR. The obtained data were normalized to the expression of the gyrA gene in the three strains. A 1.5-fold difference in transcription was interpreted as a significant difference in expression between the strains. Relative to S. agalactiae 6313, transcription of the genes cfb, lmb, cpsA, hylB, scpB, lytR, gbs1554, and gbs1556 was unaltered in the rovS mutant (data not shown).
As shown in Fig. 2, the expression of the fbsA gene was 1.8- ± 0.21-fold increased in the rovS mutant compared to the parental strain. In contrast, the transcription levels of the genes gbs0230, sodA, and rogB and the cyl operon were 1.9- ± 0.15-, 1.7- ± 0.35-, 2.2- ± 0.3-, and 4.1- ± 0.45-fold decreased in the rovS mutant compared to the parental strain 6313. Plasmid-mediated expression of rovS restored in the ΔrovS(pATrovS) strain the transcription of the above-mentioned genes to wild-type level (Fig. 2). These findings suggest that RovS represses in S. agalactiae the transcription of the fbsA gene and that it activates the expression of the genes gbs0230, rogB, and sodA and the cyl operon, respectively.
FIG. 2.
Expression analysis of the genes fbsA, gbs0230, sodA, and rogB and of the cyl operon in the S. agalactiae 6313(pAT19), ΔrovS(pAT19), and ΔrovS(pATrovS) strains. The amount of mRNA of the different genes was measured by quantitative real-time PCR in a LightCycler and normalized to the expression of the gyrA gene. The expression of the different genes is presented as the transcript copy normalized to 2.9 × 106 copies of gyrA transcript. ***, P < 0.001.
In vivo analysis of RovS-mediated regulation of fbsA expression and hemolysin formation.
To analyze the influence of RovS on fbsA expression in vivo, the S. agalactiae 6313(pAT19), ΔrovS(pAT19), and ΔrovS(pATrovS) strains were transformed with the vector pTCV-lac or alternatively with the plasmid pTfbsA, which carries the fbsA promoter in front of a promoterless lacZ gene. In pTfbsA-carrying strains, fbsA expression can be monitored by measuring the β-galactosidase activity. Strains that harbored the vector pTCV-lac did not show β-galactosidase activity during growth (data not shown). However, pTfbsA-carrying strains revealed a growth-phase-dependent expression of fbsA, peaking in the middle of the exponential growth phase with a moderate decline during late logarithmic and stationary growth. Growth experiments revealed identical growth of the S. agalactiae 6313(pAT19)(pTfbsA), ΔrovS(pAT19)(pTfbsA), and ΔrovS(pATrovS)(pTfbsA) strains (data not shown). However, in the ΔrovS(pAT19)(pTfbsA) strain, the fbsA expression was approximately 35% increased compared to the wild-type strain 6313(pAT19)(pTfbsA). These findings confirm our results of real-time PCR analysis and demonstrate fbsA repression by RovS in S. agalactiae.
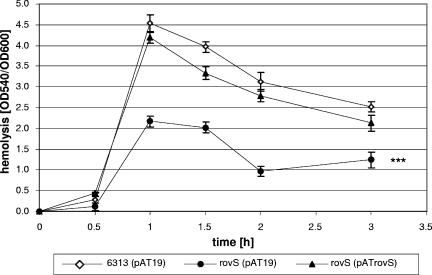
To investigate the impact of RovS on hemolysin production in S. agalactiae in vivo, the hemolytic activity of the S. agalactiae wild-type 6313(pAT19), ΔrovS(pAT19), and ΔrovS(pATrovS) strains was analyzed during growth of the bacteria in complex media. The results were plotted as quotient OD540/OD600, respectively, against time. The three strains exhibited identical growth behavior in complex medium (data not shown). In all three strains, hemolytic activity increased significantly during the early exponential growth phase, peaked in the middle of exponential growth, and decreased at the transition from the exponential to the stationary growth phase (Fig. 3). However, the hemolytic activity in the ΔrovS(pAT19) mutant was on average 50% lower than in strain 6313(pAT19). Plasmid-mediated expression of rovS in the ΔrovS(pATrovS) strain increased the hemolytic activity to values comparable to those of the wild-type strain 6313(pAT19). These findings confirm our results obtained by real-time PCR and show that the presence of RovS stimulates the transcription of the cyl operon in S. agalactiae.
FIG. 3.
