Abstract
Coccidioidomycosis is a human respiratory disease that is endemic to the southwestern United States and is caused by inhalation of the spores of a desert soilborne fungus. Efforts to develop a vaccine against this disease have focused on identification of T-cell-reactive antigens derived from the parasitic cell wall which can stimulate protective immunity against Coccidioides posadasii infection in mice. We previously described a productive immunoproteomic/bioinformatic approach to the discovery of vaccine candidates which makes use of the translated genome of C. posadasii and a computer-based method of scanning deduced sequences of seroreactive proteins for epitopes that are predicted to bind to human major histocompatibility (MHC) class II-restricted molecules. In this study we identified a set of putative cell wall proteins predicted to contain multiple, promiscuous MHC II binding epitopes. Three of these were expressed by Escherichia coli, combined in a vaccine, and tested for protective efficacy in C57BL/6 mice. Approximately 90% of the mice survived beyond 90 days after intranasal challenge, and the majority cleared the pathogen. We suggest that the multicomponent vaccine stimulates a broader range of T-cell clones than the single recombinant protein vaccines and thereby may be capable of inducing protection in an immunologically heterogeneous human population.
Coccidioidomycosis is a human respiratory disease caused by inhalation of airborne spores produced by Coccidioides spp. The pathogen is an anamorphic (asexual) ascomycetous fungus which resides in alkaline desert and semidesert soil in regions of the southwestern United States (10). The major areas where this disease is endemic include some of the most rapidly growing communities of Arizona and southern California (4), as well as numerous military bases (39) on which more than 350,000 personnel are stationed (e.g., 29 Palms, Camp Pendleton, Fort Irwin, Fort Roberts, etc.). People who are frequently exposed to dust from the soil in these regions have a high chance of infection with Coccidioides. Recurrent epidemics of coccidioidomycosis appear to be correlated with climatic conditions (6). Periods of rain in the spring enhance growth of the soilborne saprobic phase of Coccidioides, which results in “blooms” of spores that are air dispersed during the late fall and winter seasons. Risk factors known to contribute to symptomatic coccidioidal infection include pregnancy (third trimester), immunosuppression, age (>65 years), and racial or geographic origin. The attack rate for Caucasians has been estimated to be 11%, compared to reports of 54% for African-Americans, 67% for Filipinos, and 36% for Asians (5). Antifungal drug therapy for coccidioidomycosis (e.g., amphotericin B, fluconazole, or intraconazole) is typically continued for many months to years (13). Persons who contract coccidioidal meningitis require lifelong therapy (24). Coccidioides infection can be life threatening and, at the very least, is responsible for high medical costs incurred by patients as a result of hospitalization, clinic visits, lost wages, and long-term drug therapy (3).
Of the estimated 130,000 new cases of coccidioidomycosis in the United States each year, clinical manifestations occur in 40 to 50% and range from influenza-like illness to severe pneumonia and, rarely, extrapulmonary disseminated disease (5). Individuals who recover from these infections typically have lifelong immunity against a second coccidioidal infection. On this basis we have argued that development of a vaccine against coccidioidomycosis is feasible (5). Both clinical and experimental studies have demonstrated that robust and durable T-cell immunity is essential for protection against this disease (17). Our research has focused on the identification of T-cell-reactive antigens and their evaluation as vaccine candidates in cellular immunoassays and murine protection experiments. In a previous report, we introduced the application of an immunoproteomic/bioinformatic approach to the discovery of candidate vaccines against coccidioidomycosis (40). In the present study we have applied these same methods to generate a multivalent recombinant protein vaccine which protects C57BL/6 mice against a lethal intranasal challenge with Coccidioides. The level of protection provided to the vaccinated mice, as evaluated by survival and reduction of fungal burden, is superior to that conferred by the individual recombinant proteins which comprise the multivalent vaccine.
MATERIALS AND METHODS
Fungal growth conditions.
Cultures of the saprobic and parasitic phases of Coccidioides posadasii (strain C735) were grown as reported previously (19, 22). Arthroconidia produced on glucose-yeast extract agar were used to inoculate a defined glucose-salts medium for growth of the parasitic (spherule-endospore) phase (11) or for preparation of inocula to infect C57BL/6 mice as described below. First-generation cultures of the parasitic phase are fairly well synchronized, which permitted isolation of different cell types, (e.g., presegmented spherules, segmented spherules, and endosporulating spherules) produced after different times of incubation (21).
Isolation of the parasitic cell wall fraction.
The parasitic cells used for isolation of the C. posadasii wall fraction were the same as previously described (40). The cell pellet obtained by centrifugation was first washed with ice-cold disruption buffer (20 mM Tris-HCl, pH 7.4) containing 2× protease inhibitor cocktail, set IV (Calbiochem, San Diego, Calif.), and then disrupted in a Beadbeater (BioSpec Products, Inc., Bartlesville, Okla.) as described previously (40). The homogenate was centrifuged (5,000 × g), and the pellet was washed five times with cold disruption buffer as described above and then frozen and stored at −80°C until ready to use.
2D SDS-PAGE and immunoblot analysis.
Approximately 250 mg of the isolated cell wall material was suspended in a buffer used for two-dimensional (2D) polyacrylamide gel electrophoresis (PAGE) separation of the extracted protein components. The buffer composition was the same as reported previously (40). The cell wall suspension was sonicated (six 5-min bursts [50 W] on ice) using a Branson Sonifier (Branson Sonic Power Co., Danbury, Conn.) to further disrupt the wall fragments and enhance release of soluble proteins. Insoluble wall material was precipitated by centrifugation (15,000 × g, 30 min, 4°C). Determination of the protein concentration in the soluble fraction was conducted by one-dimensional PAGE separation of the sample with Coomassie blue-stained standards compared to a titration of the supernatant. The quantitative estimate was performed using a Gel Doc EQ system (Bio-Rad Laboratories, Hercules, Calif.). The average protein concentration in three separate preparations of the cell wall extract was 3 μg/μl. Aliquots (340 μl) of the supernatant were used to hydrate 18-cm Immobiline Drystrip gels (pH 3 to 10; Amersham Biosciences, Piscataway, N.J.) overnight at room temperature. The gel strips were first subjected to isoelectric focusing and then exposed to a reducing buffer, followed by electrophoresis in the second dimension in either a 10% or 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel as previously reported (40). Protein spots were visualized by Coomassie blue staining, and gel images were digitally recorded as reported previously (40). Immunoblot analysis of sister gel electrophoretic separations were conducted with either pooled human sera from 10 patients with confirmed coccidioidal infection or pooled sera from an equal number of healthy control volunteers as previously described (40). Detection of seroreactive protein spots in the 2D polyacrylamide gels was conducted using goat anti-human immunoglobulin G secondary antibody (gamma chain specific) (Southern Biotechnology Associates, Inc. Birmingham, Ala.), which was conjugated with horseradish peroxidase.
LC-MS/MS.
Coomassie blue-stained spots visible in the 2D polyacrylamide gels which were reactive with sera from patients with confirmed coccidioidomycosis were excised, destained, and subjected to in-gel digestion with sequencing-grade trypsin (Promega, Madison, Wis.) as described previously (40). Peptides were extracted from the gel, separated by reverse-phase high-pressure liquid chromatography (LC), and then introduced directly into an ion trap mass spectrometer equipped with a nanospray source (LCQ Deca XP plus; Finnigan Corp., San Jose, Calif.) as reported previously (40). Tandem mass spectrometry (MS/MS) was performed as described previously (40). Collision-induced dissociation spectra that yielded amino acid sequences of the peptides were obtained. A search for matching sequences in the translated C. posadasii (strain 735) genome database (11, 20, 40; www.tigr.org) was conducted using the TurboSEQUEST software package, version 3.0 (Finnigan). Each spectrum which matched a peptide sequence in the Coccidioides database was confirmed by manual interpretation of the spectral data. Full-length amino acid sequences of proteins were deduced from the translated genomic database for each of the peptide sequence matches. The basic local alignment search tool (BLAST) (1) was used to search the Swiss-Prot/TrEMBL database (www.us.expasy.org/toolsblast) and the National Center for Biotechnology Information (NCBI) nonredundant protein database (www.ncbi.nih.nlm.gov) for reported proteins with sequence identities/similarities to each of the deduced proteins. The WOLF PSORT II algorithm was used to examine the deduced protein sequences for prediction of signal peptides and cellular localization (http://wolfpsort.seq.cbrc.jp/), while the GPI-SOM algorithm (16) was employed to examine translated sequences for putative glycosylphosphatidylinositol anchor sites. Predictions of N-glycosylation sites were performed using the NetNGlyc algorithm (www.cbs.dtu.dk/services/NetNGlyc/), while prediction of O-glycosylation sites made use of an algorithm reported by Julenius and coworkers (25).
