Abstract
In this study, we demonstrated that the methyltransferase activity associated with Dam was essential for attenuation of Aeromonas hydrophila virulence. We mutated aspartic acid and tyrosine residues to alanine within the conserved DPPY catalytic motif of Dam and transformed the pBAD/damD/A, pBAD/damY/A, and pBAD/damAhSSU (with the native dam gene) recombinant plasmids into the Escherichia coli GM33 (dam-deficient) strain. Genomic DNA (gDNA) isolated from either of the E. coli GM33 strains harboring the pBAD vector with the mutated dam gene was resistant to DpnI digestion and sensitive to DpnII restriction endonuclease cutting. These findings were contrary to those with the gDNA of E. coli GM33 strain containing the pBAD/damAhSSU plasmid, indicating nonmethylation of E. coli gDNA with mutated Dam. Overproduction of mutated Dam in A. hydrophila resulted in bacterial motility, hemolytic and cytotoxic activities associated with the cytotoxic enterotoxin (Act), and protease activity similar to that of the wild-type (WT) bacterium, which harbored the pBAD vector and served as a control strain. On the contrary, overproduction of native Dam resulted in decreased bacterial motility, increased Act-associated biological effects, and increased protease activity. Lactone production, an indicator of quorum sensing, was increased when the native dam gene was overexpressed, with its levels returning to that of the control strain when the dam gene was mutated. These effects of Dam appeared to be mediated through a regulatory glucose-inhibited division A protein. Infection of mice with the mutated Dam-overproducing strains resulted in mortality rates similar to those for the control strain, with 100% of the animals dying within 2 to 3 days with two 50% lethal doses (LD50s) of the WT bacterium. Importantly, immunization of mice with a native-Dam-overproducing strain at the same LD50 did not result in any lethality and provided protection to animals after subsequent challenge with a lethal dose of the control strain.
Aeromonads are ubiquitous organisms which exist in aquatic environments, various types of foods, such as meat, fish and vegetables, and the intestines of apparently healthy humans with diarrhea (11). Among the various Aeromonas species, Aeromonas hydrophila is most commonly involved in human infections, such as septicemia and gastroenteritis (3, 7). The pathogenesis of A. hydrophila infection is complex and multifactorial and characterized by the involvement of a number of virulence factors (1).
Our laboratory recently extensively characterized a type 2 secretion system (T2SS)-secreted cytotoxic enterotoxin, Act, and two cytotonic enterotoxins, Alt (heat labile) and Ast (heat stable), from a diarrheal isolate, SSU, of A. hydrophila (31). We provided evidence that ferric uptake regulator (Fur) and glucose-inhibited division A protein (GidA) regulated Act levels at the transcriptional and translational levels, respectively (32, 33). All of these three enterotoxins contributed significantly to the causation of fluid secretion in both a mouse model and humans (1, 31). We also characterized a type 3 secretion system (T3SS) from the same A. hydrophila isolate and demonstrated that mutants deficient in the production of Act and an Aeromonas outer membrane protein B (AopB), which is an essential component of the T3SS needle structure, were avirulent in a mouse model (34).
More recently, we characterized the DNA adenine methyltransferase (MTase) (Dam), which methylates GATC sequences within the genomic DNA (gDNA) of A. hydrophila SSU strain (designated M.AhySSUDam based on the nomenclature of MTases), and reported its role in modulating the function of both T2SS- and T3SS-associated bacterial virulence (6). We provided evidence that Dam was required for the viability of A. hydrophila, and Dam overproduction reduced the T3SS-associated cytotoxicity and bacterial motility in in vitro models and a mouse model of lethality. In contrast, biological activities (e.g., hemolytic and cytotoxic) associated with Act and protease activity in bacterial culture supernatants were increased as a result of Dam overproduction (6).
Dam alters the interaction of some regulatory proteins with their designated gene targets by adding methyl groups to various sites along the cellular DNA and, in the process, effectively controlling the expression of those genes. Such changes can both modulate bacterial virulence and elicit protective immune responses in the host (23).
This study was undertaken as a continuation of our recently reported findings which demonstrated, by using an arabinose-inducible pBAD expression vector, that Dam overproduction in A. hydrophila attenuated the virulence of the bacterium in an in vivo model of pathogenesis (6). The intent of the present study was to show whether the decreased virulence of A. hydrophila following Dam overproduction could be attributed to a direct increase in the MTase activity associated with Dam. To address this question, we mutated aspartic acid (D) and tyrosine (Y) residues individually within the Dam catalytic DPPY motif (region IV) by in vitro site-directed mutagenesis based on earlier studies with various MTases (9, 29).
