Abstract
The progression of murine mycoplasma pneumonia is dependent on T cells and other immune cells. The role of cytokines in immunity are complex, and identifying the network of cytokines produced after infection of mice is essential in dissecting the key cytokine cascades involved mycoplasma disease pathogenesis. In the present study, mRNA expression of 143 different cytokines, chemokines, or receptors were evaluated in lung tissues from both susceptible (BALB/c and C3H/HeN) and resistant (C57BL/6) mice after Mycoplasma pulmonis infection. To accomplish this, membrane-based cDNA microarrays were used to monitor changes mRNA expression in lungs. There was a clear association with disease susceptibility and development of cytokine mRNA expression. In addition to proinflammatory cytokines, mRNA expression of an anti-inflammatory cytokine, interleukin-10, increased with disease severity, suggesting an attempt to moderate the severity of the inflammatory response. Furthermore, it is clear that an array of chemokines produced in susceptible mice could contribute to the recruitment and maintenance of inflammatory cells at the site of disease. In support of this, there was an increase in macrophage inflammatory protein 1β (MIP-1β; CCL4) and monocyte chemoattractant protein 2 (MCP-2; CCL8) mRNA levels from mycoplasma-infected mice and a corresponding accumulation of CD4+ Th cells expressing the MIP-1β/MCP-2 receptor, CCR5, in the lungs of mice. Furthermore, MIP-1β- and MCP-2-producing cells and CD4+ T cells were found to be in close association in pulmonary lesions. Thus, there was a significant cytokine response associated with disease pathogenesis, and these studies provide important leads and insights into ongoing cytokine- and chemokine-mediated processes in this persistent inflammatory disease.
Mycoplasmas are a leading cause of respiratory diseases in humans and animals worldwide. Mycoplasma pneumoniae accounts for 30% of all cases of pneumonia in humans within the United States alone (13, 14, 23) and is also associated with the exacerbation of asthma in humans (15, 26). Mycoplasma respiratory diseases are also major problem in livestock, with a significant economic impact (42). Mycoplasma pulmonis causes a naturally occurring respiratory disease in rats and mice with high morbidity and low mortality. As with other mycoplasma diseases, M. pulmonis respiratory infections are persistent and cause rhinitis, otitis media, laryngotracheitis, and bronchopneumonia. In terms of histopathology, mycoplasma diseases are characterized by the accumulation of mononuclear cells along the respiratory airways (6-8, 13, 51). This infiltrate suggests that activation and recruitment of inflammatory cells are key factors in the development of both acute and chronic disease. In fact, both innate and adaptive immune mechanisms critically affect the level of infection and the progression of disease (40); however, mycoplasma infections persist despite host immune responses. Furthermore, several studies demonstrate that a component of mycoplasma respiratory diseases is immunopathologic (4, 11, 21, 28, 45). Thus, it is critical to determine the mechanisms of immune responses that contribute to disease, as well as those that protect from infection, since this information will allow the development of new vaccines and improve existing vaccinations.
An additional complication is that the genetic background can influence immunity and the progression of mycoplasma disease in a number of species. One of the best-characterized differences is in M. pulmonis-induced disease in mice. C3H/HeN and BALB/c mice develop more severe M. pulmonis disease than C57BL/6N mice (5, 6, 9). C57BL/6N mice have a 100-fold-higher 50% lethal dose, 50% pneumonia dose, and 50% microscopic lesion dose than C3H/HeN mice (9). Both innate and adaptive immune response are associated with disease susceptibility and severity between the strains of mice (6, 16, 36).
Cytokines and chemokines are central mediators during host-pathogen interactions, including the clearance of invading microorganisms, as well as the initiation, progression, and resolution of inflammation in response to various microbes. It is clear from our previous studies that cytokine responses are associated with mycoplasma infection and pneumonia. C3H/HeN mice have higher and more persistent concentrations of tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) in bronchoalveolar lavage (BAL) fluid than C57BL/6N mice, indicating that cytokines may be an important contributor to the pathogenesis of acute M. pulmonis disease in mice (12). Recent studies from our laboratory demonstrate that both gamma interferon (IFN-γ) and IL-4 influence the host response and disease resulting from mycoplasma infection in susceptible strains of mice (48-50). We also demonstrated the production of chemokines that likely promote recruitment of different cell populations (41). Therefore, there are likely complex interactions and cascades within cytokine responses that contribute to the chronic inflammatory responses and determine the outcome of mycoplasma infection.
We believe that identifying the network of cytokines produced after the infection of resistant and susceptible strains of mice is a first and essential step in dissecting the key cytokine cascades involved in the response to mycoplasma infection. In the present study, we evaluated the mRNA expression of 143 different cytokines, chemokines, or receptors in lung tissues from both susceptible (BALB/c and C3H/HeN) and resistant (C57BL/6) mice after M. pulmonis infection. To accomplish this, we used a membrane-based cDNA microarray, followed by confirmation by quantitative real-time reverse transcription-PCR (RT-PCR), to monitor mRNA levels in lungs. There is a clear association with disease susceptibility and the development of cytokine mRNA expression. Interestingly, the mRNA expression of IL-10, an anti-inflammatory cytokine, increased with increasing disease severity, suggesting an attempt to moderate the severity of the inflammatory response by the host (46). Furthermore, it is clear that there is an array of chemokines produced in susceptible (BALB/c and C3H/HeN) mice that could contribute to the recruitment and maintenance of inflammatory cells at the site of disease. In support of this, there was an increase in macrophage inflammatory protein 1β (MIP-1β; CCL4) and monocyte chemoattractant protein 2 (MCP-2; CCL8) levels within lung lesions of infected mice, which corresponded with the accumulation of CD4+ T cells expressing the MIP-1β/MCP-2 receptor, CCR5.
MATERIALS AND METHODS
Mice.
BALB/c, C57BL/6N, and C3H/HeN mice, tested to be virus- and mycoplasma-free, were obtained from Harlan Sprague-Dawley (Indianapolis, IN). Mice were housed in sterile microisolator cages supplied with sterile bedding, food, and water given ad libitum. Mice used in the study were between 6 to 12 weeks of age. Female mice were used in all studies. Before experimental infection, mice were anesthetized with an intramuscular injection of ketamine-xylazine.
Mycoplasma.
The UAB CT strain of M. pulmonis was used in all experiments. Stock cultures were grown, as previously described (38), in mycoplasma medium and frozen in 1-ml aliquots at −80°C. For inoculation, thawed aliquots were diluted to 105 CFU/20 μl. Nasal-pulmonary inoculations of 20 μl of diluted mycoplasma were given for experimental infections.
Histopathology.
Lungs and nasal passages were fixed in alcohol formalin (4% glacial acetic acid [Fisher], 6% formaldehyde solution [Fisher], 40% deionized water, and the remaining 50% comprised 95% ethanol). Tissues were embedded in paraffin, sectioned at a thickness of 5 μm, and stained with hematoxylin and eosin for light microscopy. Each lung lobe was sectioned separately.
RNA isolation.
Total lung RNA was isolated by using the TRIzol RNA isolation reagent as recommended by the manufacture (Invitrogen/Life Technologies, Carlsbad, CA) with modification. Briefly, entire lung tissues of each mouse were homogenized in 6 ml of TRIzol reagent. A 400-μl portion of chloroform was mixed thoroughly with 1 ml of sample, and the samples were centrifuged to obtain the RNA-containing aqueous phase. To increase the quality of RNA for subsequent production of labeled cDNA probes for gene expression using microarrays, the aqueous phase was mixed with 500 μl of TRIzol again, and the second aqueous phase was extracted by addition of 500 μl of chloroform to exclude phenol. Total RNA was precipitated from aqueous phase by using isopropyl alcohol (isopropyl alcohol-aqueous phase, ≥0.7:1 [vol/vol]). The RNA was pelleted by centrifugation and washed with 75% ethanol, air dried for 20 to 30 min, and dissolved in diethyl pyrocarbonate-treated water. The concentration and integrity of RNA was assessed spectrophotometrically (GeneQuant II; Amersham Pharmacia Biotech, Piscataway, NJ) and visualized by formaldehyde RNA electrophoresis gels stained in ethidium bromide.