Prediction of promiscuous human MHC class II-restricted epitopes.
The ProPred algorithm (www.imtech.res.in/raghava/propred/) (37) was used to predict the presence of promiscuous human major histocompatibility complex (MHC) class II-restricted epitopes in the sequences of deduced seroreactive proteins as previously reported (40). Deduced proteins that were predicted to be secreted and cell wall associated were selected for epitope analysis. Promiscuous epitopes are defined as peptides that are predicted to bind to at least 80% of the MHC class II molecules expressed by the 51 human leukocyte antigen (HLA) subregion DR alleles available for analysis in the ProPred algorithm.
Real-time PCR.
Expression levels of three genes (PEP1, PLB, and AMN1) which encode putative T-cell-reactive proteins selected on the basis of methods described above were examined during the parasitic cycle of C. posadasii. Procedures used for isolation of the parasitic cells, extraction of RNA, and quantitative real-time PCR (QRT-PCR) analysis were the same as previously described (40). The primer pair and amplicon used to examine PEP1 expression has been reported previously (40). The sequences of the sense and antisense primers for PLB expression analysis were 5′-CTGGAATTGCCAACGGAACATCGTTC-3′ and 5′-GACCGACTCAAGATAGCGCATCCAA-3′, respectively. The primers amplified a 309-bp PCR product, using single-stranded template cDNA generated by reverse transcription of total RNA as reported previously (11). In the case of AMN1, the sense and antisense primers used were 5′-ATTGAGACGGGCCTATTCTGCGAT-3′ and 5′-TACGTAGCGTAGCAGCCTTCAACA-3′, respectively. This primer pair amplified a 350-bp PCR product by the same method as described above. A 191-bp amplicon of the constitutively expressed glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene of C. posadasii (GenBank accession no. AF288134) was used for normalization of the QRT-PCR assay as described previously (40). The results of real-time PCR analysis of gene expression during selected stages of the parasitic cycle of C. posadasii are presented as ratios of the amount of transcript of each target gene to the amount of GAPDH transcript in each sample, as previously reported (20). The QRT-PCR experiments were performed in triplicate.
Expression of AMN1 and PLB by Escherichia coli.
Oligonucleotide primers were designed to amplify the cDNAs of PLB and AMN1, which encode amino acids 1 to 646 and 1 to 520, respectively (predicted full-length proteins). The nucleotide sequences of the sense and antisense primers used to amplify the PLB cDNA were 5′-ATGCTAGCCATATGAGACC TATCGGGGCC-3′ and 5′-GCGGCCGCCTCGAGTGGGTGGAAATTAGTATCACCA-3′, which contained engineered NdeI and XhoI restriction sites, respectively (underlined nucleotides). The amplification parameters for PLB cDNA were as follows: an initial denaturation step at 94°C for 2 min, followed by 30 cycles which consisted of denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 3 min. The 1.9-kb amplicon was subcloned into the pGEM-TE cloning vector (Promega), and the nucleotide sequence of the insert was determined as previously reported (21). The pGEM-TE-PLB plasmid was digested with NdeI and XhoI to release the 1.9-kb insert, which was then subcloned into the pET32b expression vector (EMD Biosciences Inc., La Jolla, Calif.). The pET32b-PLB plasmid construct was used to transform E. coli strain BL21(DE3) as described previously (21). The nucleotide sequences of the sense and antisense primers used to amplify the AMN1 cDNA were 5′-GAGCTCCATATGAAGGGATCCCCCGTACTC-3′ and 5′-CTCGAGAAGCTTCTATGACAGAACGCAGGATCA-3′, which contained engineered NdeI and HindIII restriction sites, respectively (underlined nucleotides). The conditions for amplification of AMN1 cDNA were the same as described above, except that the extension step was conducted for 2.5 min. The 1.6-kb amplicon was subcloned, sequenced, and inserted into the pET32b expression vector, which was used to transform E. coli as described above.
Purification of the recombinant proteins (rAmn1 and rPlb) was conducted as reported previously (29). The pET32b vector used for expression of the recombinant proteins encoded a 109-amino-acid N-terminal Trx-Tag thioredoxin fusion peptide (EMD Biosciences), which enhanced solubility of the expressed products. The endotoxin content of the stock solutions that contained the purified recombinant proteins was determined by use of a Limulus amebocyte lyase kit (QCL-1000; BioWhittaker, Walkersville, Md.) as previously reported (29). The stock solutions contained 3.0 to 5.0 endotoxin units per μg of protein. The recombinant proteins were separated by one-dimensional SDS-PAGE and tested for seroreactivity by immunoblot assays using either pooled patient sera or pooled healthy control sera as described above. Amino acid sequence analyses of the purified proteins were conducted by LC-MS/MS as reported previously (40).
IFN-γ ELISPOT assays.
Purified recombinant protein (5 μg of either rPlb or rAmm1) plus adjuvant was used to immunize separate groups (four animals each) of either 8-week-old female C57BL/6 mice or 12-week-old HLA-DR4 (DRB1*0401) transgenic mice (a gift from Thomas Forsthuber, University of Texas at San Antonio) as described previously (40). The adjuvant used was a synthetic oligodeoxynucleotide (ODN) preparation containing unmethylated CpG dinucleotides (CpG ODN; Integrated DNA Technologies, Inc., Coralville, Iowa). The CpG ODN sequence used to immunize mice was the same as reported previously (11). The CpG ODN was solubilized in buffer (10 μg in 50 μl phosphate-buffered saline) and mixed with 50 μl of incomplete Freund's adjuvant as previously described (29). The four separate groups of mice were immunized two times (2 weeks apart) with either rPlb or rAmn1 plus adjuvant. Two weeks after the second immunization, the spleens of each group of mice were harvested, separately pooled, and macerated as described previously (29). Separation of CD90+ T cells from the cell suspension was conducted using mouse CD90 (Thy 1.2) MicroBeads (Miltenyi Biotec Inc., Auburn, Calif.) as reported previously (40). This isolation method typically yielded ≥95% CD90+ T cells, based upon results of fluorescence-activated cell sorting analysis of the isolated splenocytes. Antigen-presenting cells (APCs) were isolated from pooled splenocytes obtained from five naïve (untreated), age- and gender-matched mice. This splenocyte suspension was subjected to antibody depletion of the CD90+ T cells followed by irradiation (40). Gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assays were performed as reported previously (40). Synthetic peptides spanning each of the ProPred-predicted promiscuous human MHC class II-restricted epitopes of Plb and Amn1 (see Table 3) were synthesized and tested for their ability to stimulate immune T-cell secretion of IFN-γ as described previously (40). Peptides (20-mers) that corresponded to regions of Plb and Amn1 not predicted to contain T-cell epitopes were also synthesized and included as negative controls. Detection and analysis of activated CD90+ T cells were conducted as reported previously (40). The frequency of IFN-γ-secreting, antigen-specific CD90+ T cells on the filtration membranes was calculated as the number of spots per 4 × 105 CD90+ T cells seeded in the presence of antigen (full-length recombinant protein or synthetic peptide) minus the number of spots per equal number of CD90+ T cells in medium alone.
TABLE 3.
Amino acid sequences of synthetic peptides selected from ProPred prediction of Plb and Amn1 ligands which bind to human MHC class II molecules
| Peptide reference | Peptide sequence | Corresponding amino acids of deduced protein | % of HLA-DR alleles predicted to bind to peptidesa |
|---|---|---|---|
| Plb | |||
| P1 | MRPIGAAVNALLTAALLAGT | 1-20 | 80 |
| P2 | AIPLDSNVHIRALPNAPNGY | 25-44 | 90 |
| P3 | TFLDVLRYYAQLQSAVAGKQ | 208-227 | 96 |
| P4 | PTIFGFVPLEYLGSKFQGGV | 309-328 | 88 |
| P5 | PDVNTFVNLGLNTRPTFFGC | 480-499 | 92 |
| P6 | TPLVVYIPNYPYTTWSNIST | 508-527 | 90 |
| P7 | ARIRASPSKHSVVVFSVVVL | 621-640 | 100 |
| P8 | PTCVGCAILSRSFERTGIAM | 560-579 | 2b |
| Amn1 | |||
| P9 | LFETTIRYLGGMISAYDLLK | 125-144 | 100 |
| P10 | PAKVDVLLAQSLKLADVLKF | 154-173 | 100 |
| P11 | NGLATTGTLVLEWTRLSDIT | 199-218 | 100 |
| P12 | DSFYEYLIKMYVYDKGRFGK | 272-291 | 82 |
| P13 | YYNLRPEVIESIYYAYRMTK | 408-427 | 92 |
| P14 | ESFLFAEVMKYSYLIHSPEA | 471-490 | 98 |
| P15 | FTAIGDVNTPDGGRKYDNQE | 452-471 | 0b |
Percentage of the 51 human MHC II alleles listed in the ProPred algorithm (http://www.imtech.res.in/raghava/propred/) to which the peptide is predicted to bind at a threshold setting of 5%.