Most importantly, we showed that when mutated Dam with no MTase activity was overproduced from A. hydrophila, the latter exhibited a virulence similar to that found with the wild-type (WT) bacterium with pBAD vector alone, which served as a control strain, while overproduction of native Dam in A. hydrophila highly attenuated the virulence of this pathogen (6). Furthermore, we illustrated a link between the regulatory functions of the dam gene and the phenomenon of quorum sensing (QS). Our studies indicate that Dam enzyme overproduction led to an increase in lactone production, which returned to the levels seen in the control strain when the mutated Dam was overproduced. When overproduced from the pBAD vector in an Escherichia coli GM33 strain, additional site-directed mutants of Dam (e.g., damD71A and damE76A, in which mutations were not within the catalytic motif of Dam) exhibited an MTase activity similar to that of native Dam. Finally, we provided evidence that Dam operates via GidA in the regulation of A. hydrophila virulence.
The efficacy of Dam-based vaccines has been explored in other pathogens, such as Salmonella enterica serovar Typhimurium (5, 27) and Yersinia pseudotuberculosis (37). Our studies indicate that, in addition to causing no lethality in mice at two 50% lethal doses (LD50s) of the WT bacterium (6), the Dam-overproducing A. hydrophila SSU strain conferred protection on animals subsequently challenged with a lethal dose of the control strain. The link between the activity of Dam and its role in virulence was demonstrated by our in vivo studies, in which we showed that derivatives of an A. hydrophila SSU strain with an intact dam gene on the chromosome but overexpressing a mutated version of the dam gene (encoding mutated Dam with no detectable MTase activity) in the pBAD vector killed mice at the same rate as did the control strain. However, when the native dam gene was overexpressed in A. hydrophila from the pBAD/damAhSSU plasmid, attenuation in bacterial virulence was noted as measured by an absence of animal lethality.
Taken together, our findings provide conclusive evidence that the MTase activity associated with Dam of A. hydrophila was required to attenuate bacterial virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
The A. hydrophila SSU and E. coli strains, as well as the plasmids used in this study, are listed in Table 1. Bacterial cultures were grown at 37°C in Luria-Bertani (LB) broth and LB agar plates (6, 30). The medium was supplemented with l-arabinose (0.2%) when the native or mutated dam gene was overexpressed from the pBAD/damAhSSU, pBAD/damD/A, or pBAD/damY/A plasmid (Table 1) under the control of an araBAD promoter (10). The native dam gene from the SSU isolate of A. hydrophila was designated damAhSSU. The mutated dam gene, in which either the aspartic acid (D; amino acid position 181) or the tyrosine (Y; amino acid position 184) residue was changed to alanine (A), was designated damD/A or damY/A. The mutated dam gene, in which either the D (amino acid position 71) or the glutamic acid (E; amino acid position 76) (6) residue was changed to A, was designated damD71A or damE76A. The antibiotics ampicillin (Ap), tetracycline (Tc), and rifampin (Rif) were used at concentrations of 100, 12, and 200 μg/ml, respectively. All of the antibiotics and arabinose were obtained from Sigma, St. Louis, MO. Taq DNA polymerase, restriction endonucleases, and T4 DNA ligase were purchased from New England BioLabs, Beverly, MA. Oligonucleotides for PCR amplification of the damAhSSU, damD/A, damY/A, damD71A, and damE76A genes and mutagenic oligonucleotides that were used for the Altered Sites in vitro mutagenesis system (Promega, Madison, WI) were ordered from Integrated DNA Technologies, Inc., Coralville, IA.
TABLE 1.