Microarrays.
The mouse common cytokine and mouse chemokine and receptors GEArray gene expression arrays (SuperArray, Inc., Bethesda, MD) were used to evaluate the mRNA expression of 96 common mouse cytokine genes and 76 chemokines and receptors according to the manufacturer's instructions. In brief, biotin-cDNA probe was prepared from 3 μg of total lung RNA (pooled from three animals in an experiment) by the AmpoLabeling-LPR kit (SuperArray). The annealed RNAs were converted to cDNAs that were amplified by linear polymerase reaction. Biotin-16-2′-deoxy-uridine-5′-triphosphate (Biotin-16-dUTP; Roche Diagnostics GmbH and Molecular Biochemicals, Mannheim, Germany) was incorporated into the probes during the linear polymerase reaction process. The array membrane was prehybridized for 2 h at 60°C in GEAhyb hybridization solution with 150 μg of denatured sheared salmon sperm DNA (Sigma Chemical Co., St. Louis, MO)/ml. The denatured cDNA probe was hybridized with specific genes on the membranes at 60°C for more than 24 h. After washing and blocking, membranes were incubated for exactly 10 min at room temperature in alkaline phosphatase-conjugated streptavidin at 1:7,500 dilution in washing buffer. The signals were developed in CDP-Star chemiluminescent substrate for 3 min, and images were recorded by using a charge-coupled device camera (FluorChem 8900 imaging system; Alpha Innotech) at high resolution and low sensitivity. The image capturing time was set for half, equal, double, or triple the previewed time in order to obtain different levels of signal. The intensity of each spot of signal was transformed to digital data with auto-background subtraction during spot density analysis using the AlphaEaseFC software. The ratios of integrated density value between M. pulmonis-infected mice and relative broth control mice were analyzed by using GEArrayAnalyzer software (SuperArray), in which the results were normalized to β-actin gene expression.
Real-time RT-PCR.
RNA (1 μg per 10-μl reaction system) from individual lung samples was reverse transcribed by using Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI) at 42°C for 60 min in the presence of 5× reaction buffer (250 mM Tris-HCl [pH 8.0] at 25°C, 375 mM KCl, 15 mM MgCl2, 50 mM dithiothreitol [Promega], 500 μM deoxynucleoside triphosphate [dATP, dTTP, dCTP, and dGTP; Promega]), 25 U of Moloney murine leukemia virus reverse transcriptase, 8 to 10 U of RNAguard RNase inhibitor (Amersham Pharmacia Biotech), and 1 μM oligo(dT)16 primer.
Real-time RT-PCR was done by using the FRET probe and primer sets working with the TaqMan universal PCR master mix (Applied Biosystems, Branchburg, NJ). IL-4, IL-6, IL-12p40, IFN-γ, TNF-α, and granulocyte-macrophage colony-stimulating factor (GM-CSF) FRET probes and primers were purchased from Biosource, Inc. (Camarillo, CA). Primers and probes of IL-16, IL-17, OX40 ligand (OX40L), and CD40 ligand (CD40L) were designed by the Beacon Designer program (Applied Dynamics International, Inc., Ann Arbor, MI) and synthesized by Biosource, Inc. (Table 1) , and the specificities of the primer sets were confirmed by sequencing of amplified the product. The MIP-β (CCL4) and MCP-2 (CCL8) primer sets and the RT real-time SYBR green master mix were purchased from Superarray, Inc.
TABLE 1.
Real-time PCR primers and probes designed for this study
| Cytokine | Primer or probe | Sequence |
|---|---|---|
| IL-16 | Primer (sense) | ACAGAAGGAGAGTCAAGGAGGA |
| Primer (antisense) | GCAGTCGGAAATTCTAACCCAAG | |
| FRET probe | CCAGCCCAGAGACTCCAGCATCCC | |
| IL-17 | Primer (sense) | ATGCTGTTGCTGCTGCTGAG |
| Primer (antisense) | TTGGACACGCTGAGCTTTGAG | |
| FRET probe | CGCTGCTGCCTTCACTGTAGCCGC | |
| CD40L | Primer (sense) | CGGCAAATACCCACAGTTCCT |
| Primer (antisense) | AGCACCAGCTTGTAATTCAAACA | |
| FRET probe | CCGCCCAAGTGAACAGACTGCTGC | |
| OX40L | Primer (sense) | ACGGATCAAGGCCAAGATTCAA |
| Primer (antisense) | TGAAGCACAGAAGCATCCCTG | |
| FRET probe | CCCAGAGACCACCAGCCTTAGCGT |
PCRs were performed in 25-μl SmartCycler tubes (Cepheid, Sunnyvale, CA) containing 0.5 μM concentrations of the primer pairs, 0.02 nM FRET probe, and 2.5 μl of cDNA template. The real-time PCR products were amplified by using a SmartCycler system (Cepheid) at 95.0°C for 10 min, followed by 50 cycles of 95.0°C for 15 s and 60.0°C for 60 s. The threshold of the growth curve (CT) was set at a value of 30 by the SmartCycler software. The changes of gene expression levels were evaluated by relative quantification between cDNA templates from M. pulmonis-infected mice and broth-inoculated control mice. The expression of the housekeeping gene, β-actin, was used to normalize the data between samples. The formula for the difference (ΔCT) between the amplified cytokine gene and the normalizer (β-actin) is ΔCT = CT (cytokine) − CT (β-actin). The value of cDNA amplification from broth control was considered as reference for baseline levels of cytokine mRNA. The comparative ΔΔCT value between M. pumonis and broth control mice was calculated by the following formula: ΔΔCT = ΔCT (mycoplasma infection) − ΔCT (control). The formula for calculation of absolute comparative expression level was −2−ΔΔCT which showed the fold difference of cytokine mRNA between the infection group and the broth control group.
CCR5 expression on pulmonary T-cell populations.
Mononuclear cells were isolated from lungs as previously described (41). Three-color immunofluorescent staining was performed to identify CCR5 expression on both major T-cell populations using phycoerythrin-labeled anti-mouse CCR5 monoclonal antibody (MAb) (C34-3448; BD Pharmingen, San Diego, CA), PerCP-labeled anti-CD4 MAb (L3T4, RM4-5; BD Pharmingen, San Diego, CA), and fluorescein isothiocyanate-labeled anti-CD8 MAb (Lyt-2, 53-6.7; BD Pharmingen). Briefly, 2 × 106 cells per tube were incubated with purified 2.4G2 MAb (BD Pharmingen) for 5 min at 4°C to reduce nonspecific binding of FcII/III receptors prior to fluorescent MAb staining. The cells were incubated for 30 min at 4°C with 250 μl of fluorescent MAb (2 μg/ml). The cells were washed in staining buffer (Mg2+-free Ca2+-free phosphate-buffered saline [PBS] with 0.05% sodium azide and 1% fetal bovine serum [HyClone]) and fixed with 4% paraformaldehyde solution for 30 min, and the cells were then suspended in staining buffer until analysis.
The cells were analyzed by using an EPICS XL-MCL flow cytometer (Beckman Coulter). The data collection was done by using system 2 software (Beckman Coulter), with further analysis done by using Expo 2 analysis software (Beckman Coulter). Lymphocyte gates and detector voltages were set by using unstained (control) lung and splenic cells. The proportion of each cell population was expressed as the percentage of the number of stained cells.
Lung frozen section preparation.
Lungs of mice were infiltrated with 2% paraformaldehyde (Sigma Chemical Co., St. Louis, MO) through the trachea and fixed in the solution for ca. 2 to 4 h. The lungs were then placed in 30% sucrose overnight. Separated lung lobes were embedded in Tissue-Tek OCT compound (Sakura Finetechnical, Torrance, CA) on dry ice and kept at −80°C. Lungs were sectioned at 15 μm by using an Ultrapro 5000 cryostat (Vibratome, St. Louis, MO) and mounted on PEN membrane glass slides (Arcturus, Moutain View, CA) for further laser capture microdissection (LCM) or on FisherBrand SuperFrost/plus microscope slides (Fisher Scientific, Hampton, NH) for further immunofluorescence staining.