Peptides predicted by ProPred algorithm not to bind to human MHC class II molecules served as a controls in the IFN-γ ELISPOT assays of immune T cells obtained from C57BL/6 or HLA-DR4 transgenic mice.
Vaccination, animal challenge, and evaluation of protection.
Imunoprotection experiments with C57BL/6 mice (females, 8 weeks old) were performed as previously described (40). Mice (15 per group) were immunized subcutaneously with either the purified, bacterially expressed recombinant phospholipase (rP1b; 1 μg or 5 μg per dose), recombinant alpha-mannosidase (rAmn1; 1 μg or 5 μg per dose), the previously described recombinant aspartyl protease (rPep1; 1 μg per dose) (40), or a combination of all three recombinant antigens at an immunization dose of 1 μg each. The same CpG ODN preparation as described above was used as the adjuvant in each case, and the same immunization protocol was employed. Control mice were immunized with adjuvant alone. Mice were challenged by the intranasal (i.n.) route with 80 viable arthroconidia of C. posadasii (strain C735) 4 weeks after the second immunization. The animals were scored for survival and evaluated for fungal burden in their lungs and spleens at 90 days postchallenge as previously described (29). Survival differences between groups were analyzed for statistical significance by the Kaplan-Meier method, and differences in fungal burden were analyzed by the Mann-Whitney U test as described previously (11).
Nucleotide sequence accession numbers.
The genomic and cDNA sequences of the PLB and AMN1 genes identified in the C. posadasii database, and confirmed by cloning and nucleotide sequence analysis, have been deposited in GenBank (accession no. DQ188099 and DQ176863, respectively).
RESULTS
2D PAGE/immunoblot analyses of parasitic cell wall extracts reveal multiple seroreactive proteins.
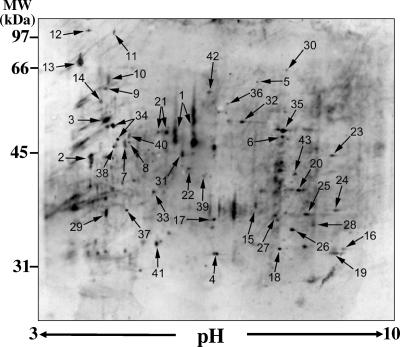
Immunoblots of 2D PAGE separations of multiple cell wall extracts of C. posadasii using the same protocol as described in this paper showed similar patterns of protein spots (Fig. 1). Several prominent patient seroreactive proteins identified by LC-MS/MS were previously observed in SDS-PAGE separations of culture filtrates and isolated parasitic cell wall extracts (40) and served as references in the electrophoretic separations. These included the previously reported C. posadasii chitinase 1 (Cts1) (35), aspartyl protease (Pep1), alpha-mannosidase (Amn1), proline/threonine-rich protein, and spherule outer wall glycoprotein (SOWgp) (22) (Fig. 1; Table 1). The estimated molecular masses and isoelectric points (pI) of these reference proteins were in the range of 43 to 70 kDa and pH 4.5 to 6.0, respectively, which included most of the seroreactive proteins of interest in this study. Approximately 98 seroreactive spots were distinguished in the immunoblot, but only 43 of these, which are numbered in Fig. 1, were reproducibly correlated with Coomassie blue-strained spots in the 2D PAGE sister gels. Each protein in the sister gel was excised and sequenced by LC-MS/MS. All 43 spots yielded peptide sequences which exactly matched translated protein sequences in the C. posadasii genome database. Putative protein identifications were conducted by BLAST searches of the Swiss-Prot/TrEMBL and NCBI databases for amino acid sequences which were homologous to the deduced full-length C. posadasii proteins, and the results are reported in Table 1. Each of the deduced, full-length protein sequences was deposited in the GenBank database. Fifteen of the 43 Coccidioides proteins in the cell wall extract were predicted to be cell wall associated or extracellular, and three of these polypeptides were suggested to be glycosylphosphatidylinositol anchored. The remaining 28 deduced proteins were predicted to be plasma membrane bound, cytosolic, or mitochondrion associated. Several of these 28 proteins were deduced from sequences of peptides derived from more than one 2D PAGE gel spot. However, in each case, the peptide sequences derived from trypsin digests of multiple gel spots revealed amino acid sequence matches with a single polypeptide deduced from the translated C. posadasii genome database. Electrophoretic separation and detection of components of the same protein are probably the result of proteolytic digestion, which may have occurred naturally in vitro and/or during preparation of the cell wall extract. We determined that the majority of proteins suspected to have undergone proteolysis, even in the presence of a broad-spectrum protease inhibitor cocktail, were predicted to be intracellular. In addition, sequence analyses of these putative cytosolic and mitochondrial proteins predicted essentially no N- or O-glycosylation linkages. We suggest that most of the membrane-bound, cytosolic and mitochondrial proteins are contaminants of the spherule homogenization and wall isolation procedures. Examination of the amino acid sequences of the putative wall-associated proteins in Table 1, on the other hand, typically revealed one or more predicted glycosylation sites. For example, two N-glycosylation sites were predicted for Pep1, at amino acids 138 and 338. The deduced phospholipase B was predicted to have six N-glycosylation sites (amino acids 176, 293, 453, 472, 503, and 534), while Amn1 has two predicted sites of N-linked glycosylation (amino acids 187 and 444). No O-glycosylation sites were predicted for these three deduced protein, but chitinase 1 (Cts1) had a predicted O-glycosylation linkage to amino acid 258. Linkage of sugar residues to the proteins may help to protect these cell wall-associated polypeptides from digestion during exposure to proteases which are secreted during parasitic cell development (32).
FIG. 1.
Immunoblot of 2D PAGE separation (10% gel) of the parasitic cell wall extract. Numbers correspond to protein spots identified by LC-MS/MS (see Table 1). MW, molecular mass.
TABLE 1.