Strains and plasmids used in the study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| A. hydrophila strains | ||
| SSU | CDC, Atlanta, Ga.a | |
| SSU-R | Rifampin-resistant (Rifr) strain of A. hydrophila SSU | Laboratory stock |
| Control | A. hydrophila SSU-R harboring pBAD, Rifr Apr | 6 |
| Dam-overproducing strain | A. hydrophila SSU-R harboring pBAD-damAhSSU, Rifr Apr | 6 |
| D/A Dam mutant | A. hydrophila SSU-R harboring pBAD-damD/A, Rifr Apr | This study |
| Y/A Dam mutant | A. hydrophila SSU-R harboring pBAD-damY/A, Rifr Apr | This study |
| MgidA | gidA isogenic mutant of A. hydrophila SSU-R, Rifr Kmr | 31 |
| MgidA/pBAD | gidA isogenic mutant of A. hydrophila SSU-R harboring pBAD plasmid, Rifr Kmr Apr | This study |
| MgidA/pBAD-damAhSSU | gidA isogenic mutant of A. hydrophila SSU-R harboring pBAD-damAhSSU plasmid, Rifr Kmr Apr | This study |
| E. coli strains | ||
| ES1301 mutS | lacZ53 mutS201::Tn5 thyA36 rha-5 metB1 deoC | Promega |
| JM109 | endA1 recA1 gyrA96 thi hsdR17 relA1 supE44 λ− Δ(lac-proAB) [F′ traD36 proA+B+laclZΔM15] | Promega |
| GM33 | F−dam-3 sup-85 (Am) | 25 |
| C. violaceum CV026 | Mini-Tn5 mutant of ATCC 31532 | 26 |
| Plasmids | ||
| pALTER-1 | Vector for the damAhSSU gene cloning, Tcr | Promega |
| pALTER-1/damAhSSU | dam gene of A. hydrophila cloned at the EcoRI/BamHI sites of pALTER-1 vector for mutagenesis, Tcr | This study |
| pALTER-1/damD/A | pALTER-1 vector containing damD/A mutation, Apr | This study |
| pALTER-1/damY/A | pALTER-1 vector containing damY/A mutation, Apr | This study |
| pBAD/Thio-E | araBAD promoter expression vector, Apr | Invitrogen |
| pBAD | Control vector, Apr | 6 |
| pBAD/damAhSSU | dam gene of A. hydrophila cloned into pBAD/Thio-E, Apr | 6 |
| pBAD/damD/A | pBAD vector carrying damD/A mutation, Apr | This study |
| pBAD/damY/A | pBAD vector carrying damY/A mutation, Apr | This study |
| pBAD/damD71A | pBAD vector carrying damD71A mutation, Apr | This study |
| pBAD/damE76A | pBAD vector carrying damE76A mutation, Apr | This study |
CDC, Centers for Disease Control and Prevention.
Generation of mutated dam genes by site-directed mutagenesis.
To obtain the pALTER-1/damD/A and pALTER-1/damY/A plasmids (Tcs and Apr) (Table 1) with a mutated dam gene, first, the PCR-amplified native damAhSSU gene was cloned into the pALTER-1 vector with A. hydrophila SSU gDNA and the damN-EcoRI/damC-BamHI primers (5′-CCGGAATTCATGAAAAAAACACGCGCTTTTTTAAA-3′/5′-CGCGGATCCTCAGCCGAGTGGCGCCAGTTCGGCGT-3′), with underlined bases representing the restriction enzyme sites. Subsequently, E. coli JM109 cells containing the pALTER-1/damAhSSU recombinant plasmid (Tcr and Aps) were infected with the R408 helper phage, and the phagemid single-stranded DNA was isolated as described by the manufacturer. This DNA was used as a template for the mutagenesis reaction with the mutagenic damD/A and damY/A primers (5′-GTCATCTATTGCGCTCCGCCCTATGCGCCGCTCTC-3′ and 5′-GTCATCTATTGCGATCCGCCCGCTGCGCCGCTCTC-3′). The underlined bases represented nucleotides that were changed, resulting in the mutations of D and Y to A (6). Each mutagenesis reaction was transformed into E. coli ES1301 mutS-competent cells. The plasmid DNA isolated from this bacterial strain was then transformed into the E. coli JM109 strain, and transformants were screened for tetracycline sensitivity. The mutations within the dam gene were confirmed by DNA sequence analysis. All procedures were performed according to Promega's protocol. Competent E. coli cells (JM109 and ES1301 mutS) were prepared using a Z-Competent E. coli transformation kit from Zymo Research, Orange, CA.
For generation of mutated damD71A and damE76A genes, we used the 5′-GCGGACGCCGGCCAGCTTCATCGCCGAGGCGCGCA-3′ and 5′-CGCCGGACAGCTTCATCGCCGCGGCGCGCAAGCTG-3′ mutagenic (D71A and E76A) primers, respectively. The underlined bases represent nucleotides that were changed, resulting in the mutations of D to A and E to A at amino acid positions 71 and 76 of Dam. Thus, by using an Altered Sites in vitro mutagenesis system, we mutated D, Y, and E residues to A within and outside the conserved DPPY catalytic motif of Dam.