LCM and RNA extraction.
Membrane slides were dried for 10 min, and the tissues were stained with HistoGene LCM frozen section staining kit (Arcturus). Briefly, the slides were placed on a slide jar containing 75% ethanol for 30 s and nuclease-free water for 30 s. Tissues were stained with 50 μl of hematoxylin for 15 s and rinsed in nuclease-free water for 30 s. Tissues were dehydrated sequentially in 75, 95, and 100% ethanol for 30 s for each wash. To complete the dehydration, slides were placed in xylene for 5 min and then air dried prior to microdissection.
LCM was performed by using the Veritas laser capture microdissection and laser cutting system (Arcturus). The slides were viewed under a Veritas laser capture microdissection and laser cutting system microscope under a ×4 objective lens. The regions with or without significant cell infiltration were selected. Similar regions in the lungs of broth-inoculated mice were also selected as controls. The infrared capture laser settings were as follows: spot size, 10.0 μm; power, 70 to 100 mW; pulse, 2,500 μs; and intensity, 100 to 200 mV. The UV cutting laser was set at ca. 30 to 35 mV and at a 2.0-μm spot size. Selected areas were cut by the UV cutting laser and collected by infrared capture laser onto CapSure LCM Macrocaps (Arcturus). The efficiency of laser cutting and capture was evaluated by examining the LCM Macrocap after capture and by examining the tissue before and after lifting off the cap. After tissue collection, the cap was incubated in 50 μl of PicoPure RNA extraction buffer supplied with the PicoPure RNA isolation kit (Arcturus) for 30 min at 42°C. Cell extracts were collected by centrifugation into a 0.5-ml plastic tube. RNAs were extracted according to the manufacturer's instructions. RNAs were eluted in 20 μl of elution buffer.
Immunofluorescence staining.
Tissue sections mounted on SuperFrost glass slides were fixed in acetone for 5 min, rinsed with PBS, and blocked by 10% fetal bovine serum in PBS. Biotinylated goat anti-mouse MIP-1β (CCL4) or MCP-2 (CCL8) antibody (R&D Systems, Minneapolis, MN) was coincubated with Alexa Fluor 488-conjugated rat anti-mouse CD4 antibody (Invitrogen/Caltag Laboratories, Carlsbad, CA) on tissue, followed by incubation with anti-biotin Alexa Flour 594 secondary antibody (Invitrogen/Molecular Probes, Carlsbad, CA). An isotype control antibody to confirm specificity of chemokine stain was used to stain lungs as a control. The images were observed under an AX70 Olympus fluorescence microscope and captured by using a DP70 digital camera (Olympus, Center Valley, PA).
Data analysis.
Changes in mRNA expression using the arrays were considered significant if there was at least an average twofold change with no individual value being ≤1.5-fold. Real-time PCR data were evaluated by analysis of variance, followed by a Tukey's honestly significant difference multigroup comparison. These analyses were performed by using the JMP 5.01 (SAS Institute, Cary, NC) computer program. A P value of ≤0.05 was considered statistically significant. All experiments were repeated at least twice.
RESULTS
Cytokine and chemokine transcription profiles during M. pulmonis infection in susceptible and resistant strains of mice.
The expression of 143 different cytokines, chemokines, or receptors mRNAs were determined in lung tissues from both susceptible (BALB/c and C3H/HeN) and resistant (C57BL/6) mice after M. pulmonis infection. To accomplish this, BALB/c, C3H/HeN, and C57BL/6 mice were infected with M. pulmonis, and lungs were collected 3, 14, and 28 days later. As controls, groups of mice were inoculated with mycoplasma broth.
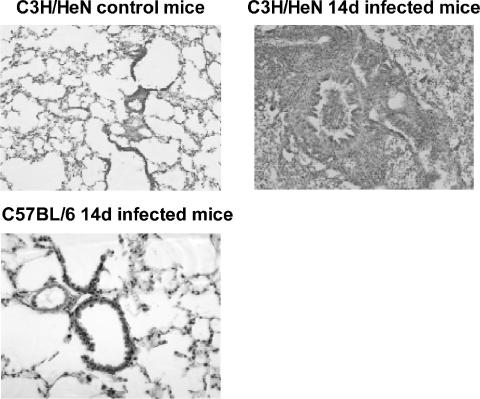
C57BL/6N mice developed little to no clinical M. pulmonis disease, whereas BALB/c and C3H/HeN mice had clinical disease. M. pulmonis-infected BALB/c and C3H/HeN mice showed significant weight loss compared to broth-inoculated controls (approximately 2 g at 14 day after infection [P ≤ 0.05]), whereas infected C57BL/6N mice had weights comparable to those of the broth-inoculated controls. In addition, inflammatory lesions developed in the lungs of BALB/c and C3H/HeN mice but not in C57BL/6N mice (Fig. 1). These results are consistent with previously published studies on strain differences in disease severity (5, 6, 9).
FIG. 1.
C3H/HeN and BALB/c mice develop inflammatory lesions that are more severe than do C57BL/6 mice after mycoplasma infection. Mice were inoculated with either M. pulmonis or sterile broth, and 14 days later the lungs were collected for histological examination. There were no cellular infiltrates in the lungs of C3H/HeN mice inoculated with sterile broth, whereas severe lesions developed in these mice after mycoplasma infection. The lungs of BALB/c mice were very similar to those of the C3H/HeN mice (not shown). In contrast, C57BL/6 mice did not have any evidence of inflammatory disease in their lungs.
To examine the changes in cytokine mRNA in lungs, total RNA was isolated from the lungs of individual mice, and pooled RNA from each experiment (n = 3 per experiment [two experiments]) was used to probe membrane-based cDNA cytokine and chemokine microarrays at 3, 14, and 28 days after infection.