Summary of deduced seroreactive proteins of C. posadasii identified in 2D PAGE separations of parasitic cell wall-extracted proteins
| Gel spot no. | Putative protein identificationa | GenBank accession no. of deduced C. posadasii protein | Predicted localization and GPI anchorb |
|---|---|---|---|
| 1 | Chitinase 1 (Cts1) | P54196 | Cw, extracellular |
| 2 | Aspartyl protease (Pep1) | DQ164306 | Cw, extracellular, GPI |
| 3 | Alpha-mannosidase (Amn1) | DQ176863 | Cw, extracellular |
| 4 | Metalloprotease 1 (Mep1) | AAQ07436 | Cw, extracellular |
| 5 | Endo-1,3-beta-glucanase | DQ674533 | Cw, extracellular |
| 6 | Endo-1,3-beta-glucanase | DQ674533 | Cw, extracellular |
| 7 | Carboxypeptidase Y | DQ176864 | Cw, extracellular |
| 8 | Exo-1,3-beta-d-glucanase | DQ674534 | Cw, extracellular |
| 9 | Protein disulfide isomerase | AAL50638 | Cw, extracellular |
| 10 | Proline/threonine-rich protein | DQ176865 | Cw, extracellular |
| 11 | Beta-glucosidase 5 (Bgl5) | AAL09829 | Cw, extracellular |
| 12 | Beta-glucosidase 2 (Bgl2) | AAF21242 | Cw, extracellular |
| 13 | Spherule outer wall glycoprotein (SOWgp) | AAL09436 | Cw, extracellular, GPI |
| 14 | Phospholipase B (Plb) | DQ188099 | Cw, extracellular, GPI |
| 15 | Beta-glucosidase 4 (Bgl4) | AAL09828 | Cw, ER |
| 16 | Porin | DQ674535 | Plasma membrane |
| 17 | Elongation factor 2 | DQ674536 | Cytosolic |
| 18 | Elongation factor 2 | DQ674536 | Cytosolic |
| 19 | Elongation factor 2 | DQ674536 | Cytosolic |
| 20 | Alcohol dehydrogenase I | DQ674537 | Cytosolic |
| 21 | Enolase | DQ674538 | Cytosolic |
| 22 | Fructose biphosphate aldolase | DQ674539 | Cytosolic |
| 23 | Citrate synthase | DQ674540 | Cytosolic |
| 24 | Citrate synthase | DQ674540 | Cytosolic |
| 25 | Malate dehydrogenase | DQ674541 | Cytosolic |
| 26 | Malate dehydrogenase | DQ674541 | Cytosolic |
| 27 | Malate dehydrogenase | DQ674541 | Cytosolic |
| 28 | 3-Hydroxybutyryl coenzyme A dehydrogenase | DQ674542 | Mitochondria |
| 29 | 70-kDa heat shock protein (HSP70) | DQ674543 | Mitochondria |
| 30 | Aconitase | DQ674544 | Mitochondria |
| 31 | Aconitase | DQ674544 | Mitochondria |
| 32 | Aconitase | DQ674544 | Mitochondria |
| 33 | Aconitase | DQ674544 | Mitochondria |
| 34 | ATP synthase(beta chain) | DQ674545 | Mitochondria |
| 35 | Dihydrolipoamide dehydrogenase | DQ674546 | Mitochondria |
| 36 | Electron transfer flavoprotein (alpha-subunit) | DQ674547 | Mitochondria |
| 37 | Electron transfer flavoprotein (alpha-subunit) | DQ674547 | Mitochondria |
| 38 | 60-kDa heat shock protein (HSP60) | AAD00521 | Mitochondria |
| 39 | Ketol-acid reductoisomerase | DQ674548 | Mitochondria |
| 40 | Mitochondrial processing peptidase subunit | DQ674549 | Mitochondria |
| 41 | NADH-ubiquinone oxidoreductase subunit | DQ674550 | Mitochondria |
| 42 | Succinate dehydrogenase | DQ674551 | Mitochondria |
| 43 | Processing/enhancing protein precursor | DQ674552 | Mitochondria |
Protein identity based on homology to reported fungal proteins determined by BLAST searches (1).
Based on amino acid sequence analysis using WoLF PSORT II and GPI-SOM algorithms. Cw, cell wall associated; ER, endoplasmic reticulum; GPI, glycosylphosphatidylinositol anchored.
Deduced proteins predicted to be cell wall associated include candidate T-cell-reactive antigens.
The amino acid sequences of the 15 deduced full-length, seroreactive proteins which were identified in the 2D PAGE gels and predicted to be cell wall associated were examined using the ProPred algorithm. The results of this search for promiscuous epitopes that bind to human MHC class II molecules are presented in Table 2. All but three of the putative cell wall proteins contain multiple promiscuous epitopes. None of these epitopes were predicted to include N- or O-glycosylation sites discussed above. We generated a rank order of the candidate T-cell-reactive antigens based on the ratio of the number of ProPred-predicted promiscuous T-cell epitopes to the molecular mass of the deduced full-length protein. We reasoned that cell wall-associated proteins with the highest number of promiscuous epitopes on a comparative molecular mass basis are candidate antigens worthy of further evaluation in cellular immunoassays and protection experiments. We selected the top three deduced proteins (Pep1, Plb, and Amn1) in Table 2. The aspartyl protease (Pep1) of C. posadasii was previously identified as an immunogenic protein and candidate vaccine against coccidioidomycosis (40).
TABLE 2.
Deduced proteins of C. posadasii selected on the basis of predicted cell wall association and analyzed by the ProPred algorithm for presence of epitopes which bind to human MHC class II-restricted molecules
| Gel spot no. | Selected seroreactive proteina | Predicted molecular mass (kDa) | No. of predicted promiscuous T-cell-reactive epitopesb | Ratio of no. of predicted promiscuous epitopes to molecular mass (kDa) of deduced protein |
|---|---|---|---|---|
| 2 | Aspartyl protease (Pep1) | 43.5 | 5 | 0.115 |
| 14 | Phospholipase B (Plb) | 68.6 | 7 | 0.107 |
| 3 | Alpha-mannosidase (Amn1) | 56.9 | 6 | 0.105 |
| 1 | Chitinase 1 (Cts1) | 47.4 | 5 | 0.104 |
| 15 | Beta-glucosidase 4 (Bgl4) | 32.9 | 3 | 0.091 |
| 8 | Exo-1,3-beta-d-glucanase | 93.1 | 8 | 0.085 |
| 5, 6 | Endo-1,3-beta-glucanase | 96.3 | 8 | 0.083 |
| 7 | Carboxypeptidase Y | 60.3 | 5 | 0.083 |
| 12 | Beta-glucosidase 2 (Bgl2) | 92.8 | 5 | 0.054 |
| 11 | Beta-glucosidase 5 (Bgl5) | 56.7 | 3 | 0.053 |
| 9 | Protein disulfide isomerase | 57.3 | 2 | 0.035 |
| 13 | Spherule outer wall glycoprotein (SOWgp) | 46.3 | 1 | 0.021 |
| 4 | Metalloprotease 1 (Mep1) | 29.7 | 0 | 0 |
| 10 | Proline/threonine-rich protein | 29.0 | 0 | 0 |
PEP1, PLB, and AMN1 are expressed at maximum levels during different stages of the parasitic cycle.
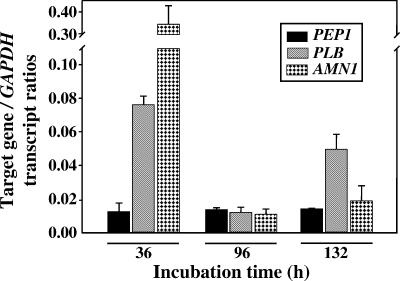
We have previously reported, on the basis of quantitative real-time PCR analysis of PEP1 expression, that approximately equal amounts of the gene transcript are detected in total mRNAs isolated from each of the same developmental stages of the parasitic cycle examined in this study (40) (Fig. 2). We also pointed out that in spite of the comparatively low, constitutive expression of this gene, the Pep1 protein was a prominent component of 2D PAGE separations of the parasitic cell wall extract. We speculated that the PEP1 transcript is stable throughout the parasitic cycle and/or that the cell wall-associated Pep1 accumulates and persists during spherule development. PLB and AMN1 expression during the parasitic cycle was also compared to temporal expression of the constitutive GAPDH gene. The largest amounts of transcript of both genes were detected during the early, presegmented phase of spherule development (36 h of incubation). The amount of AMN1 transcript decreased sharply during subsequent stages of the parasitic cycle, while PLB expression remained elevated compared to PEP1 and AMN1 expression during the endosporulation phase (132 h). Although the parasitic cell wall isolate was derived from a mixture of segmented spherules (96 h) and endosporulation phase parasitic cells (132 h), Amn1 was observed to be a prominent protein in the Coomassie blue-stained 2D PAGE separation of the cell wall extract, suggesting that Amn1, like Pep1, accumulates and persists in association with the spherule wall during parasitic cell development. The Plb protein, on the other hand, appeared to be a minor protein in the cell wall extract or was not well stained with Coomassie blue.
FIG. 2.
QRT-PCR of PEP1, PLB, and AMN1 gene expression in vitro during the parasitic cycle compared to expression levels of the constitutive GAPDH gene. Error bars indicate standard deviations.
Bacterium-expressed recombinant Plb and Amn1 are recognized by sera from patients with coccidioidomycosis.
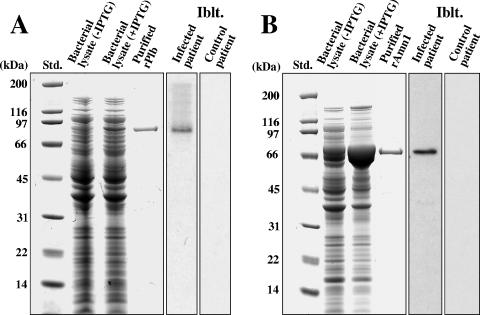
Recombinant Plb was produced by E. coli transformed with the pET32b-PLB plasmid (Fig. 3A). Moderate amounts of rPlb were produced in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside). The predicted and observed molecular masses of the recombinant protein in the SDS-polyacrylamide gel are 86 kDa, which includes the vector-encoded fusion peptide that contained the His tag at its N terminus. The 86-kDa rPlb was isolated by nickel affinity chromatography, subjected to electrophoresis, excised from the gel, trypsin digested, and sequenced by LC-MS/MS analysis. Five peptides were obtained, and their sequences matched that of the single translated PLB gene in the C. posadasii genome database. Immunoblot assays of the bacterial lysate which contained the rPlb were conducted with either pooled sera from patients with confirmed coccidioidal infection or sera from control patients without fungal infections. The recombinant protein was recognized only by sera from Coccidioides-infected patients (Fig. 3A).