Hemolytic activity of S. agalactiae 6313(pAT19), ΔrovS(pAT19), and ΔrovS(pATrovS) strains. The quotient OD540/OD600 represents the hemolytic activity of each strain during growth. Each assay was performed at least three times in triplicate.
RovS specifically binds to the promoter of some of the RovS-regulated genes.
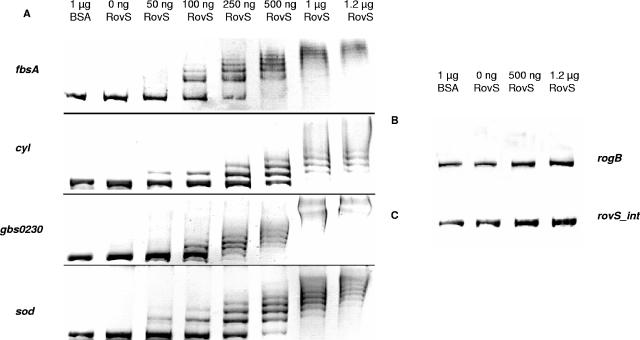
To characterize the interaction of RovS with RovS-regulated genes on the molecular level, EMSAs were performed with purified RovS protein and respective DNA probes. These probes were amplified by PCR, purified, incubated with different amounts of RovS protein, and separated on agarose gels. The relevant results of all EMSAs are shown in Fig. 4. As depicted in Fig. 4A the purified RovS protein showed a concentration-dependent binding to the promoter fragments of the genes fbsA (−706 to +41), gbs0230 (−890 to +115), and sodA (−448 to +52) and the cyl operon (−645 to +326). All these fragments were completely retarded by incubation with 1.0 μg of RovS. However, RovS showed no binding to a probe, containing the promoter region of the gene rogB (−245 to +89) (Fig. 4B). Incubation of the fragments with 1 μg of bovine serum albumin did not result in retardation (Fig. 4A, line 2). Conversely, unspecific DNA (i.e., an internal fragment of rovS) was not shifted by up to 1 μg of RovS protein (Fig. 4C). These results indicate that RovS in fact binds to the promoter/operator regions of the genes fbsA, gbs0230, and sodA and the cyl operon and that this binding is specific.
FIG. 4.
(A) EMSAs using purified RovS protein to analyze the binding to promoter fragments of the genes fbsA, gbs0230, and sodA and the cyl operon of S. agalactiae. (B) EMSAs using purified RovS protein to analyze the binding to the rogB promoter region. (C) Control EMSAs using purified RovS protein and a rovS gene internal DNA fragment. BSA, bovine serum albumin.
Identification of the RovS binding site within the fbsA promoter region.
By using primer extension analysis, we first located the transcription start point of the fbsA gene of S. agalactiae 59 nucleotides upstream from the initiation codon with T as the first transcribed nucleotide (data not shown). From this site the promoter structure TTGACA(17 bp)TAATAT was identified, which shows high homology to the consensus sequences of promoters in gram-positive bacteria and in E. coli.
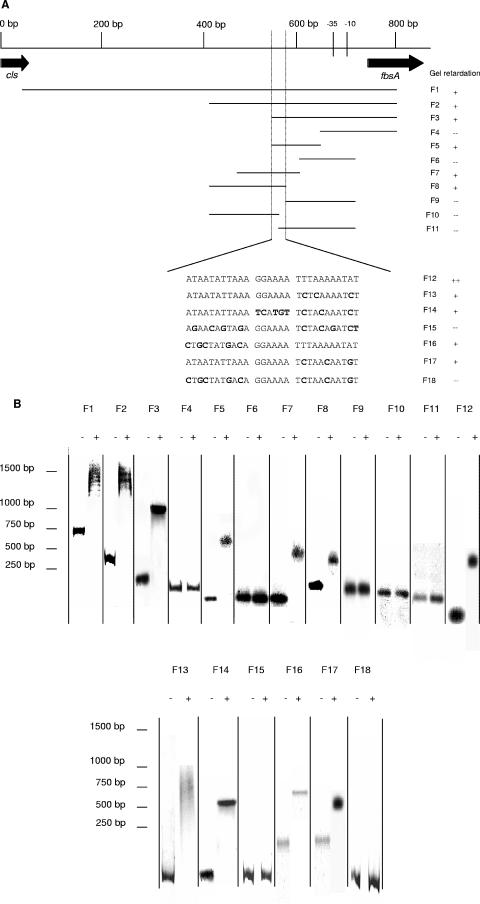
Subsequently, to precisely localize the RovS binding site within the fbsA promoter, EMSAs were performed with 18 different DNA fragments of this promoter region. The localization of these fragments within the fbsA promoter is shown in Fig. 5A. The results obtained with the fragments F1 to F12 (Fig. 5B) showed that the binding site of RovS is located within a 28-bp region, situated 111 to 139 bp upstream of the fbsA transcriptional start. Fragments containing this 28-bp region (F1, F2, F3, F5, F7, and F8) were completely shifted whereas fragments lacking this region (F4, F6, F9, F10, and F11) were not shifted at all (Fig. 5B).