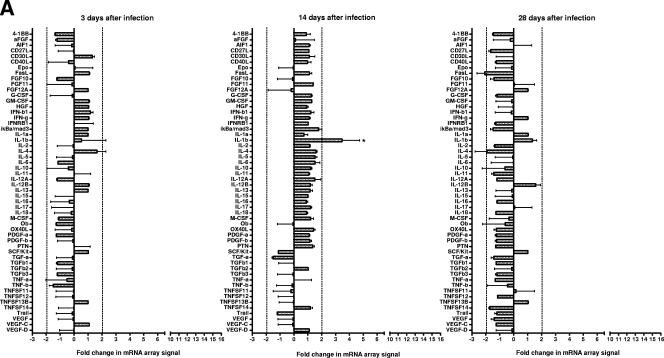
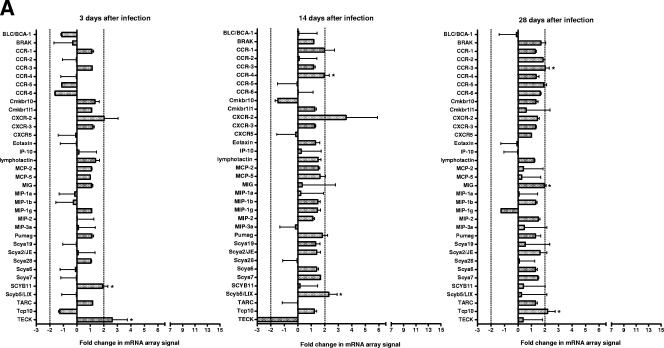
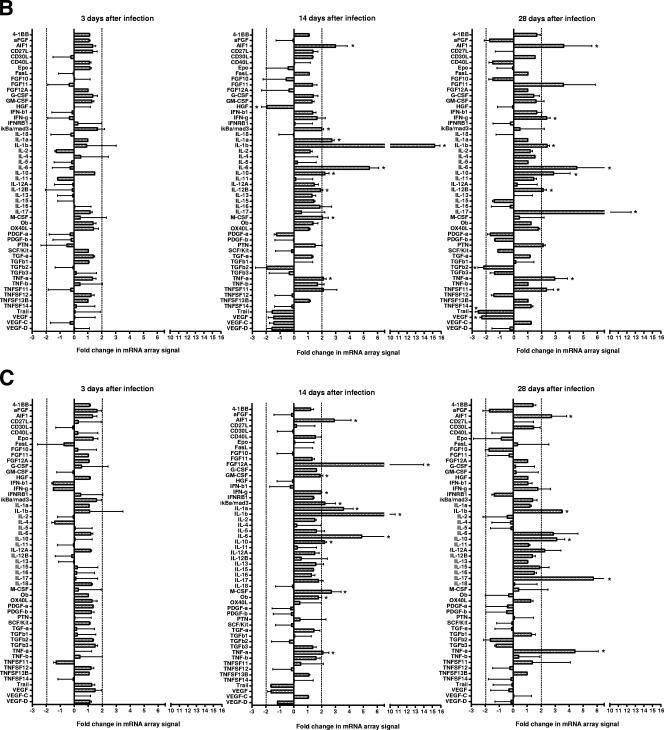
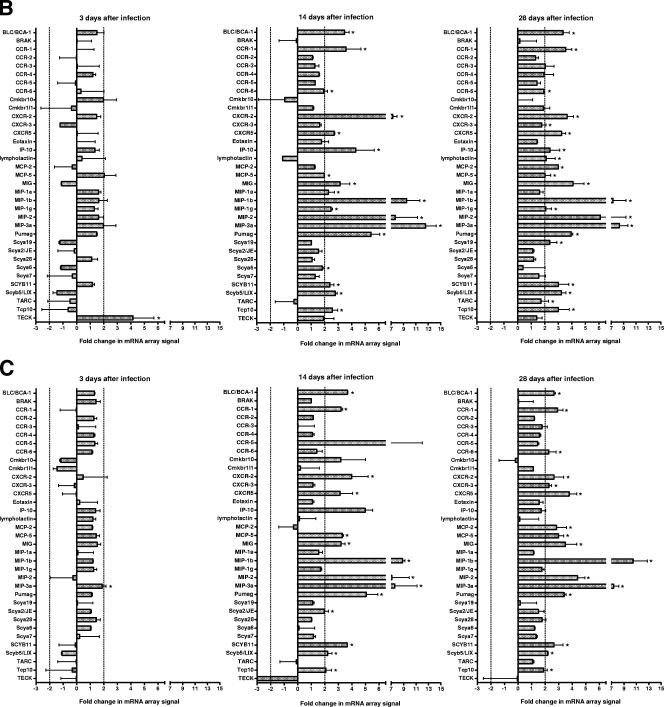
C3H/HeN and BALB/c mice expressed similar cytokine (Fig. 2) and chemokine (Fig. 3) profiles. On day 3 after infection, there were few consistent changes in cytokine and chemokine mRNA expression. However, on days 14 and 28 after infection, an increase was detected in the mRNA levels of a number of cytokines and chemokines in these susceptible mouse strains. In contrast, there was a very limited response in resistant C57/BL6N mice. There were also a number of cytokine and chemokine mRNAs that were not detected using the arrays in any strain of mice throughout the course of disease pathogenesis (Table 2).
FIG.2.
Cytokine mRNA changes in the lungs of susceptible (C3H/HeN and BALB/c) and resistant (C57BL/6N) mice as determined by using arrays. Total RNA was isolated from the lungs of individual C57BL/6N (A), BALB/c (B), and C3H/HeN (C) mice, and pooled RNA from each experiment (n = 3 per experiment for each time point [two experiments]) was used to probe membrane-based cDNA chemokine microarrays at 3, 14, and 18 days after infection. The intensities of the spots were normalized using the intensity of the reactivity to the housekeeping gene, β-actin, and the results from infected mice were compared to those obtained from RNA collected from lungs of control mice inoculated with mycoplasma broth medium. Asterisks indicate cytokines where changes in mRNA expression as determined by spot intensity were at least an average of 2-fold, with no individual value being ≤1.5-fold.
FIG.3.
Chemokine mRNA changes in the lungs of susceptible (C3H/HeN and BALB/c) and resistant (C57BL/6N) mice as determined by using arrays. Total RNA was isolated from lungs of individual C57BL/6N (A), BALB/c (B), and C3H/HeN (C) mice, and pooled RNA from each experiment (n = 3 per experiment for each time point [two experiments]) was used to probe membrane-based cDNA chemokine microarrays at 3, 14, and 18 days after infection. The intensities of the spots were normalized using the intensity of the reactivity to the housekeeping gene, β-actin, and the results from infected mice were compared to those obtained from the RNA collected from the lungs of control mice inoculated with mycoplasma broth medium. Asterisks indicate cytokines where changes in mRNA expression as determined by spot intensity were at least an average of 2-fold, with no individual value being ≤1.5-fold.
TABLE 2.
Cytokine and chemokine mRNAs not detected in murine lungs using microarrays
| Cytokine or chemokine | mRNAs not detected |
|---|---|
| Cytokines | G-CSF, FGF11, FGF14 (FHF4), FGF16, FGF17, FGF18, FGF2, FGF20, FGF22, FGF5, FGF6, FGF9, IL-11, IL-17B, IL-20, IL-3, IL-4, thrombopoietin, and TNFSF14 (HVEM-L) |
| Chemokines | Gro1, Ltb4r2, TCA-3, and JE |
Real-time RT-PCR supports the array results and demonstrates a progressive increase in IL-10, MIP-1β (CCL4), and MCP-2 (CCL8) mRNA expression after infection in susceptible mice.
Real time RT-PCR was used to examine whether changes in IL-4, IL-6, IL-10, IL-12p40, IL-16, IL-17, GM-CSF, TNF-α, IFN-γ, CD40L, and OX40L mRNA levels were found in samples collected from the control (broth-inoculated) or infected (day 14 postinfection) lungs from individual mice (n = 6 for each experimental group). The results from the cDNA array analysis were highly correlated with the data obtained by real-time RT-PCR (P < 0.0001), which supports the results obtained by using microarrays.
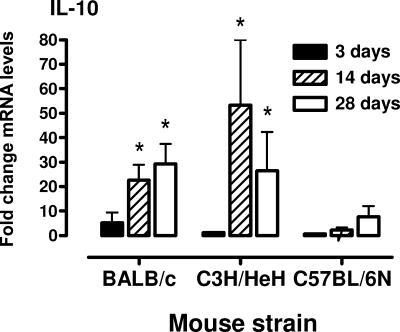
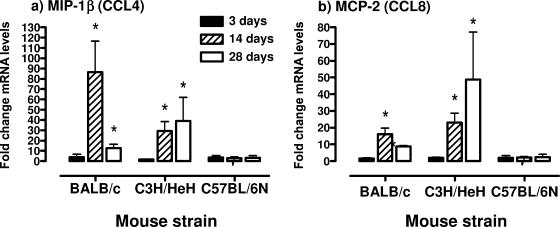
Using real-time RT-PCR, the levels of mRNAs encoding the anti-inflammatory cytokine, IL-10, and two chemokines (MIP-1β [CCL4] and MCP-2 [CCL8]) that could participate in the recruitment of T cells were determined in the lungs of mice at 3, 14, and 28 days after infection. The results from the cytokine arrays indicated there was an increase in IL-10 mRNA levels in C3H/HeN and BALB/c mice in response to mycoplasma infection, and this was confirmed by using real-time RT-PCR (Fig. 4). There was a progressive increase in IL-10 mRNA levels in lung tissue from day 7 and later after infection (P ≤ 0.05). In addition, there was an increase in MIP-1β (CCL4) and MCP-2 (CCL8) mRNA levels in lung tissues by day 14 after mycoplasma infection of BALB/c and C3H/HeN mice (P ≤ 0.05) but not C57BL/6N mice (Fig. 5).
FIG. 4.
mRNA levels of IL-10 in lungs of C3H/HeN, BALB/c, and C57BL/6N mice after mycoplasma infection. At 3, 14, and 28 days after infection, lungs were collected, and total RNA was isolated. IL-10 transcript levels were measured by real-time RT-PCR. The levels of mRNA were standardized to the β-actin housekeeping gene. The vertical bars represent the means ± the standard error of the mean (SEM; n = 6 for each time point and mouse strain) of the fold difference in IL-10 mRNA levels relative to the average baseline values of samples from uninfected (control, inoculated with mycoplasma broth medium) mice. An asterisk denotes a statistical difference (P ≤ 0.05) compared to uninfected mice of the same mouse strain.