FIG. 3.
SDS-PAGE separation and immunoblot (Iblt.) analysis of E. coli-expressed rPlb (A) and rAmn1 (B). Shown are standards (Std.), lysates of bacteria transformed with either the pET32b-PLB (A) or pET32b-AMN1 (B) plasmid vector in the presence (+) or absence (−) of IPTG, and the nickel affinity-isolated rPlb and rAmn1, which were subsequently purified by electroelution from SDS-polyacrylamide gels as previously reported (11). The immunoblots of the bacterial lysates were incubated with either pooled sera from patients with confirmed coccidioidal infection or pooled sera from control patients.
E. coli transformed with the pET32b-AMN1 plasmid showed a marked increase in production of rAmn1 in the presence of IPTG compared to in the absence of the induction reagent (Fig. 3B). Isolation and sequence analysis of the 74-kDa recombinant protein were conducted as described above. Only pooled sera from patients with coccidioidomycosis recognized rAmn1 in the immunoblot of the SDS-PAGE separation of the bacterial lysate.
Synthetic peptides whose sequences correspond to the epitopes predicted to bind to human MHC II complexes stimulate immune T-cell response in vitro.
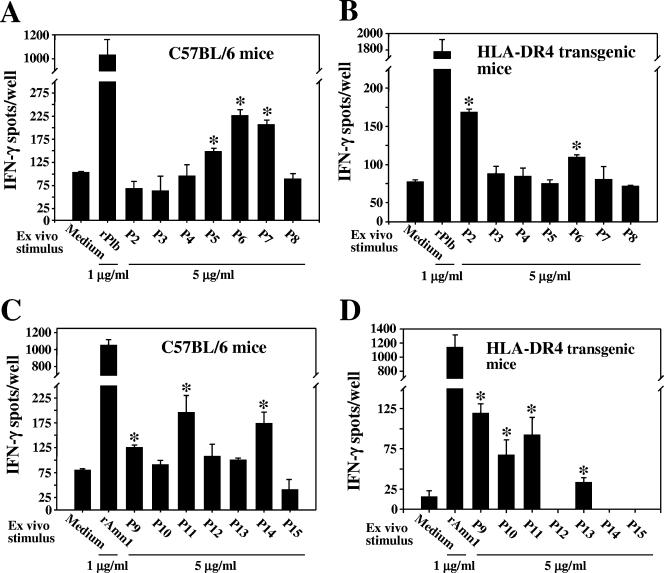
Amino acid sequence analysis of Pep1, Plb, and Amn1 with the ProPred algorithm identified five, seven, and six regions (18- to 32-mers) of the respective full-length proteins that were predicted to contain ligands which can bind to human MHC class II molecules (40) (Table 3). Synthetic peptides corresponding to six of the seven epitopes of Plb (P2 to P7; Table 3) were used separately to test the in vitro response of immune T cells obtained from rPlb-immunized C57BL/6 and HLA-DR4 transgenic mice (Fig. 4A and B, respectively). The peptide corresponding to P1 was not synthesized. A single synthetic peptide (P8), which was not predicted to bind to any of the 51 HLA-DR molecules examined in the ProPred algorithm, was included as a negative control. IFN-γ ELISPOT assays of the immune T-cell response to each of the synthetic peptides at a concentration of 5 μg/ml and to the full-length recombinant proteins at 1 μg/ml were compared to the background response of the T lymphocytes in growth medium alone. T cells derived from both immunized C57BL/6 and transgenic mice showed significant responses to the presence of the full-length rPlb and peptide P6 compared to the background response (P = 0.0012 and 0.0011 and P = 0.001 and 0.0013, respectively). The immune T lymphocytes obtained from the inbred C57BL/6 and transgenic mice were otherwise distinguished by their response to different Plb epitopes (P5/P7 and P2, respectively). Immune T cells from both strains of mice showed no significant response to the other synthetic peptides (P3 and P4) or to the control peptide, P8.
FIG. 4.
Assessment of IFN-γ production by immune CD90+ T cells derived from C57BL/6 or HLA-DR4 (DRB1*0401) transgenic mice, conducted by IFN-γ ELISPOT assays. Asterisks indicate statistically significant differences between responses of immune T cells in the presence of peptides and those in the presence of medium alone. Peptides P8 and P15 correspond to regions of the respective proteins not predicted to bind to MHC II complexes and served as negative controls. These results are representative of three separate experiments. Error bars indicate standard deviations.
Sequence analysis of Amn1 identified six regions of the deduced protein predicted to be T-cell epitopes (Table 3). In this case, greater similarity was observed between the response of immune T cells obtained from C57BL/6 and transgenic mice to the presence of the same set of synthetic peptides (P9 to P14; Fig. 4C and D). The full-length rAmn1 as well as peptides P9 and P11 stimulated significant response of immune T cells derived from the two strains of mice (P = 0.0042 and 0.0004, P = 0.3392 and 0.0035, and P = 0.0043 and 0.0035, respectively). On the other hand, immune T cells expressing the murine MHC II complex (C57BL/6) versus the humanized complex (DRB1*0401 strain) were distinguished by their response to P14 and P13, respectively. Immune T cells from the two mouse strains failed to respond to the control peptide P15. We have also shown that CD90+ T cells isolated from mice immunized with phosphate-buffered saline plus adjuvant failed to produce significant amounts of IFN-γ upon exposure to either recombinant Plb or Amn1 or to the respective synthetic peptides in ELISPOT assays (data not shown). We proposed in our previous report (40) that induction of greater cellular responses to the peptides representing putative T-cell epitopes may have been obtained with higher concentrations of the synthetic products. These assays are planned for future studies. However, the results shown in Fig. 4 A to D suggest that both rPlb and rAmn1 are T-cell-reactive proteins and confirm the ProPred prediction that each of these polypeptides contains multiple epitopes that can activate murine and HLA-DR4 immune T cells. As previously reported (40), we also concluded that selected synthetic epitopes of Pep1 induce immune T cells in vitro to produce significant amounts of IFN-γ, and on this basis, the recombinant Pep1 protein was selected for protection experiments in our murine model of coccidioidomycosis.
We conducted BLASTp searches (1) of the murine and human genome databases, using each of the peptides of Plb, Amn1, and Pep1 (40) that elicited a positive in vitro response of immune T cells derived from either C57BL/6 or transgenic mice, to determine percent amino acid sequence homology with mammalian proteins. None of the peptides of Plb showed homology to murine or human proteins. Two of the immunoreactive peptides of Amn1 (P9 and P13) revealed 65% and 70% sequence identity, respectively, to a human alpha-mannosidase homolog. None of the other immunoreactive peptides of Amn1 or Pep1 (40) showed sequence homology with mammalian proteins.
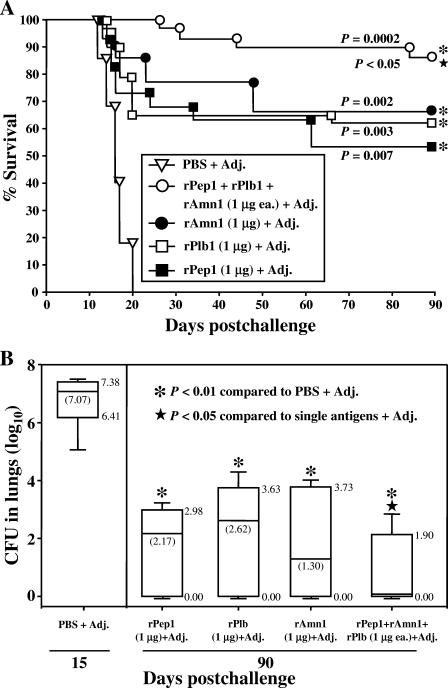
Vaccination with the combination of rPep1, rPlb, and rAmn1 provides enhanced protection against a lethal intranasal challenge of Coccidioides.