Cloning of the mutated dam gene into the pBAD/Thio-E vector.
To overexpress mutated dam genes, the pBAD/damD/A and pBAD/damY/A plasmids were constructed in a manner similar to that for the pBAD/damAhSSU plasmid, as recently described (6), using the same primers, damN-NcoI/damC-PmeI (5′-ATGCCATGGATGAAAAAAACACGCGCTTTTTTAAAATGG-3′/5′-AGCTTTGTTTAAACGCCGAGTGGCGCCAGTTCGGCGTCGC-3′), with underlined bases representing restriction enzyme sites. We used pALTER-1/damD/A and pALTER-1/damY/A plasmids as templates for PCRs. The PCR conditions were the same as those described recently (6), and the PCR products were digested with NcoI-PmeI restriction endonucleases and ligated to the pBAD/Thio-E vector cut with the compatible restriction enzymes. The ligation reaction was transformed into E. coli JM109-competent cells. The pBAD/damD71A and pBAD/damE76A plasmids were constructed similarly. The recombinant plasmids were then isolated from E. coli, and to generate A. hydrophila SSU with the mutated dam gene, the pBAD/damD/A, pBAD/damY/A, pBAD/damD71A, and pBAD/damE76A plasmids were transformed into bacterial cells by electroporation, and these A. hydrophila SSU mutated strains were used for the study of bacterial virulence. The levels of native Dam versus mutated Dam production from the pBAD vector system after induction of appropriate A. hydrophila cultures with arabinose were analyzed by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE) of the whole cells, and the gels were stained with Coomassie blue (6).
Dam activity.
To examine MTase activity associated with the native or mutated Dam, the pBAD/damD/A, pBAD/damY/A, pBAD/damD71A, and pBAD/damE76A plasmids as well as the pBAD/damAhSSU plasmid (with the native dam gene to serve as a positive control) were transformed into the E. coli GM33 (dam-deficient) strain (Table 1) (25). These cultures were grown overnight in the presence of arabinose at 37°C with shaking (180 rpm). The gDNA was then isolated and subjected to digestion with DpnI (which cuts methylated DNA) and DpnII (which cuts nonmethylated DNA) restriction enzymes. As a negative control, we used gDNA isolated from the original E. coli GM33 (dam-deficient) strain. Dam activity for some Dam mutants was also confirmed by transfer of the 3H-methyl group from [methyl-3H]AdoMet (S-adenosyl-l-methionine) by Dam onto N6-methyladenine-free lambda DNA as described previously (6).
Measurement of motility, Act-associated biological activities, and protease activity.
We determined the motility of A. hydrophila SSU strains carrying the pBAD vector alone, pBAD/damAhSSU, pBAD/damD/A, or the pBAD/damY/A plasmid, as recently described, using LB agar (0.35%) plates (6). Likewise, the culture supernatants from bacteria were used to measure Act-associated hemolytic and cytotoxic activities as well as the protease activity (6). The hemolytic activity was measured by examining the release of hemoglobin (optical density at 540 nm [OD540]) from rabbit erythrocytes (3%), while cytotoxic activity associated with Act was determined by measuring the percentage of lactate dehydrogenase (LDH) enzyme released from RAW264.7 murine macrophages treated with bacterial culture supernatants for 2 h with a CytoTox96 kit from Promega (6). The protease activity was measured by examining the release of blue color (OD595) from Hide azure powder. The enzyme-linked immunosorbent assay (ELISA) for the detection of Act antigen levels in the culture supernatants was performed as described previously (6).
Lactone production.