FIG. 5.
mRNA levels of MIP-1β (CCL4) and MCP-2 (CCL8) in the lungs of C3H/HeN, BALB/c, and C57BL/6N mice after mycoplasma infection. At 3, 14, and 28 days after infection, lungs were collected, and total RNA was isolated. MIP-1β (CCL4) (a) and MCP-2 (CCL8) (b) transcript levels were measured by using real-time RT-PCR. The levels of mRNA were standardized to the β-actin housekeeping gene. The vertical bars represent the means ± the SEM (n = 6 for each time point and mouse strain) of the fold difference in MIP-1β (CCL4) and MCP-2 (CCL8) mRNA levels, relative to the average baseline values of samples from uninfected (control, inoculated with mycoplasma broth medium) mice. An asterisk denotes a statistical difference (P ≤ 0.05) compared to uninfected mice of the same mouse strain.
There is an increase in CCR5-expressing T cells in mice after mycoplasma infection.
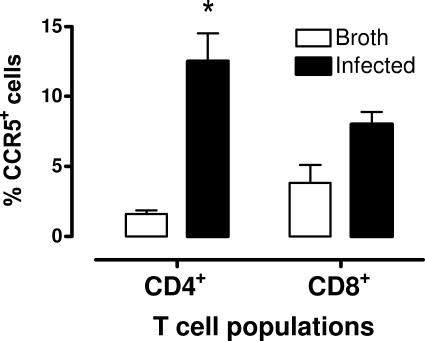
The increase in MIP-1β (CCL4) and MCP-2 (CCL8) mRNA levels suggests their involvement in the recruitment of cells to the lungs in response to mycoplasma infection. To further address this possibility, we examined the expression of CCR5 protein, a receptor for MIP-1β (CCL4) and MCP-2 (CCL8), on the surface of pulmonary T-cell populations from the lungs of mice infected with M. pulmonis. BALB/c mice were inoculated with either M. pulmonis or mycoplasma broth. After 14 days, pulmonary mononuclear cells were collected and immunofluorescently stained for CD8 and CD4 expression, as well as CCR5, and analyzed by flow cytometry.
There was an increase in the numbers of CCR5+ T cells in the lungs of infected BALB/c mice (Fig. 6). There was a fivefold increase in the percentage of CCR5+ CD4+ T cells in the lungs of mice at 14 days after infection (P ≤ 0.05). There was also about a twofold increase in the percentage of CCR5+ CD8+ T cells, but this was not a statistically significant difference.
FIG. 6.
Increase in CCR5-expressing Th cells in lungs of mycoplasma-infected mice. C3H/HeN mice were infected with mycoplasmas, and 14 days later, lung cells were isolated and stained for CD4 (Th), CD8, and CCR5 and analyzed by flow cytometry. Shown are the populations of CD4+ Th and CD8+ T cells and the percentage of CCR5+ cells in each T-cell population in uninfected and infected lungs. Vertical bars represent mean ± the SE (n = 3). An asterisk denotes a statistical difference (P ≤ 0.05) compared to uninfected mice.
MIP-1β (CCL4) and MCP-2 (CCL8) mRNA levels are preferentially elevated at the sites of inflammatory lesions.
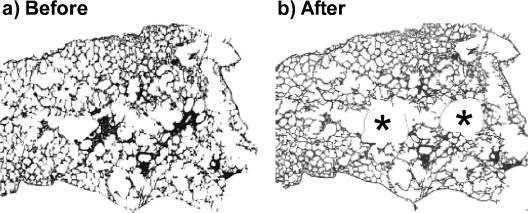
To determine whether the expression of MIP-1β (CCL4) and MCP-2 (CCL8) mRNA is associated with the cell infiltration, we took advantage of laser cutting and capture to isolate cells from the inflammatory lesions. To accomplish this, BALB/c mice were infected with M. pulmonis and, 14 days later, frozen sections from lung lobes were prepared. Inflammatory lesions from the lungs were dissected (Fig. 7), and the total RNA was isolated. We also collected similar regions from lungs that did not have any histological evidence of cell infiltration. As controls, groups of mice were inoculated with mycoplasma broth, and similar regions were collected. The levels of MIP-1β (CCL4) and MCP-2 (CCL8) mRNA were monitored by using real-time RT-PCR.
FIG. 7.
Pulmonary cellular infiltrates collected by laser cutting and capture. At 14 days after infection of BALB/c mice, the lungs were collected, and areas of cell infiltrates were collected by laser cutting and capture. An example of a lung section with some isolated lesions is shown with a lung section before (a) and after (b) laser cutting and capture. The asterisks in panel b denote the areas from which the tissue was collected.
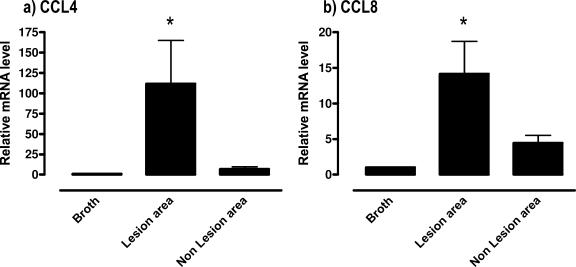
There was a clear association of increased MIP-1β (CCL4) and MCP-2 (CCL8) mRNA expression at the sites of cell infiltration. There was a >100-fold increase in CCL4 mRNA in the lung samples collected from areas of cell infiltration due to mycoplasma disease (P ≤ 0.05) compared to lungs from control mice (Fig. 8), and there was a slightly higher, but insignificant, increase in MIP-1β (CCL4) mRNA expression in areas in the same lung without visible cell infiltration. There was also an ∼14-fold increase in MCP-2 (CCL8) mRNA expression at the sites of cell infiltration (P ≤ 0.5), but there was a much lower and insignificant increase in mRNA expression at nonlesion sites.
FIG. 8.
MIP-1β (CCL4) and MCP-2 (CCL8) mRNA levels were preferentially increased at sites of cell infiltration. At 14 days after infection of BALB/c mice, lungs were collected, and areas of cell infiltrates were collected by laser cutting and capture. As controls, regions without visible infiltrates in the same lung were also collected, along with similar areas from the lungs of sterile mycoplasma broth medium-inoculated mice. Total RNA was isolated from the captured regions. MIP-1β (CCL4) (a) and MCP-2 (CCL8) (b) transcript levels were measured by using real-time RT-PCR. The levels of mRNA were standardized to the β-actin housekeeping gene. The vertical bars represent the means ± the SEM (n ≥ 5) of the fold difference in MIP-1β (CCL4) and MCP-2 (CCL8) mRNA levels relative to the average baseline values of samples from uninfected (inoculated with sterile mycoplasma broth medium) mice. An asterisk denotes the statistical difference (P ≤ 0.05) from uninfected mice of the same mouse strain.
MIP-1β (CCL4)- and MCP-2 (CCL8)-producing cells are associated with clusters of CD4+ cells within cell infiltrates.
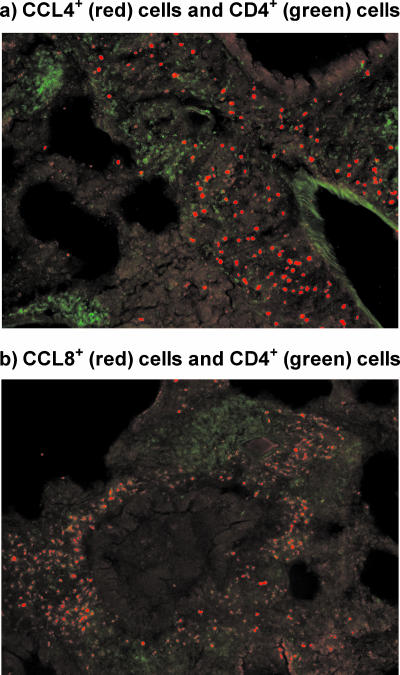
In the previous studies, we found that there was a dramatic increase in the levels of CCL4 and CCL8 mRNA at the sites of cell infiltration. In addition, CD4+ T cells were shown to have an increased expression of the chemokine receptor, CCR5, after mycoplasma infection. Using immunofluorescence microscopy, the association of MIP-1β (CCL4) and MCP-2 (CCL8) proteins with CD4+ T cells in inflammatory lesions was further determined. BALB/c mice were infected with M. pulmonis and, 14 days later, frozen sections from lung lobes were prepared and stained with anti-MIP-1β (CCL4) or anti-MCP-2 (CCL8) antibodies, plus anti-CD4 fluorescence-labeled antibodies.