C57BL/6 mice inoculated intranasally with a lethal dose of C. posadasii arthroconidia (approximately 80 viable spores) show a highly reproducible mortality profile over a period of 20 to 25 days postchallenge (11, 40) (Fig. 5A). Separate groups of mice (15 per group) were vaccinated with one of three recombinant proteins (rPep1, rPlb, or rAmn1) at doses of either 1 μg (Fig. 5A) or 5 μg (not shown). We previously reported that C57BL/6 mice immunized with rPep1 showed a marked increase in percent survival after i.n. challenge with Coccidioides compared to nonvaccinated mice and that the difference in percent survival between groups of mice vaccinated with either 1-μg or 5-μg doses of the recombinant protein was not statistically significant (40). Comparison of survival plots of rPlb- and rAmn1-vaccinated mice, using doses of 1 μg or 5 μg, also revealed no statistically significant difference by Kaplan-Meier analysis (P = 0.79 and 0.85, respectively) (data not shown). We also compared the protective efficacies of the single recombinant protein vaccines at the 1-μg dose to that of the multivalent vaccine composed of all three proteins (rPep1, rPlb, and rAmn1), each at a 1-μg dose. Figure 5A is a representative survival plot from two separate experiments using the same vaccination and challenge protocols. A statistically significant difference (P < 0.05) was observed between the survival plots of mice immunized with the multivalent vaccine versus the single recombinant protein vaccines. On the basis of this observation, we also examined whether a significant difference was evident in clearance of the pathogen from the spleens and lungs of infected C57BL/6 mice which had been vaccinated with the single recombinant proteins versus the combined proteins (Fig. 5B). Control mice sacrificed at 15 days postchallenge showed high numbers of CFU in their lungs (mean of 107.07) (Fig. 5B) and spleens (103) (not shown). None of the vaccinated mice showed detectable CFU in their spleens between 15 and 90 days after intranasal challenge. Each group of mice vaccinated either with the single recombinant protein (rPep1, rPlb, or rAmn1) or the combined recombinant protein vaccine showed a significant reduction of the fungal burden in their lungs at 90 days postchallenge compared to the control mice at 15 days (P = 0.001, 0.004, 0.001, and 0.0001, respectively). In support of the trend observed in Fig. 5A, mice vaccinated with the three recombinant proteins in combination showed significantly better clearance of the pathogen from their lungs at 90 days than mice vaccinated with either rPep1, rPlb, or rAmn1 alone (P = 0.362, 0.016, and 0.045, respectively).
FIG. 5.
(A) Representative evaluation of the protective efficacy of single recombinant protein vaccines (rPep1, rPlb, and rAmn1) compared to that of the multivalent vaccine. In each case, CpG ODN was used as the adjuvant (Adj.), and C57BL/6 mice were challenged by the intranasal route with a potentially lethal inoculum of C. posadasii arthroconidia. Each recombinant protein was administered singly or in combination at 1 μg per dose. Statistical significances (P values) of the differences in survival plots for the vaccinated versus nonvaccinated mice (asterisks) are shown. The P value for difference between the survival plot for mice immunized with the combined recombinant vaccine versus the single recombinant vaccines (star) is also shown. The results are representative of two separate vaccination/survival experiments using the same immunization and challenge protocols. PBS, phosphate-buffered saline. (B) Representative box plots of CFU detected in dilution plate cultures of lung homogenates obtained from nonvaccinated, i.n.-inoculated control mice at 15 days postchallenge and from vaccinated, i.n.-inoculated mice at 90 days postchallenge. The boxes indicate the 25th and 75th percentiles, and the bars show the 5th and 95th percentiles. The horizontal lines within the boxes indicate the medians. The median value for CFU in the lungs of mice vaccinated with the combined vaccine was less than 1. The asterisks indicate statistically significant differences between the fungal burdens in lungs of vaccinated and nonvaccinated mice. The P values are provided in the text. The star indicates a significant difference between mice immunized with the combined vaccine versus each of the single recombinant protein vaccines. The results are representative of two separate fungal burden assays.
DISCUSSION
Animal model studies of coccidioidomycosis have demonstrated that protective immunity against this disease requires the production of interleukin-12, IFN-γ, and MHC class II-restricted T cells (26). Fierer and coworkers (17) have shown that CD4+ T-cell knockout mice vaccinated with a live attenuated strain of Coccidioides were protected against a lethal intraperitoneal infection of the pathogen, suggesting that CD8+ T cells also play a role in vaccine-induced immunity. We have argued that coccidioidal antigens that contain epitopes which bind in a promiscuous manner to murine or human MHC class II molecules and activate immune T cells to secrete significant amounts of IFN-γ are candidate vaccines against coccidioidomycosis (40). Furthermore, use of an appropriate adjuvant, such as CpG ODN, which stimulates immunity in an interleukin-12/IFN-γ-dependent manner, can enhance the efficacy of the vaccine to induce a protective host response to coccidioidal infection (11, 29, 40). Our laboratory and others have demonstrated that the parasitic cell wall of Coccidioides is a reservoir of immunoreactive macromolecules, some of which are vaccine candidates (36). Recent studies have shown that soluble extracts from spherule homogenates can promote human dendritic cell maturation and activation, a pivotal event during the cell-mediated immune response to coccidioidal infection (12). Two-dimensional electrophoresis, combined with immunoblot analysis and bioinformatics, has been used to identify fungal proteins in parasitic cell wall extracts which are worthy of evaluation as vaccine components (8, 33, 40). We have also incorporated into this approach to the discovery of candidate vaccines the use of the ProPred algorithm (37) to screen amino acid sequences of immunoreactive proteins for predicted promiscuous HLA-DR binding epitopes. This algorithm is a matrix-based prediction that relies on mathematical processing of individual peptides. The sum matrix values for each peptide frame yield a peptide score which is suggested to correlate with the binding affinity of the HLA ligands. Peptides predicted to bind to MHC II complexes have been confirmed to be T-cell epitopes by tetramer-guided epitope mapping, which supports the biological validation of the ProPred algorithm (27). T-cell epitope prediction has recently been used to design microbial and cancer vaccines (7, 9). A prototype tuberculosis vaccine which contains epitopes derived from mapping selected Mycobacterium tuberculosis genome open reading frames has been generated (9). The M. tuberculosis epitope selections were validated by measuring T-cell responses of human peripheral blood mononuclear cells obtained from tuberculin skin test-positive donors. Twenty-four validated M. tuberculosis epitopes were selected for inclusion in the recombinant protein vaccine used to immunize HLA-DRB*0101 transgenic mice. Eight of the 24 epitopes induced a potent IFN-γ ELISPOT T-cell recall response 1 week after the last immunization. The HLA-DR4 transgenic mice used in the study by Ito et al. were genetically engineered to express the extracellular α1 and β1 domains of HLA-DR4, which form the peptide binding groove that defines peptide binding specificity in conjunction with the murine α2 and β2 domains of the CD4 molecules (23). The transgenic mice maintain their IE cytoplasmic domains, which are essential for signaling and sorting of the MHC class II molecules and for performing their normal APC functions. The transgenic mice were backcrossed to MHC-deficient IE mice to eliminate the expression of endogenous murine MHC class II molecules (30). The HLA-DR allele (DRB1*0401) is the most prevalent DR4 allele in humans (38), and we argue that the transgenic mouse strain employed in these studies is appropriate for identification of Coccidioides-derived T-cell-reactive antigens and for evaluation of candidate vaccines against coccidioidomycosis. The combined applications of immunoproteomics and bioinformatics to the discovery of T-cell-reactive proteins is a promising approach to the development of microbial vaccines (9).