FIG. 5.
EMSAs using purified RovS protein to identify the RovS binding site in the fbsA promoter region. (A) DNA fragments used for EMSAs, containing different parts of the fbsA promoter region, were named F1 to F12. Fragments F13 to F18 are derivatives of fragment F12 containing different mutations, shown in boldface. The black dashed lines indicate the region required for a shift, and the sequence of this 28-bp region is shown below. (B) About 100 ng of each DNA fragment was incubated either without (lanes marked with −) or with (lanes marked with +) 50 pmol RovS for 20 min at room temperature. Binding or nonbinding of RovS is indicated by + or −, respectively, in panel A.
Interestingly, this 28-bp region comprises a palindromic sequence, containing a spacer of 6 bp and carrying two mismatches. To confirm the importance of this 28-bp region for the binding of RovS to the fbsA promoter, 28-bp oligonucleotides containing different mutations (F13 to F18) were synthesized and tested in EMSAs. All of the tested oligonucleotides, except fragments F15 and F18, were completely shifted. Oligonucleotide F12 contains the original DNA sequence and was already shifted at a RovS concentration of 0.1 μg, whereas the oligonucleotides F13 and F14 and oligonucleotides F16 and F17, containing mutations on one or the other end of the binding region, were shifted at a higher protein concentration (0.5 μg) (data not shown). A complete change (e.g., F15 and F18) or a cleavage in the middle of the palindromic sequence (F10 and F11) resulted in the loss of the binding of RovS. All EMSA results suggest that this palindromic sequence is of great importance for the binding of RovS to the fbsA promoter.
As EMSAs had demonstrated binding of RovS to the upstream region of the structural genes fbsA, gbs0230, and sodA and the cyl operon, the promoter regions of these genes were aligned to identify a putative consensus sequence for RovS binding. In the upstream regions of the analyzed genes the putative RovS binding sites ATAAAAATGAA-N7-ATGATAAGTAT (gbs0230), AAAATCCTAAT-N6-TTTTAAAAAAG (sodA), and ATAATGTTTAT-N6-TTTAAACTAAT (cyl operon) (boldface indicates conserved nucleotides) could be identified, resulting in the consensus sequence AWAAWVHTDAW-N6/7-WTKWWAMDWAK.
The identified RovS binding motif was used to search the S. agalactiae NEM316 genome sequence with the program Predict Regulon (51) for further putative RovS binding sites. Allowing two mismatches within the whole RovS binding motif identified 25 further putative RovS binding sites in the promoter region of S. agalactiae genes (data not shown). Among these genes five code for proteins of the central metabolism (gbs0091, gbs0871, gbs1088, gbs1376, and gbs2122), eight code for unknown proteins (gbs0399, gbs0662, gbs0729, gbs0828, gbs0967, gbs0980, gbs1202, and gbs2112), five code for putative transcriptional regulators or proteins that are part of the transcriptional apparatus (gbs0094, gbs0425, gbs1381, gbs1509, and gbs1712), four code for putative transporters (gbs0119, gbs1332, gbs2007, and gbs2017), and three code for surface proteins (gbs0451, gbs1478, and gbs1998). These findings indicate that RovS controls the expression of a variety of different genes in S. agalactiae.
Under native conditions RovS is existent as a dimer.
To identify the native molecular mass of RovS, gel filtration chromatography was performed. Using cytochrome C1 (12.5 kDa), chymotrypsinogen (25 kDa), albumin (45 kDa), bovine serum albumin (68 kDa), aldolase (158 kDa), katalase (240 kDa), and ferritin (450 kDa) as protein standards, the native molecular mass of RovS was determined as approximately 58 kDa (data not shown). As a monomer of RovS has a molecular mass of 32 kDa, this result indicates that RovS is a dimer in its native form.