To detect acylhomoserine lactones (AHLs), we used the WT A. hydrophila SSU strain (a positive control), A. hydrophila strains containing the pBAD, pBAD/damAhSSU, pBAD/damD/A, or pBAD/damY/A plasmid, and the gidA isogenic mutant of A. hydrophila (32) harboring the pBAD or the pBAD/damAhSSU plasmid. We employed the method of McClean et al. (26) with modifications and used the Chromobacterium violaceum CV026 strain for lactone production (Table 1). This method allowed us to quantitate the amount of lactone produced, compared to the original qualitative method for the detection of lactones (36). Briefly, Aeromonas strains were grown overnight in LB medium (pH 7.0) with appropriate antibiotics at 37°C, and the C. violaceum CV026 strain was grown in LB broth for 24 h at 28°C with shaking (150 rpm). The test strains of A. hydrophila were reinoculated in the morning using 3 ml of the LB medium with arabinose and incubated for 4 h (an optimal time interval that was standardized empirically). The OD600 and the number of CFU were determined for each culture, which was centrifuged at 13,000 × g for 10 min. The culture supernatants were then filter sterilized (0.22 μm). Subsequently, the C. violaceum culture was mixed with the supernatant from each of the A. hydrophila cultures at a 1:10 ratio and incubated for 24 h at 28°C with shaking. The cultures were centrifuged and pellets suspended in 50% ethanol and 1% SDS solution. The mixture was vortexed, and the bacterial sediment was centrifuged. The deep blue color in the supernatant was measured at OD590 using a VERSAmax reader (Molecular Devices Corporation, Sunnyvale, CA). The use of C. violaceum CV026 culture for lactone production is by far the best for Aeromonas and Pseudomonas species and was discussed in our recent publication (34) as well as in other studies (26, 36). C. violaceum specifically detects AHLs with N-acyl side chains of four to eight carbons, especially BHLs (N-[butanoyl]-l-homoserine lactones), which are produced predominantly by Aeromonas species.
Animal experiments.
Eight-week-old female Swiss Webster mice (Taconic Farms, CA) were used to determine whether animals immunized with the A. hydrophila Dam-overproducing strain would protect mice against lethal infection with the WT A. hydrophila harboring the pBAD plasmid alone. Forty mice (group 1) were inoculated with Dulbecco's phosphate-buffered saline (DPBS) and served as controls. A second group (10 mice) was infected intraperitoneally (i.p.) with 1 × 107 CFU of WT A. hydrophila (with pBAD vector alone), and group 3 (40 mice) was infected with 1 × 107 CFU of an A. hydrophila (pBAD/damAhSSU) strain that overproduced Dam. Animals were observed for mortality for 1 week. The animals were provided with 0.2% arabinose and Ap (40 mg/kg of body weight/day) in their drinking water over the first 2 to 3 days to retain the plasmid in bacteria. At 1, 3, 5, and 6 weeks postinoculation, mice (n = 10) in groups 1 and 3 were challenged with two LD50s (1 × 107 CFU) of the WT A. hydrophila SSU strain containing pBAD vector alone without the dam gene and observed for mortality.
To define whether mutations within the catalytic domain of Dam affected lethality in mice, we infected another four groups of mice (each group contained 10 mice) i.p. with a lethal dose (5 × 106 to 1 × 107 CFU) of the WT A. hydrophila (pBAD), the Dam-overproducing strain (pBAD/damAhSSU), and the mutated-Dam-overproducing strains (with pBAD/damD/A or pBAD/damY/A plasmid). Mice were observed daily for 2 weeks for mortality.
Statistical analysis.
Wherever appropriate, the data were analyzed using Student's t test or the Fisher exact test, and P values of ≤0.05 were considered significant. At least three independent experiments were performed for statistical analysis of data.
RESULTS
To define precisely the role of Dam activity in the virulence potential of A. hydrophila, we constructed A. hydrophila strains in which the mutated dam gene was expressed from the pBAD vector system. All MTases possess a similar catalytic domain and D/NPPY/F sequence, which represents one of nine described motifs (I to VIII and X) and is the common conserved catalytic motif in region IV of adenine-N6 and cytosine-C4 DNA MTases (21, 29, 43). Residues located in motifs IV (Asp193 and Tyr196), V (Asp211), VI (Ser229 and Trp231), and VIII (Tyr258) of EcoRV adenine-N6-methyltransferase (Dam) are involved in catalysis, some of them presumably in the binding of the flipped target base, because mutations in these residues fail to significantly interfere with DNA and AdoMet binding but strongly reduce catalysis (29). To verify that alteration of amino acid residues within or outside the conserved catalytic DPPY active domain abolishes the MTase activity of A. hydrophila, we examined E. coli GM33 strains containing pBAD/dam plasmids with different amino acid mutations within the dam gene.
The mutated Dam in the DPPY motif did not possess MTase activity.