We have previously reported that a Triton X-114 detergent extract of the isolated parasitic cell wall was able to protect C57BL/6 mice against a potentially lethal pulmonary infection with Coccidioides (approximately 90% survival beyond 50 days postchallenge). We focused on a single, prominent cell wall-associated antigenic component of the Triton X-114 extract, which showed 93% sequence similarity with a cell wall-bound aspartyl protease of Aspergillus fumigatus (35). Using the same methods as described in this paper, we reported that the C. posadasii protein (Pep1) contains multiple predicted T-cell epitopes and that the recombinant protein (rPep1) both stimulates in vitro production of IFN-γ by immune T lymphocytes and protects C57BL/6 mice against a lethal intranasal infection of C. posadasii arthroconidia (40). On the basis of these results, we argued that the bacterium-expressed recombinant protein is a vaccine candidate. The immunoblots of 2D-PAGE separations of the crude cell wall extract used to detect Pep1 also revealed numerous other patient seroreactive proteins, including a putative phospholipase B (Plb) and alpha-mannosidase (Amn1). The results of protein sequence analyses, cellular immunoassays, and protection experiments using these two polypeptides, either singly or in combination with recombinant Pep1, have confirmed that recombinant Plb and Amn1 are additional T-cell-reactive antigens that can be incorporated into a vaccine against coccidioidomycosis. Necropsied mice that had been vaccinated with either a single recombinant protein or the three combined proteins, and which survived to 90 days after intranasal challenge, all showed that a granulomatous response had occurred in their lungs. Histopathological examinations of the lung tissue of these mice revealed well-differentiated granulomata, indicative of a protective response to coccidioidal infection (31). Shubitz and coinvestigators (36) have proposed that, in humans, a multicomponent vaccine against coccidiodomycosis has the potential to stimulate a broader range of T-cell clones than a single recombinant protein vaccine and thus may be more efficacious in evoking protection in immunologically heterogenous human populations. For example, combining antigens from the sporozote, intrahepatic, asexual erythrocytic, and even sexual stages of Plasmodium falciparum into a multivalent vaccine against malaria has been suggested to provide the greatest likelihood of achieving formidable protection against this insidious organism (14). Vaccination of C57BL/6 mice with a multivalent vaccine that consists of recombinant Pep1, Plb, and Amn1, together with CpG ODN as the adjuvant, resulted in increased survival and better pathogen clearance in our murine model of coccidioidomycosis than immunization with the single recombinant proteins. Even a modest increase in protection with the multicomponent vaccine could translate into improved disease prevention in the clinical setting (36). As in the malarial vaccine cited above, we suggest that our multivalent vaccine against coccidioidomycosis contains T-cell antigens expressed at maximum levels during different stages of the parasitic cycle. In vitro quantitative real-time PCR analyses of gene expression indicated that production of peak amounts of PLB and AMN1 transcripts occurs during initiation of the parasitic cycle, while the PEP1 gene is expressed constitutively. Immunoblot assays of protein production during parasitic cell development are necessary to determine whether there is a correlation between the transcriptional and translational data. Nevertheless, expression of the vaccine candidates during initial spherule formation (i.e., 36-h in vitro incubation period) suggests that activation of the host immune response to coccidioidal infection in vaccinated mice may occur prior to metastasis (endosporulation) of the pathogen. Polymorphonuclear neutrophils, macrophages, and dendritic cells are likely to play key roles in defense against Coccidioides infections (2, 15). However, mature spherules are typically 30 to 80 μm or larger in diameter, and single phagocytes are unable to engulf these fungal cells. The opportunity for activated phagocytes to engulf the pathogen is restricted to periods of spherule initial formation and endosporulation. Although PLB expression is up-regulated during endosporulation, inclusion of an additional immunogenic protein in the multivalent vaccine which is produced during spherule rupture and endospore release may further enhance protection. The chitinase 1 gene of Coccidioides (CTS1) (34) has been shown to be expressed at a sixfold-higher level during the endosporulation phase than during earlier stages of the parasitic cycle (unpublished data). We have shown in this study that Cts1 is predicted to contain multiple promiscuous human T-cell epitopes. Vaccination of C57BL/6 mice with recombinant Cts1 has been reported to result in moderate but significant protection against a potentially lethal intraperitoneal infection of C. posadasii (T. N. Kirkland, personal communication). We suggest that the addition of rCts1 to the multivalent vaccine may further improve its protective efficacy.
Shubitz and coworkers (36) have pointed out that although a multicomponent vaccine may have an immunologic benefit to the host, the cost of its production is prohibitively higher than that of a single-component vaccine. Future studies in our laboratory will be directed to the design of a chimeric protein which contains validated T-cell epitopes of the selected vaccine candidates described in this report. Epitope-driven, chimeric vaccines offer several advantages over vaccines that encode whole-protein antigens (9). The use of epitopes overcomes potential safety concerns associated with whole recombinant proteins. Epitope-based vaccines also appear to be capable of inducing more potent responses than whole-protein vaccines by simultaneously directing the host immune response to multiple dominant and subdominant epitopes. The selection of epitopes which would be incorporated into a human chimeric vaccine against coccidioidomycosis will require additional evaluations of immunoreactivity and HLA binding affinity. We have initiated in vitro binding assays of selected synthetic peptides of Pep1, Plb, and Amn1 which showed the highest immunoreactivity in IFN-γ ELISPOT assays of CD90+ T cells derived from HLA-DR4 transgenic mice. The method employed determines relative affinities of selected peptides for the HLA-DR4 molecules by an inhibition enzyme-linked immunosorbent assay based on a dissociation-enhanced lanthanide fluoroimmunoassay (38). Preliminary results showed that the 50% inhibitory concentrations of the selected peptides are in the range of 60 nM to 1600 nM, indicating high to moderate affinity for HLA-DR4 molecules. Details of these assays will be presented in a future report. Finally, the use of eukaryotic expression systems (i.e., Saccharomyces cerevisiae, [36] or Uncinocarpus reesii [41]) for production of the chimeric vaccine overcomes the potential adverse effects of endotoxin contamination associated with bacterially expressed proteins (18). The eukaryotic expression system also introduces the option of modifying the chimeric constructs to include amino acid sequences with predicted O-glycosylation linkages. For example, mannosylation may significantly augment vaccine immunogenicity by targeting antigens to receptors on macrophages and dendritic cells (e.g., mannose receptors and CD260 of macrophages and DC-SIGN and CD290 of dendritic cells) (12, 28), resulting in more efficient activation and antigen presentation by these APCs.
Acknowledgments
Support for this study was provided by Public Health Service grants AI19149 and U01 AI50910 (Coccidioides genome sequencing project) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. Support was also provided by the Margaret Batts Tobin Foundation of San Antonio, Texas.
Editor: A. Casadevall
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awasthi, S., V. Awasthi, D. M. Magee, and J. J. Coalson. 2005. Efficacy of antigen 2/proline-rich antigen cDNA-transfected dendritic cells in immunization of mice against Coccidioides posadasii. J. Immunol. 175:3900-3906. [DOI] [PubMed] [Google Scholar]
- 3.Barnato, A. E., G. D. Sanders, and D. K. Owens. 2001. Cost-effectiveness of a potential vaccine for Coccidioides immitis. Emerg. Infect. Dis. 7:797-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2005. Summary of notifiable diseases—United States, 2003. Morb. Mortal. Wkly. Rep. 52:1-88. [PubMed] [Google Scholar]