DISCUSSION
S. agalactiae is a commensal bacterium colonizing the gastrointestinal tract of a considerable part of the human population. On the other site, this organism is also the leading cause of invasive infections in neonates. The colonization and infection of different regions within the human host require a regulatory network in S. agalactiae that senses environmental stimuli and controls the formation of specific virulence factors. In the present study, RovS, a novel transcriptional regulator for expression control of virulence genes in S. agalactiae, was identified and characterized. RovS reveals similarity to members of the Rgg-like protein family of transcriptional regulators, well known and well studied in S. pyogenes (5, 6, 7, 8) and S. gordonii (43, 49). In S. pyogenes, the rgg gene product is involved in the synthesis of the erythrogenic toxin B (SpeB), also known as streptococcal pyrogenic exotoxin B (10, 28). Inactivation of rgg leads to a decreased synthesis of SpeB (6). A number of reports suggest that SpeB plays an important role in the virulence of S. pyogenes (3, 24, 25). Because of its proteolytic activity, SpeB has been suggested to enable S. pyogenes to affect the interaction with the human host during infection. SpeB is able to cleave the streptococcal adhesin M1 protein from the bacterial surface (3) and to degrade host cell extracellular matrix factors, including fibronectin and vitronectin (24). In S. gordonii, Rgg is a positive transcriptional regulator that controls the expression of the glycosyltransferase gene gtfG (43). The glucan polymers synthesized by streptococcal glucosyltransferase enzymes enable bacterial accumulation on surfaces, such as the microbial biofilm formation on tooth surfaces. Regulation of glycosyltransferase enzymes affects the microbial composition of the biofilm and influences the state of health or disease. All these studies suggest an involvement of Rgg proteins in the virulence of streptococci. However, nothing is known about Rgg proteins in S. agalactiae. There is no gene homologous to speB of S. pyogenes and only two genes, encoding putative glycosyltransferase enzymes, with no similarity to gtfG of S. gordonii in the genome of S. agalactiae. Therefore, RovS in S. agalactiae must have different functions from those of RopB of S. pyogenes and Rgg of S. gordonii.
In S. agalactiae the deletion of rovS resulted in an increased binding of the bacteria to immobilized fibrinogen and to the human lung epithelial cell line A549. This suggests that RovS negatively controls the attachment of the bacteria to the ECM and to host cells. The ECM of mammalian tissues is a stable macromolecular structure underlying epithelial and endothelial cells and is composed of structural glycoproteins such as collagen, laminin, fibronectin, and fibrinogen (19). The interaction of bacteria with proteins of the ECM is frequently required for the successful colonization of the human host. In S. pyogenes it was shown that bacterial binding to ECM proteins mediates adherence to and invasion in eukaryotic cells (29, 45). In S. agalactiae, the fibrinogen-binding protein FbsA plays a prominent role in the bacterial adherence to host cells (39). As demonstrated by real-time PCR and in vivo expression analysis, the increased fbsA expression in the rovS mutant may be the molecular basis for its increased adherence to host cells.
Real-time PCR analysis revealed, aside from fbsA control, a RovS-dependent effect on the expression of other known and putative virulence genes in S. agalactiae. RovS was shown to stimulate the expression of gbs0230, encoding an Rgg-like transcriptional regulator; of gbs0644, encoding cylE, the first gene of the cyl operon, which is essential for the production of hemolysin (42); and of gbs0808, encoding the manganese-dependent superoxide dismutase of S. agalactiae. The significance of the regulator Gbs0230 is not yet known; however, it can be speculated that it influences the expression of other virulence genes. Superoxide dismutases convert superoxide anions to molecular oxygen and hydrogen peroxide, which in turn are metabolized by catalases and peroxidases. These enzymes constitute one of the major defense mechanisms of cells against oxidative stress and therefore play a role in the pathogenesis of certain bacteria. It was demonstrated that S. agalactiae possesses a single Mn-cofactored superoxide dismutase, SodA. A sodA mutant showed an increased sensitivity to killing by macrophages, and the survival of the mutant in the blood and the brain of infected mice was markedly reduced in comparison to that of the parental strain (32). The importance of the cytolysin CylE of S. agalactiae for the bacterial invasion in epithelial cells was shown by Doran et al. (13). Recently, also in vivo evidence for a critical role of beta-hemolysin in S. agalactiae neonatal pneumonia was provided (20). Rabbits infected with wild-type S. agalactiae developed focal pneumonia and had about 100-fold more bacteria in lung tissue than did rabbits infected with a hemolysin-negative mutant. Accordingly, mortality and development of bacteremia were significantly higher in rabbits challenged with S. agalactiae wild type than in those challenged with the mutant strain.