As shown in Fig. 9, MIP-1β (CCL4)+ cells, as well as MCP2 (CCL8)+ cells, were found in the lungs of infected mice and were grouped around areas of clusters of CD4+ T cells. Lung sections stained with isotype control antibodies did not show a reaction (not shown). In addition, there was little staining of lungs sections from uninfected mice (not shown). Thus, there is a clear association of CCL4 and CCL8 production and the accumulation of Th cells.
FIG. 9.
MIP-1β (CCL4)- and MCP-2 (CCL8)-producing cells are colocalized with CD4+ T cells within pulmonary infiltrates due to mycoplasma infection. At 14 days after infection of BALB/c mice, lungs were collected. The lungs were stained for either for CCL4 (red) and CD4 (green) (a) or for CCL8 (red) and CD4 (green) (b). Lung sections stained with isotype control antibodies did not show a reaction (not shown). In addition, there was little staining of lung sections from uninfected mice (data not shown).
DISCUSSION
In the present study, we evaluated the mRNA expression of 143 different cytokines, chemokines, or receptors in lung tissues from both susceptible (BALB/c and C3H/HeN) and resistant (C57BL/6) mice after M. pulmonis infection. There was a clear association between disease pathogenesis and the development of cytokine mRNA expression. The susceptible strains of mice, C3H/HeN and BALB/c, expressed similar patterns of cytokine gene expression as determined by array analysis by 14 days after infection. In contrast, there was a very limited cytokine response in resistant C57BL/6N mice. The results from the cDNA array analysis were highly correlated with data obtained by real-time RT-PCR, supporting our overall conclusion. Thus, the susceptible mouse strains (C3H/HeN and BALB/c) produced a wide assortment of cytokines in their lungs in response to mycoplasma infection, whereas the resistant C57BL/6N mice had a limited reaction.
In addition to proinflammatory cytokines, IL-10 (a product of Th cells, macrophages, and other cells) mRNA expression in the lungs increased with increasing mycoplasma disease severity. The results from the cytokine arrays indicated that there was an increase in the IL-10 mRNA levels in C3H/HeN and BALB/c mice in response to mycoplasma infection, and this was confirmed by using real-time RT-PCR. There was a progressive increase in IL-10 mRNA levels in lung tissue from day 7 onward after infection. In a previous study (50), increases in IL-10 mRNA expression were not found at 3 days after M. pulmonis infection in BALB/c mice, a finding consistent with the current data, but IL-10 mRNA and protein were increased at the same time point in IFN-γ-knockout mice. Since IFN-γ-knockout mice develop more severe mycoplasma respiratory disease (49, 50), an association between increased inflammation and IL-10 production has been demonstrated previously, as well as in the present study. A major function of IL-10 is to limit damage from an out-of-control inflammatory response associated with infection (46). IL-10 dampens the activation of macrophages and related production of inflammatory cytokines. In doing this, however, IL-10 also has an effect on the bactericidal activity of macrophages, which can allow the survival of intracellular pathogens (19, 37) and can promote persistent infection and chronic inflammation in some infectious diseases (3, 17, 29, 30, 35). Thus, along with increases in proinflammatory cytokines, IL-10 production corresponds with increasing severity of mycoplasma pulmonary disease; this IL-10 response may be an attempt to limit damage from the associated inflammatory responses but may inadvertently interfere with mechanisms that are effective in eliminating the mycoplasma infection. Although an attractive hypothesis, further studies are needed to explore the role of IL-10 in mycoplasma disease pathogenesis.
There is clearly a robust chemokine response in the lungs during disease pathogenesis. In both BALB/c and C3H/HeN mice, there were increases in several chemokine mRNA levels by 14 days after infection, whereas changes in chemokine mRNA levels were limited in C57BL/6N mice. Chemokines are a subset of cytokines whose characteristic function is the recruitment of other cells (34). Many of the chemokines found to increase in C3H/HeN and BALB/c mice by day 14 after infection are chemotactic for inflammatory cells, including neutrophils (Scyb5/LIX [47] and MIP-2 [1]), monocytes/macrophages/dendritic cells (MIP-3α [20] and MCP-5 [39]), and lymphocytes (SCYB11 [27], MIP-3α [25], MIP-1β [43], MIG [22], and BLC/BCA-1 [24]). In addition, there were increases in the mRNA levels of a number of chemokine receptors. Because M. pulmonis has mitogenic activity for lymphocytes (31, 33), it was long hypothesized that the accumulation of mononuclear cells in the lung was due to local nonspecific proliferation of cells (10, 32); however, demonstration of vigorous chemokine responses suggests that these chemotactic factors are responsible for the recruitment of cells and determines the severity and the type of inflammatory disease due to mycoplasma infection. Thus, there are a number of chemokines produced that probably play a role in the recruitment and maintenance of immune cells involved in the immunopathology of mycoplasma pulmonary disease.
Based on our previous studies demonstrating the contribution of T cells to disease pathogenesis (18), the recruitment of T cells by chemokines elicited at the site of infection would likely contribute to the pathogenesis of mycoplasma disease. To further examine this possibility, we selected two key chemokines, MIP-1β (CCL4) and MCP-2 (CCL8), whose increase in expression after infection was apparent in the arrays and confirmed by real-time RT-PCR. Furthermore, the mRNA expression of both these chemokines was preferentially associated with the sites of pulmonary lesions characterized by mononuclear cell infiltrates. Furthermore, immunofluorescence staining of lung sections from infected mice demonstrated the presence of MIP-1β (CCL4)- and MCP-2 (CCL8)-producing cells in association with clusters of CD4+ T cells. MIP-1β (CCL4) and MCP-2 (CCL8) chemokines are chemotactic to T cells and monocyte MIP-1β (43, 44); interestingly, MIP-1β (CCL4) is associated with the preferential recruitment of Th1 cells (43), which we previously demonstrated to appear in mycoplasma-infected lungs (41). These data suggest an association between the production of these chemokines and the recruitment of T cells and monocytes. Importantly, previous studies from our laboratory demonstrated that the numbers of T cell, predominantly CD4+ Th cells, increase in the lungs of infected mice during the time frame of the observed increases (18). To further address this possibility, we examined the expression of CCR5 protein, a receptor for MIP-1β (CCL4) and MCP-2 (CCL8), on the surface of pulmonary T-cell populations from lungs of mice infected with M. pulmonis 14 days earlier using immunofluorescent staining and flow cytometry. There was an increase in the numbers of CCR5+ CD4+ T cells in the lungs of infected BALB/c mice. Thus, CCR5 was expressed on higher numbers of Th cells in lungs, along with corresponding increases in the amounts of chemokines that utilize CCR5 as a receptor to mediate chemotactic activity. This is compelling evidence that these chemokine responses likely contribute to the recruitment and possible retention of Th cells into infected lungs, which subsequently participate in disease pathogenesis.
In the present study, there was a clear association with disease severity and development of cytokine mRNA expression in lungs. Microarrays were useful in establishing the cytokine profiles in total lungs of susceptible and resistant mouse strains at different times in disease pathogenesis, and there were also a number of cytokine and chemokine mRNAs that were not detected using the arrays in any strain of mice throughout the course of disease pathogenesis. However, microarrays can have limited sensitivity and will likely overlook some localized cytokine responses within focal inflammatory lesions in the lungs. In fact, in a previous study from our laboratory, JE (MCP-1) mRNA and protein was detected in the lungs of C3H/HeN mice with mycoplasma pneumonia by using RT-PCR (41), whereas it was not detected using arrays (Table 2). In the present study, we found a dramatic increase in the levels of two chemokines at the lesion sites. This demonstrates that other techniques, such as laser cutting and capture, can be used to examine localized responses and further augment our understanding of the localized events in disease pathogenesis.