- 5.Cole, G. T., J. Xue, C. Okeke, E. J. Tarcha, V. Basrur, R. A. Schaller, R. A. Herr, J.-J. Yu, and C.-Y. Hung. 2004. A vaccine against coccidioidomycosis is justified and attainable. Med. Mycol. 42:189-216. [DOI] [PubMed] [Google Scholar]
- 6.Comrie, A. C. 2005. Climate factors influencing coccidioidomycosis seasonality and outbreaks. Environ. Health Persp. 113:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consogno, G., S. Manici, V. Facchinetti, A. Bachi, J. Hammer, B. M. Conti-Fine, C. Rugarli, C. Traversari, and M. P. Protti. 2003. Identification of immunodominant regions among promiscuous HLA-DR-restricted CD4+ T-cell epitopes on the tumor antigen MAGE-3. Blood 101:1038-1044. [DOI] [PubMed] [Google Scholar]
- 8.da Fonseca, C. A., R. S. Jesuino, M. S. Felipe, D. A. Cunha, W. A. Brito, and C. M. Soares. 2001. Two-dimensional electrophoresis and characterization of antigens from Paracoccidioides brasiliensis. Microbes Infect. 3:535-542. [DOI] [PubMed] [Google Scholar]
- 9.De Groot, A. S., J. McMurry, L. Marcon, J. Franco, D. Rivera, M. Kutzler, D. Weiner, and B. Martin. 2005. Developing an epitope-driven tuberculosis (TB) vaccine. Vaccine 23:2121-2131. [DOI] [PubMed] [Google Scholar]
- 10.De Hoog, G. S., B. Bowman, Y. Graser, G. Haase, M. El Fari, A. H. Gerrits van den Ende, B. Melzer-Krick, and W. A. Untereiner. 1998. Molecular phylogeny and taxonomy of medically important fungi. Med. Mycol. 36(Suppl. 1):52-56. [PubMed] [Google Scholar]
- 11.Delgado, N., J. Xue, J.-J. Yu, C.-Y. Hung, and G. T. Cole. 2003. A recombinant beta-1,3-glucanosyltransferase homolog of Coccidioides posadasii protects mice against coccidioidomycosis. Infect. Immun. 71:3010-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dionne, S. O., A. B. Podany, Y. W. Ruiz, N. M. Ampel, J. N. Galgiani, and D. F. Lake. 2006. Spherules derived from Coccidioides posadasii promote human dendritic cell maturation and activation. Infect. Immun. 74:2415-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dismukes, W. E. 2006. Antifungal therapy: lessons learned over the past 27 years. Clin. Infect. Dis. 42:1289-1296. [DOI] [PubMed] [Google Scholar]
- 14.Doolan, D. L., and S. L. Hoffman. 1997. Multi-gene vaccination against malaria: a multistage, multi-immune response approach. Parasitol. Today 13:171-178. [DOI] [PubMed] [Google Scholar]
- 15.Drutz, D. J., and M. Huppert. 1983. Coccidioidomycosis: factors affecting the host-parasite interaction. J. Infect. Dis. 147:372-390. [DOI] [PubMed] [Google Scholar]
- 16.Fankhauser, N., and P. Maser. 2005. Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics 21:1846-1852. [DOI] [PubMed] [Google Scholar]
- 17.Fierer, J., C. Waters, and L. Walls. 2006. Both CD4+ and CD8+ T cells can mediate vaccine-induced protection against Coccidioides immitis infection in mice. J. Infect. Dis. 193:1323-1331. [DOI] [PubMed] [Google Scholar]
- 18.Geier, D. A., and M. R. Geier. 2002. Clinical implications of endotoxin concentrations in vaccines. Ann. Pharmacother. 36:776-780. [DOI] [PubMed] [Google Scholar]
- 19.Hung, C.-Y., N. M. Ampel, L. Christian, K. R. Seshan, and G. T. Cole. 2000. A major cell surface antigen of Coccidioides immitis which elicits both humoral and cellular immune responses. Infect. Immun. 68:584-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung, C.-Y., K. R. Seshan, J.-J. Yu, R. Schaller, J. Xue, V. Basrur, M. J. Gardner, and G. T. Cole. 2005. A metalloproteinase of Coccidioides posadasii contributes to evasion of host detection. Infect. Immun. 73:6689-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung, C.-Y., J.-J. Yu, P. F. Lehmann, and G. T. Cole. 2001. Cloning and expression of the gene which encodes a tube precipitin antigen and wall-associated beta-glucosidase of Coccidioides immitis. Infect. Immun. 69:2211-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung, C.-Y., J.-J. Yu, K. R. Seshan, U. Reichards, and G. T. Cole. 2002. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70:3443-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, K., H. J. Bian, M. Molina, J. Han, J. Magram, E. Saar, C. Belunis, D. R. Bolin, R. Arceo, R. Campbell, F. Falcioni, D. Vidovic, J. Hammer, and Z. A. Nagy. 1996. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. J. Exp. Med. 183:2635-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, R. H., and H. E. Einstein. 2006. Coccidioidal meningitis. Clin. Infect. Dis. 42:103-107. [DOI] [PubMed] [Google Scholar]
- 25.Julenius, K., A. Molgaard, R. Gupta, and S. Brunak. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153-164. [DOI] [PubMed] [Google Scholar]
- 26.Kirkland, T. N., E. Raz, and S. K. Datta. 2006. Molecular and cellular mechanisms of protective immunity to coccidioidomycosis. Vaccine 24:495-500. [DOI] [PubMed] [Google Scholar]
- 27.Kwok, W. W., J. A. Gebe, A. Liu, S. Agar, N. Ptacek, J. Hammer, D. M. Koelle, and G. T. Nepom. 2001. Rapid epitope identification from complex class-II-restricted T-cell antigens. Trends Immunol. 22:583-588. [DOI] [PubMed] [Google Scholar]
- 28.Lam, J. S., M. K. Mansour, C. A. Specht, and S. M. Levitz. 2005. A model vaccine exploiting fungal mannosylation to increase antigen immunogenicity. J. Immunol. 175:7496-7503. [DOI] [PubMed] [Google Scholar]
- 29.Li, K., J.-J. Yu, C.-Y. Hung, P. F. Lehmann, and G. T. Cole. 2001. Recombinant urease and urease DNA of Coccidioides immitis elicit an immunoprotective response against coccidioidomycosis in mice. Infect. Immun. 69:2878-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madsen, L. S., E. C. Andersson, L. Jansson, M. Krogsgaard, C. B. Andersen, J. Engberg, J. L. Strominger, A. Svejgaard, J. P. Hjorth, R. Holmdahl, K. W. Wucherpfennig, and L. Fugger. 1999. A humanized model for multiple sclerosis using HLA-DR2 and a human T-cell receptor. Nat. Genet. 23:343-347. [DOI] [PubMed] [Google Scholar]
- 31.Mirbod-Donovan, F., R. Schaller, C.-Y. Hung, J. Xue, U. Reichard, and G. T. Cole. 2006. Urease produced by Coccidioides posadasii contributes to the virulence of this respiratory pathogen. Infect. Immun. 74:504-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monod, M., S. Capoccia, B. Lechenne, C. Zaugg, M. Holdom, and O. Jousson. 2002. Secreted proteases from pathogenic fungi. Intern. J. Med. Microbiol. 292:405-419. [DOI] [PubMed] [Google Scholar]
- 33.Orsborn, K. I., L. F. Shubitz, T. Peng, E. M. Kellner, M. J. Orbach, P. A. Haynes, and J. N. Galgiani. 2006. Protein expression profiling of Coccidioides posadasii by two-dimensional differential in-gel electrophoresis and evaluation of a newly recognized peroxisomal matrix protein as a recombinant vaccine candidate. Infect. Immun. 74:1865-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pishko, E. J., T. N. Kirkland, and G. T. Cole. 1995. Isolation and characterization of two chitinase-encoding genes (cts1, cts2) from the fungus Coccidioides immitis. Gene 167:173-177. [DOI] [PubMed] [Google Scholar]
- 35.Reichard, U., G. T. Cole, R. Ruchel, and M. Monod. 2000. Molecular cloning and targeted deletion of PEP2 which encodes a novel aspartic proteinase from Aspergillus fumigatus. Intern. J. Med. Microbiol. 290:85-96. [DOI] [PubMed] [Google Scholar]
- 36.Shubitz, L. F., Yu J-J, C.-Y. Hung, T. N. Kirkland, T. Peng, R. Perrill, J. Simons, J. Xue, R. A. Herr, G. T. Cole, and J. N. Galgiani. 2006. Improved protection of mice against lethal respiratory infection with Coccidioides posadasii using two recombinant antigens expressed as a single protein. Vaccine 24:5904-5911. [DOI] [PubMed] [Google Scholar]
- 37.Singh, H., and G. P. Raghava. 2001. ProPred: prediction of HLA-DR binding sites. Bioinformatics 17:1236-1237. [DOI] [PubMed] [Google Scholar]
- 38.Southwood, S., J. Sidney, A. Kondo, M.-F. del Guercio, E. Appella, S. Hoffman, R. T. Kubo, R. W. Chesnut, H. M. Grey, and A. Sette. 1998. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 160:3363-3373. [PubMed] [Google Scholar]
- 39.Standaert, S. M., W. Schaffner, J. N. Galgiani, R. W. Pinner, L. Kaufman, E. Durry, and R. H. Hutcheson. 1995. Coccidioidomycosis among visitors to a Coccidioides immitis-endemic area: an outbreak in a military reserve unit. J. Infect. Dis. 171:1672-1675. [DOI] [PubMed] [Google Scholar]
- 40.Tarcha, E. J., V. Basrur, C.-Y. Hung, M. J. Gardner, and G. T. Cole. 2006. A recombinant aspartyl protease of Coccidioides posadasii induces protection against pulmonary coccidioidomycosis in mice. Infect. Immun. 74:516-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, J.-J., T. N. Kirkland, L. K. Hall, J. Wopschall, R. C. Smith, C.-Y. Hung, X. Chen, E. Tarcha, P. W. Thomas, and G. T. Cole. 2005. Characterization of a serodiagnostic complement fixation antigen of Coccidioides posadasii expressed in the nonpathogenic fungus Uncinocarpus reesii. J. Clin. Microbiol. 43:5462-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]