Our EMSAs showed that RovS binds directly to the promoter regions of the genes fbsA, gbs0230, and sodA and the cyl operon, thereby directly controlling the transcription of these genes. As increasing RovS concentrations resulted in the formation of a banding pattern, it can be speculated that RovS bound to its target sequence in different multimeric forms. Although real-time PCR experiments clearly showed an effect of RovS on rogB expression, the RovS protein did not bind to the rogB promoter in EMSAs. This suggests that the transcription of rogB obviously is stimulated by RovS indirectly, perhaps by influencing the expression of another regulatory protein.
Although some Rgg-like regulators have been characterized in function and structure, to our knowledge no general consensus sequence has been proposed for the DNA-binding site of this family of transcriptional regulators. To identify the RovS binding site, EMSAs were performed with truncated fbsA promoter regions, resulting in the identification of the palindromic sequence ATAATATTAAA-N6-TTTAAAAATAT, which carries two mismatches. Previous studies demonstrated that the regulation of different genes or operons in gram-positive bacteria may involve the binding of regulator proteins to palindromic domains in their control regions. Jakubovics et al. (23) could identify the ScaR DNA-binding site that was shown to consist of one complete and one incomplete palindromic sequence. ScaR is a key regulator of scaA expression in S. gordonii; scaA encodes a cell surface lipoprotein antigen implicated in interbacterial adhesion. Transcription factors often bind as dimers to specific promoter regions. Thereby, two subunits make contacts with conserved base pairs within repeated elements or palindromic sequences and hence regulate transcription in a cooperative manner (4, 22). This also seems to be the case for RovS, which could be detected as a dimer under native conditions and which specifically binds to a region of the fbsA promoter containing an imperfect palindromic sequence. An alignment of the promoter regions of fbsA, gbs0230, sodA, and the cyl operon revealed that this sequence can also be found upstream of the transcriptional start of gbs0230 (−706 to −678), sodA (−201 to −173), and the cyl operon (−240 to −212), resulting in the consensus sequence AWAAWVHTDAW-N6/7-WTKWWAMDWAK (boldface indicates conserved nucleotides) for the RovS binding motif. Therefore, it can be speculated that one subunit of the RovS dimer is able to bind to one part of the palindromic sequence and to regulate the expression of these genes, because RovS can also bind to DNA sequences with mutations in one site of the palindromic domain. Furthermore, in accordance with the finding that putative RovS binding sites can also be found upstream of 25 other genes, it might be speculated that the RovS motif identified in this study has broader significance as a determinant in a global regulatory system of S. agalactiae. Further studies are required to address this hypothesis.
In summary, we have identified and characterized a novel regulatory gene, termed rovS, encoding a regulator of virulence in S. agalactiae, which is involved in the expression control of known and putative virulence genes in these bacteria, influencing the attachment to human fibrinogen and the binding to the human lung epithelial cell line A549. With EMSAs the direct binding of RovS to the promoter regions of different virulence genes could be demonstrated. We were also able to propose a consensus sequence for the DNA-binding site of RovS, an Rgg-like transcriptional regulator.
.
Acknowledgments
We thank Axel Schubert for performing the primer extension analysis. We thank Christian Riedel for his assistance in statistical analysis of our results.
Editor: J. B. Bliska
REFERENCES
- 1.Baker, C. J., and M. S. Edwards. 1995. Group B streptococcal infections, p. 980-1054. In J. S. Remington and J. O. Klein (ed.), Infectious disease of the fetus and newborn infant. The W. B. Saunders Co., Philadelphia, Pa.