In summary, there were significant cytokine and chemokine responses associated with disease pathogenesis. These results provide groundwork on the cytokines and chemokines produced in response to mycoplasma infection and disease, and this by itself has provided important leads and insights into ongoing processes. There was an array of chemokines produced that could contribute to the recruitment and maintenance of inflammatory cells at the site of disease. Based on our previous studies demonstrating the contribution of T cells to disease pathogenesis (18) and the results from the present studies, chemokine receptor (e.g., CCR5)-bearing T cells are likely recruited and retained in the lungs by chemokines (e.g., MIP-1β and MCP-2) elicited at the site of infection. These data suggest the intriguing possibility that the Th1 cell responses in the lungs of mycoplasma-infected mice (18) may be in part due to the recruitment of Th1 cells to the lung, and modulation of the chemokine responses could alter the predominant T-cell responses effective due to the pathogenesis of chronic inflammatory lung disease due to mycoplasma infection. Furthermore, IL-10 (an anti-inflammatory cytokine) mRNA expression increased, along with a number of proinflammatory cytokines, with increasing disease severity, suggesting an attempt to moderate the severity of the inflammatory response by the host (37). It is possible that IL-10 may also dampen immune mechanisms responsible for mycoplasma clearance, leading to persistence of the infection, and additional studies should examine whether CD4+ CD25+ Treg cells are a source of IL-10 and participate in dampening the inflammatory responses (2). Because of the persistence of mycoplasma infection and associated immunopathology, further studies that address the functions of cytokine and chemokines will undoubtedly provide fascinating information about the relationship between the host and mycoplasma infections.
FIG. 2—
Continued.
FIG. 3—
Continued.
Acknowledgments
We thank Anthony Cheung for help with these studies.
This study was supported by U.S. Public Health Service grants HL069431-01A2, AI042075-06A1, and AI055907-01 (to J.W.S.).
Editor: J. L. Flynn
REFERENCES
- 1.Appelberg, R. 1992. Macrophage inflammatory proteins MIP-1 and MIP-2 are involved in T cell-mediated neutrophil recruitment. J. Leukoc. Biol. 52:303-306. [DOI] [PubMed] [Google Scholar]
- 2.Belkaid, Y. 2003. The role of CD4+CD25+ regulatory T cells in Leishmania infection. Expert Opin. Biol. Ther. 3:875-885. [DOI] [PubMed] [Google Scholar]
- 3.Belkaid, Y., K. F. Hoffmann, S. Mendez, S. Kamhawi, M. C. Udey, T. A. Wynn, and D. L. Sacks. 2001. The role of interleukin (IL)-10 in the persistence of Leishmania major in the skin after healing and the therapeutic potential of anti-IL-10 receptor antibody for sterile cure. J. Exp. Med. 194:1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartner, S. C., J. R. Lindsey, J. Gibbs-Erwin, G. H. Cassell, and J. W. Simecka. 1998. Roles of innate and adaptive immunity in respiratory mycoplasmosis. Infect. Immun. 66:3485-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartner, S. C., J. W. Simecka, D. E. Briles, G. H. Cassell, and J. R. Lindsey. 1996. Resistance to mycoplasmal lung disease in mice is a complex genetic trait. Infect. Immun. 64:5326-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cartner, S. C., J. W. Simecka, J. R. Lindsey, G. H. Cassell, and J. K. Davis. 1995. Chronic respiratory mycoplasmosis in C3H/HeN and C57BL/6N mice: lesion severity and antibody response. Infect. Immun. 63:4138-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassel, G. H., J. R. Lindsey, R. G. Overcash, and H. J. Baker. 1973. Murine mycoplasma respiratory disease. Ann. N. Y. Acad. Sci. 225:395. [Google Scholar]
- 8.Dajani, A. S., W. A. J. Clyde, and F. W. Denny. 1965. Experimental infection with Mycoplasma pneumoniae (Eaton's agent). J. Exp. Med. 121:1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, J. K., R. F. Parker, H. White, D. Dziedzic, G. Taylor, M. K. Davidson, N. R. Cox, and G. H. Cassell. 1985. Strain differences in susceptibility to murine respiratory mycoplasmosis in C57BL/6 and C3H/HeN mice. Infect. Immun. 50:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, J. K., J. W. Simecka, J. S. Williamson, S. E. Ross, M. M. Juliana, R. B. Thorp, and G. H. Cassell. 1985. Nonspecific lymphocyte responses in F344 and LEW rats: susceptibility to murine respiratory mycoplasmosis and examination of cellular basis for strain differences. Infect. Immun. 49:152-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evengard, B., K. Sandstedt, G. Bolske, R. Feinstein, I. Riesenfelt-Orn, and C. I. Smith. 1994. Intranasal inoculation of Mycoplasma pulmonis in mice with severe combined immunodeficiency (SCID) causes a wasting disease with grave arthritis. Clin. Exp. Immunol. 98:388-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faulkner, C. B., J. W. Simecka, M. K. Davidson, J. K. Davis, T. R. Schoeb, J. R. Lindsey, and M. P. Everson. 1995. Gene expression and production of tumor necrosis factor alpha, interleukin 1, interleukin 6, and gamma interferon in C3H/HeN and C57BL/6N mice in acute Mycoplasma pulmonis disease. Infect. Immun. 63:4084-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foy, H. M. 1993. Infections caused by Mycoplasma pneumoniae and possible carrier state in different populations of patients. Clin. Infect. Dis. 17(Suppl. 1):S37-S46. [DOI] [PubMed] [Google Scholar]
- 14.Foy, H. M., G. E. Kenny, M. K. Cooney, and I. D. Allan. 1979. Long-term epidemiology of infections with Mycoplasma pneumoniae. J. Infect. Dis. 139:681-687. [DOI] [PubMed] [Google Scholar]
- 15.Freymuth, F., A. Vabret, J. Brouard, F. Toutain, R. Verdon, J. Petitjean, S. Gouarin, J. F. Duhamel, and B. Guillois. 1999. Detection of viral, Chlamydia pneumoniae, and Mycoplasma pneumoniae infections in exacerbations of asthma in children. J. Clin. Virol. 13:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickman-Davis, J. M., S. M. Michalek, J. Gibbs-Erwin, and J. R. Lindsey. 1997. Depletion of alveolar macrophages exacerbates respiratory mycoplasmosis in mycoplasma-resistant C57BL mice but not mycoplasma-susceptible C3H mice. Infect. Immun. 65:2278-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs, M., N. Brown, N. Allie, R. Gulert, and B. Ryffel. 2000. Increased resistance to mycobacterial infection in the absence of interleukin-10. Immunology 100:494-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones, H., L. Tabor, X. Sun, M. Woolard, and J. Simecka. 2002. Depletion of CD8+ T cells exacerbates CD4+ Th cell associated inflammatory lesions during murine mycoplasma respiratory disease. J. Immunol. 168:3493-3501. [DOI] [PubMed] [Google Scholar]
- 19.Kane, M. M., and D. M. Mosser. 2001. The role of IL-10 in promoting disease progression in leishmaniasis. J. Immunol. 166:1141-1147. [DOI] [PubMed] [Google Scholar]
- 20.Kellermann, S. A., S. Hudak, E. R. Oldham, Y. J. Liu, and L. M. McEvoy. 1999. The CC chemokine receptor-7 ligands 6Ckine and macrophage inflammatory protein-3β are potent chemoattractants for in vitro- and in vivo-derived dendritic cells. J. Immunol. 162:3859-3864. [PubMed] [Google Scholar]
- 21.Keystone, E. C., D. Taylor-Robinson, M. F. Osborn, L. Ling, C. Pope, and V. Fornasier. 1980. Effect of T-cell deficiency on the chronicity of arthritis induced in mice by Mycoplasma pulmonis. Infect. Immun. 27:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koga, S., M. B. Auerbach, T. M. Engeman, A. C. Novick, H. Toma, and R. L. Fairchild. 1999. T-cell infiltration into class II MHC-disparate allografts and acute rejection is dependent on the IFN-gamma-induced chemokine Mig. J. Immunol. 163:4878-4885. [PubMed] [Google Scholar]
- 23.Krause, D. C., and D. Taylor-Robinson (ed.). 1993. Mycoplasma which infect humans. American Society for Microbiology, Washington, D.C.