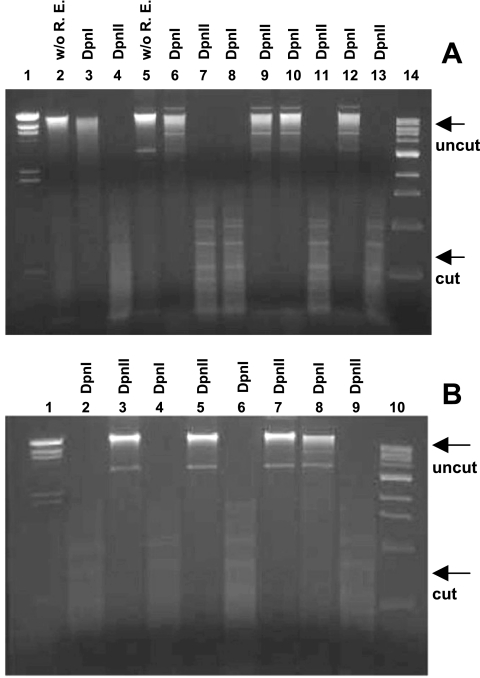
To test the GATC DNA methylation activity of the mutated A. hydrophila Dam enzyme, we used the E. coli GM33 (dam-deficient) strain (Table 1), which is deficient in N6-adenine MTase activity (chromosomal dam gene mutation of the WT E. coli GM1 strain) (25). The pBAD/damD/A and pBAD/damY/A plasmids were transformed into Z-competent E. coli GM33 cells, and isolated gDNA from these strains was digested with DpnI, which digests methylated DNA, and the DpnII restriction endonuclease, which cuts nonmethylated DNA. As controls, we prepared gDNA from the E. coli GM33 strain, which either did not harbor any plasmid or harbored the pBAD vector alone. The gDNA from the E. coli GM33 strain containing the pBAD/damAhSSU plasmid, which encoded native and functionally active Dam, was also used as a positive control. As shown in Fig. 1A, the gDNA isolated from either the E. coli GM33 (pBAD/damD/A) or the E. coli GM33 (pBAD/damY/A) strain grown in the presence of arabinose was resistant to DpnI (which cuts methylated DNA) digestion (Fig. 1A, lanes 10 and 12) and sensitive to DpnII (which cuts nonmethylated DNA) restriction endonuclease cutting (Fig. 1A, lanes 11 and 13). These data confirmed that the gDNA from these E. coli GM33 strains, with mutated, overproduced A. hydrophila Dam, was not methylated. These data were similar to those obtained when the gDNA of the E. coli GM33 (dam-deficient) strain was digested with DpnI and DpnII restriction enzymes (Fig. 1A, lanes 3 and 4), indicating the nonmethylated status of the gDNA in this strain. A similar digestion pattern with enzymes DpnI and DpnII was noted when gDNA from the E. coli GM33 strain with pBAD vector alone was used (Fig. 1A, lanes 6 and 7). The additional lower molecular bands in the vicinity of the gDNA (Fig. 1A, lanes 5, 6, 9, 10, and 12) represented the plasmid DNA. Opposite cleavage patterns were noted when gDNA from the E. coli GM33 strain with the pBAD/damAhSSU plasmid was digested with DpnI and DpnII enzymes. Isolated gDNA from this strain was sensitive to DpnI (Fig. 1A, lane 8) and resistant to DpnII (Fig. 1A, lane 9) digestion. These data signified that the overproduction of native Dam from pBAD/damAhSSU plasmid in the E. coli GM33 strain methylated its gDNA, while the mutated Dam could not, thus indicating that mutations of D and Y residues within the DPPY motif resulted in no detectable Dam activity.
FIG. 1.
DpnI and DpnII cleavage patterns of gDNA isolated from the E. coli GM33 (dam-deficient) strain containing pBAD plasmid with the native or mutated dam gene. (A) Lane 1, lambda DNA/HindIII markers (Promega); lanes 2, 3, and 4, gDNA from the E. coli GM33 strain (without any plasmid as a control) either without any restriction enzyme digestion (w/o R. E.) (lane 2) or treated with DpnI (lane 3) or DpnII (lane 4); lanes 5, 6, and 7, gDNA from E. coli GM33 containing the pBAD plasmid alone either with no enzyme digestion (w/o R. E.) (lane 5) or treated with DpnI (lane 6) or DpnII (lane 7); lanes 8 and 9, gDNA from E. coli GM33 containing the pBAD/damAhSSU recombinant plasmid treated with DpnI or DpnII enzymes; lanes 10 and 11, gDNA from the E. coli GM33 strain with the pBAD/damD/A plasmid treated with DpnI or DpnII; lanes 12 and 13, gDNA from E. coli GM33 strain carrying the pBAD/damY/A plasmid treated with DpnI and DpnII, respectively; and lane 14, 1-kb DNA ladder (New England BioLabs). (B) Lanes 1 and 10, lambda DNA/HindIII markers (Promega) and 1-kb DNA ladder (New England BioLabs), respectively; lanes 2 and 3, gDNA from E. coli GM33 containing the pBAD/damAhSSU recombinant plasmid treated with DpnI or DpnII enzymes; lanes 4 and 5, gDNA from the E. coli GM33 strain with the pBAD/damD71A plasmid treated with DpnI or DpnII; lanes 6 and 7, gDNA from the E. coli GM33 strain carrying the pBAD/damE76A plasmid treated with DpnI and DpnII, respectively; lanes 8 and 9, gDNA from the E. coli GM33 strain with the pBAD/damD/A plasmid treated with DpnI or DpnII. All cultures which were used for the isolation of gDNA were grown in the presence of arabinose. The gDNA was digested at 37°C for 1 h and subjected to 1% agarose gel electrophoresis. The gels were visualized under UV light after being stained with ethidium bromide.