- 2.Beckmann, C., J. D. Waggoner, T. O. Harris, G. S. Tamura, and C. E. Rubens. 2002. Identification of novel adhesins from group B streptococci by use of phage display reveals that C5a peptidase mediates fibronectin binding. Infect. Immun. 70:2869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berge, A., and L. Björck. 1995. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem. 270:9862-9867. [DOI] [PubMed] [Google Scholar]
- 4.Busby, S. A., and R. H. Ebright. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79:743-746. [DOI] [PubMed] [Google Scholar]
- 5.Chaussee, M. A., E. A. Callegari, and M. S. Chaussee. 2004. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol. 186:7091-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 67:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaussee, M. S., G. L. Sylva, D. E. Sturdevant, L. M. Smoot, M. R. Graham, R. O. Watson, and J. M. Musser. 2002. Rgg influences the expression of multiple regulatory loci to coregulate virulence factor expression in Streptococcus pyogenes. Infect. Immun. 70:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 185:6016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaussee, M. S., D. Gerlach, C.-E. Yu, and J. J. Ferretti. 1993. Inactivation of the streptococcal erythrogenic toxin B gene (speB) in Streptococcus pyogenes. Infect. Immun. 61:3719-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, Q., D. Stafslien, S. S. Purushothaman, and P. Cleary. 2002. The group B streptococcal C5a peptidase is both a specific protease and an invasin. Infect. Immun. 70:2408-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cieslewicz, M. J., D. L. Kasper, Y. Wang, and M. R. Wessels. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J. Biol. Chem. 276:139-146. [DOI] [PubMed] [Google Scholar]
- 13.Doran, K. S., J. C. Chang, V. M. Benoit, I. Eckmann, and V. Nizet. 2002. Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J. Infect. Dis. 185:196-203. [DOI] [PubMed] [Google Scholar]
- 14.Dubendorff, J. W., and F. W. Studier. 1991. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J. Mol. Biol. 219:45-59. [DOI] [PubMed] [Google Scholar]
- 15.Eickhoff, T. C., J. O. Klein, K. L. Daly, D. I. Ingall, and M. Finland. 1964. Neonatal sepsis and other infections due to group B beta-hemolytic streptococci. N. Engl. J. Med. 271:1221-1228. [DOI] [PubMed] [Google Scholar]
- 16.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 17.Gutekunst, H., B. J. Eikmanns, and D. J. Reinscheid. 2003. Analysis of RogB-controlled virulence mechanisms and gene repression in Streptococcus agalactiae. Infect. Immun. 71:5056-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan, D. 1985. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Hay, E. D. 1991. Cell biology of extracellular matrix. Plenum Press, New York, N.Y.
- 20.Hensler, M. E., G. Y. Liu, S. Sobczak, K. Benirschke, V. Nizet, and G. P. Heldt. 2005. Virulence role of group B Streptococcus beta-hemolysin/cytolysin in a neonatal rabbit model of early-onset pulmonary infection. J. Infect. Dis. 191:1287-1291. [DOI] [PubMed] [Google Scholar]
- 21.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishihama, A. 1993. Protein-protein communication within the transcription apparatus. J. Bacteriol. 175:2483-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakubovics, N. S., A. W. Smith, and H. F. Jenkinson. 2000. Expression of the virulence-related Sca (Mn2+) permease in Streptococcus gordonii is regulated by a diphtheria toxin metallorepressor-like protein ScaR. Mol. Microbiol. 38:140-153. [DOI] [PubMed] [Google Scholar]
- 24.Kapur, V., S. Topouzis, M. W. Jajesky, L.-L. Li, M. R. Hamrick, R. J. Patti, and J. M. Musser. 1993. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb. Pathog. 15:327-346. [DOI] [PubMed] [Google Scholar]
- 25.Kapur, V., J. T. Maffei, R. S. Greer, L.-L. Li, G. J. Adams, and J. M. Musser. 1994. Vaccination with streptococcal extracellular cysteine protease (interleukin-1 beta convertase) protects mice against challenge with heterologous group A streptococci. Microb. Pathog. 16:443-450. [DOI] [PubMed] [Google Scholar]
- 26.Kellmann, J.-W., E. Piechersky, and B. Piechulla. 1990. Analysis of the diurnal expression patterns of the tomato chlorophyll a/b binding protein genes. Influence of light and characterization of the gene family. Phytochem. Phytobiol. 52:35-41. [DOI] [PubMed] [Google Scholar]