- 24.Legler, D. F., M. Loetscher, R. S. Roos, I. Clark-Lewis, M. Baggiolini, and B. Moser. 1998. B cell-attracting chemokine 1, a human CXC chemokine expressed in lymphoid tissues, selectively attracts B lymphocytes via BLR1/CXCR5. J. Exp. Med. 187:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao, F., R. L. Rabin, C. S. Smith, G. Sharma, T. B. Nutman, and J. M. Farber. 1999. CC-chemokine receptor 6 is expressed on diverse memory subsets of T cells and determines responsiveness to macrophage inflammatory protein 3 alpha. J. Immunol. 162:186-194. [PubMed] [Google Scholar]
- 26.Martin, R. J., M. Kraft, H. W. Chu, E. A. Berns, and G. H. Cassell. 2001. A link between chronic asthma and chronic infection. J. Allergy Clin. Immunol. 107:595-601. [DOI] [PubMed] [Google Scholar]
- 27.Meyer, M., M. Erdel, H. C. Duba, E. R. Werner, and G. Werner-Felmayer. 2000. Cloning, genomic sequence, and chromosome mapping of Scyb11, the murine homologue of SCYB11 (alias betaR1/H174/SCYB9B/I-TAC/IP-9/CXCL11). Cytogenet. Cell Genet. 88:278-282. [DOI] [PubMed] [Google Scholar]
- 28.Mizutani, H., T. Kitayama, A. Hayakawa, and E. Nagayama. 1971. Delayed hypersensitivity in Mycoplasma pneumoniae infections. Lancet i:186-187. [DOI] [PubMed] [Google Scholar]
- 29.Murray, H. W., C. M. Lu, S. Mauze, S. Freeman, A. L. Moreira, G. Kaplan, and R. L. Coffman. 2002. Interleukin-10 (IL-10) in experimental visceral leishmaniasis and IL-10 receptor blockade as immunotherapy. Infect. Immun. 70:6284-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray, H. W., A. L. Moreira, C. M. Lu, J. L. DeVecchio, M. Matsuhashi, X. Ma, and F. P. Heinzel. 2003. Determinants of response to interleukin-10 receptor blockade immunotherapy in experimental visceral leishmaniasis. J. Infect. Dis. 188:458-464. [DOI] [PubMed] [Google Scholar]
- 31.Naot, Y. 1982. In vitro studies on the mitogenic activity of mycoplasmal species toward lymphocytes. Rev. Infect. Dis. 4(Suppl.):S205-S209. [DOI] [PubMed] [Google Scholar]
- 32.Naot, Y., S. Davidson, and E. S. Lindenbaum. 1984. Role of mitogenicity in pathogenicity of mycoplasmas for murine hosts. Ann. Microbiol. 135A:95-101. [DOI] [PubMed] [Google Scholar]
- 33.Naot, Y., S. Merchav, E. Ben-David, and H. Ginsburg. 1979. Mitogenic activity of Mycoplasma pulmonis. I. Stimulation of rat B and T lymphocytes. Immunology 36:399-406. [PMC free article] [PubMed] [Google Scholar]
- 34.Oppenheim, J. J., C. O. Zachariae, N. Mukaida, and K. Matsushima. 1991. Properties of the novel proinflammatory supergene “intercrine” cytokine family. Annu. Rev. Immunol. 9:617. [DOI] [PubMed] [Google Scholar]
- 35.Padigel, U. M., J. Alexander, and J. P. Farrell. 2003. The role of interleukin-10 in susceptibility of BALB/c mice to infection with Leishmania mexicana and Leishmania amazonensis. J. Immunol. 171:3705-3710. [DOI] [PubMed] [Google Scholar]
- 36.Parker, R. F., J. K. Davis, D. K. Blalock, R. B. Thorp, J. W. Simecka, and G. H. Cassell. 1987. Pulmonary clearance of Mycoplasma pulmonis in C57BL/6N and C3H/HeN mice. Infect. Immun. 55:2631-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redpath, S., P. Ghazal, and N. R. Gascoigne. 2001. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 9:86-92. [DOI] [PubMed] [Google Scholar]
- 38.Reiner, S. L. 2001. Helper T-cell differentiation, inside and out. Curr. Opin. Immunol. 13:351-355. [DOI] [PubMed] [Google Scholar]
- 39.Sarafi, M. N., E. A. Garcia-Zepeda, J. A. MacLean, I. F. Charo, and A. D. Luster. 1997. Murine monocyte chemoattractant protein (MCP)-5: a novel CC chemokine that is a structural and functional homologue of human MCP-1. J. Exp. Med. 185:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simecka, J. W. 2003. Contrasting roles of immunity in mycoplasma respiratory disease, p. 203-219. In S. G. Pandalai (ed.), Recent research developments in infection and immunity. Transworld Research Network.
- 41.Simecka, J. W. 1999. β-chemokines are produced in lungs of mice with mycoplasma respiratory disease. Curr. Microbiol. 39:163-167. [DOI] [PubMed] [Google Scholar]
- 42.Simecka, J. W., J. K. Davis, M. K. Davidson, S. E. Ross, C. Stadtlander, and G. H. Cassell. 1992. Mycoplasma diseases of animals, p. 391-415. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas: molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 43.Siveke, J. T., and A. Hamann. 1998. T helper 1 and T helper 2 cells respond differentially to chemokines. J. Immunol. 160:550-554. [PubMed] [Google Scholar]
- 44.Taub, D. D., P. Proost, W. J. Murphy, M. Anver, D. L. Longo, J. van Damme, and J. J. Oppenheim. 1995. Monocyte chemotactic protein-1 (MCP-1), -2, and -3 are chemotactic for human T lymphocytes. J. Clin. Investig. 95:1370-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, G., D. Taylor-Robinson, and G. W. Fernald. 1974. Reduction in the severity of Mycoplasma pneumoniae-induced pneumonia in hamsters by immunosuppressive treatment with antithymocyte sera. J. Med. Microbiol. 7:343-348. [DOI] [PubMed] [Google Scholar]
- 46.Trinchieri, G. 2001. Regulatory role of T cells producing both interferon gamma and interleukin 10 in persistent infection. J. Exp. Med. 194:F53-F57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Den Steen, P. E., A. Wuyts, S. J. Husson, P. Proost, J. Van Damme, and G. Opdenakker. 2003. Gelatinase B/MMP-9 and neutrophil collagenase/MMP-8 process the chemokines human GCP-2/CXCL6, ENA-78/CXCL5, and mouse GCP-2/LIX and modulate their physiological activities. Eur. J. Biochem. 270:3739-3749. [DOI] [PubMed] [Google Scholar]
- 48.Woolard, M. D., R. Doug Hardy, and J. W. Simecka. 2004. IL-4-independent pathways exacerbate methacholine-induced airway hyperreactivity during mycoplasma respiratory disease. J. Allergy Clin. Immunol. 114:645-649. [DOI] [PubMed] [Google Scholar]
- 49.Woolard, M. D., L. M. Hodge, H. P. Jones, T. R. Schoeb, and J. W. Simecka. 2004. The upper and lower respiratory tracts differ in their requirement of IFN-gamma and IL-4 in controlling respiratory mycoplasma infection and disease. J. Immunol. 172:6875-6883. [DOI] [PubMed] [Google Scholar]
- 50.Woolard, M. D., D. Hudig, L. Tabor, J. A. Ivey, and J. W. Simecka. 2005. NK cells in gamma-interferon-deficient mice suppress lung innate immunity against Mycoplasma spp. Infect. Immun. 73:6742-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yancey, A. L., H. L. Watson, S. C. Cartner, and J. W. Simecka. 2001. Gender is a major factor in determining the severity of mycoplasma respiratory disease in mice. Infect. Immun. 69:2865-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]