However, production of MTase from pBAD/damD71A and pBAD/damE76A plasmids in the E. coli GM33 strain methylated its gDNA, showing the expected phenotype of native Dam (expressed from the pBADdamAhSSU plasmid), i.e., digestion with DpnI but not with DpnII (Fig. 1B, lanes 4, 5, 6, and 7 versus lanes 2 and 3), while mutated Dam from the pBAD/damD/A plasmid exhibited the opposite pattern (digestion with DpnII but not DpnI) (Fig. 1B, lanes 8 and 9), demonstrating an original Dam− phenotype for the E. coli GM33 strain. These data indicate that mutations of amino acid residues outside the catalytic motif of Dam did not affect MTase activity associated with Dam.
To demonstrate production of native and mutated Dam proteins, we performed SDS-PAGE with E. coli whole cells containing tested recombinant plasmids with the dam gene under the araBAD promoter (data not shown). The gels were visualized after being stained and destained. All of the mutants tested produced similar levels of Dam after induction with arabinose, as noted, with E. coli overproducing native Dam. These data suggested possibly no or minimal alteration in the conformation of the mutated Dam after site-directed mutagenesis that could affect MTase activity. We expected minimal conformational changes, as similar amino acid residues were mutated in other MTases with no significant alterations in the levels of the mutated protein produced. Further, these mutated MTases behaved biochemically in a manner predicted from their structural models (18, 29, 43).
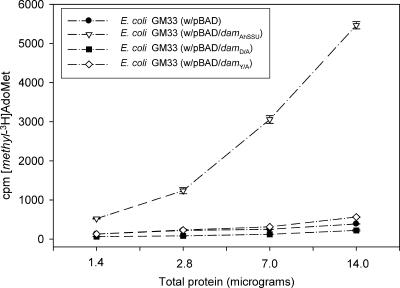
The inability of mutated Dam in the DPPY motif (pBAD/damD/A and pBAD/damY/A) to transfer methyl groups from [methyl-3H]AdoMet to N6-methyladenine-free lambda DNA was confirmed quantitatively by using a radioactivity incorporation assay (Fig. 2). As noted from Fig. 2, cell extracts from the E. coli GM33 strain with native Dam (pBAD/damAhSSU) showed a concentration-dependent increase in MTase activity. The MTase activity associated with the mutated Dam in the E. coli cell extracts was similar to that in E. coli with pBAD vector alone.
FIG. 2.
MTase activity associated with native and mutated Dam of A. hydrophila SSU in the E. coli GM33 strain by the AdoMet assay. The cell extracts from E. coli strains containing pBAD plasmid (as a control), the pBAD/damAhSSU recombinant plasmid, pBAD/damD/A, and the pBAD/damY/A plasmid were prepared, and the ability of Dam to transfer methyl-3H groups from AdoMet to N6-methyladenine-free lambda DNA was measured (6). Three independent experiments were performed, and the arithmetic means ± standard deviations were plotted. The increase in MTase activity in the cell extracts of E. coli overproducing native Dam was statistically significant at all of the protein concentrations according to Student's t test, compared to the activity in E. coli cell extracts overproducing mutated Dam. cpm, counts per minute.
Overproduction of inactive mutated Dam in A. hydrophila SSU causes its virulence level to revert to that of WT A. hydrophila.
In our recently published studies, we showed that overproduction of native Dam in A. hydrophila altered its virulence potential (6). In particular, our data demonstrated that native Dam overproduction decreased the motility and the T3SS-associated cytotoxicity of the bacterium and increased the biological activities associated with Act and protease production.