- 27.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neely, M. N., W. R. Lyon, D. L. Runft, and M. Caparon. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 185:5166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okada, N., M. Watarai, V. Ozeri, E. Hanski, M. Caparon, and C. Sasakawa. 1997. A matrix form of fibronectin mediates enhanced binding of Streptococcus pyogenes to host tissue. J. Biol. Chem. 272:26978-26984. [DOI] [PubMed] [Google Scholar]
- 30.Pospiech, A., and B. Neumann. 1995. A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet. 11:217-218. [DOI] [PubMed] [Google Scholar]
- 31.Poyart, C., M. C. Lamy, C. Boumaila, F. Fiedler, and P. Trieu-Cuot. 2001. Regulation of d-alanyl-lipoteichoic acid biosynthesis in Streptococcus agalactiae involves a novel two-component regulatory system. J. Bacteriol. 183:6324-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poyart, C., E. Pellegrini, O. Gaillot, C. Boumaila, M. Baptista, and P. Trieu-Cuot. 2001. Contribution of Mn-cofactored superoxide dismutase (SodA) to the virulence of Streptococcus agalactiae. Infect. Immun. 69:5098-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 34.Qi, F., P. Chen, and P. W. Caufield. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and J. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Samen, U., B. Gottschalk, B. J. Eikmanns, and D. J. Reinscheid. 2004. Relevance of peptide uptake systems to the physiology and virulence of Streptococcus agalactiae. J. Bacteriol. 186:1398-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 38.Schubert, A., K. Zakikhany, M. Schreiner, R. Frank, B. Spellerberg, B. J. Eikmanns, and D. J. Reinscheid. 2002. A fibrinogen receptor from group B Streptococcus interacts with fibrinogen by repetitive units with novel ligand binding sites. Mol. Microbiol. 46:557-569. [DOI] [PubMed] [Google Scholar]
- 39.Schubert, A., K. Zakikhany, G. Pietrocola, A. Meinke, P. Speziale, B. J. Eikmanns, and D. J. Reinscheid. 2004. The fibrinogen receptor FbsA promotes adherence of Streptococcus agalactiae to human epithelial cells. Infect. Immun. 72:6197-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, and R. Lütticken. 2002. rgf encodes a novel two-component signal transduction system of Streptococcus agalactiae. Infect. Immun. 70:2434-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spellerberg, B., E. Rozdzinski, S. Martin, J. Weber-Heynemann, N. Schnitzler, R. Lütticken, and A. Podbielski. 1999. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect. Immun. 67:871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spellerberg, B., B. Pohl, G. Haase, S. Martin, J. Weber-Heynemann, and R. Lütticken. 1999. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J. Bacteriol. 181:3212-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulavik, M. C., and D. B. Clewell. 1996. Rgg is a positive transcriptional regulator of the Streptococcus gordonii gtfG gene. J. Bacteriol. 178:5826-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sulavik, M. C., G. Tardif, and D. B. Clewell. 1992. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 174:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talay, S. R., A. Zock, M. Rohde, G. Molinari, M. Oggioni, G. Pozzi, C. A. Guzman, and G. S. Chhatwal. 2000. Co-operative binding of human fibronectin to Sfbl protein triggers streptococcal invasion into respiratory epithelial cells. Cell. Microbiol. 2:521-535. [DOI] [PubMed] [Google Scholar]
- 46.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZ alpha gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 48.Valentin-Weigand, P., P. Benkel, M. Rohde, and G. S. Chhatwal. 1996. Entry and intracellular survival of group B streptococci in J774 macrophages. Infect. Immun. 64:2467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vickerman, M. M., and P. E. Minick. 2002. Genetic analysis of the rgg-gtfG junctional region and its role in Streptococcus gordonii glucosyltransferase activity. Infect. Immun. 70:1703-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waite, D. C., E. J. Alper, and B. J. Mady. 1996. Adult group B streptococcal disease. Ann. Intern. Med. 125:152-153. [DOI] [PubMed] [Google Scholar]
- 51.Yellboina, S., J. Seshadri, M. S. Kumar, and A. Ranjan. 2004. PredictRegulon: a web server for the prediction of the regulatory protein binding sites and operons in prokaryote genomes. Nucleic Acids Res. 32:W318-W320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zangwill, K. M., A. Schuchat, and J. D. Wenger. 1992. Group B streptococcal disease in the United States, 1990: report from a multistate active surveillance system. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 41:25-32. [PubMed] [Google Scholar]