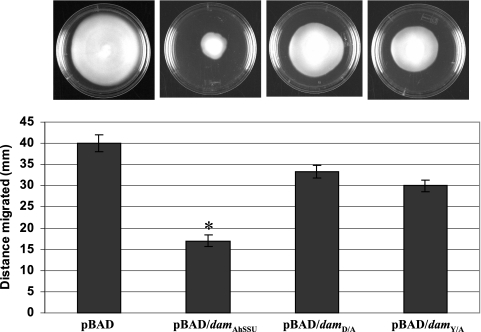
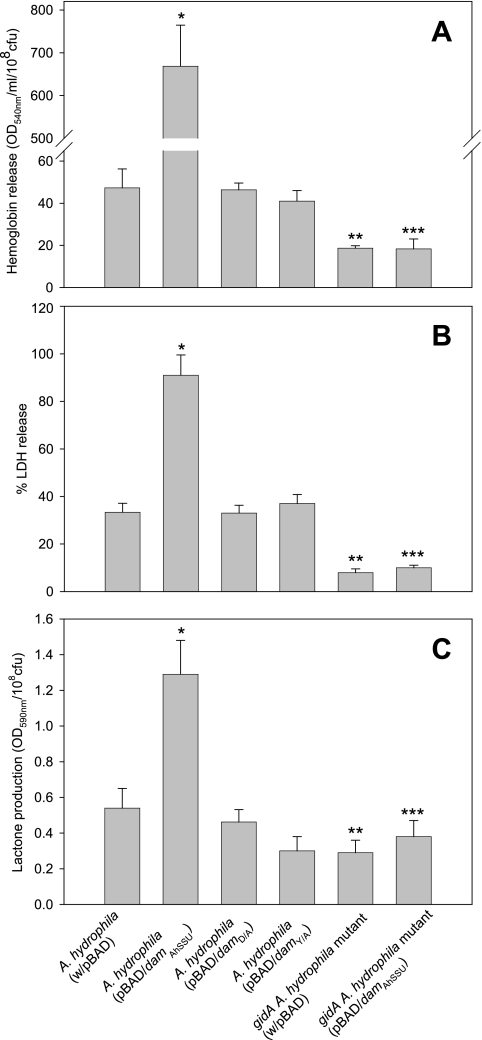
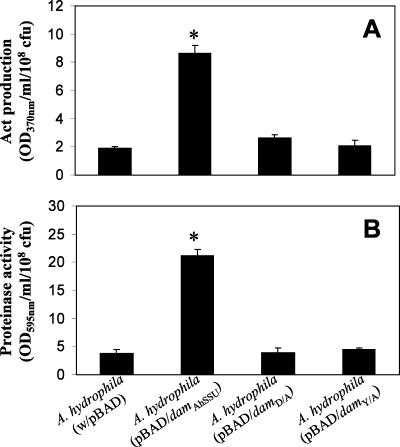
To define the role of Dam (both enzymatically active and enzymatically inactive forms) overproduced from A. hydrophila SSU in terms of its effect on overall bacterial virulence, we used arabinose-induced native and mutated dam genes that were expressed from the araBAD promoter in vector pBAD. We used A. hydrophila SSU with pBAD vector alone as a control. Based on SDS-PAGE and the stained gels, the levels of native and mutated Dam, when Dam was overproduced from the pBAD vector after arabinose induction, were similar (data not shown). As noted from Fig. 3, the native Dam-overproducing strain was much less motile (by 57%) than the control strain (P = 0.0008), confirming our previous data (6). When the dam gene was mutated, the bacterium showed a significantly higher motility than A. hydrophila expressing the native dam gene, although it did not reach the level for WT A. hydrophila with the pBAD vector alone (P = 0.05). These data might indicate some residual MTase activity associated with mutated Dam. The hemolytic activity associated with Act of A. hydrophila was significantly increased (at least 14-fold) when the native dam gene was overexpressed compared to that of the control A. hydrophila strain with the pBAD vector alone. The hemolytic activity in the culture filtrates of A. hydrophila with mutated Dam was similar to that in the control strain (Fig. 4A).
FIG. 3.
(Top) A. hydrophila with overproducing mutated Dam (with pBAD/damD/A or pBAD/damY/A plasmid) exhibits a motility phenotype similar to that of the A. hydrophila SSU strain containing pBAD vector alone. The motility of the A. hydrophila strain with pBAD/damAhSSU plasmid was significantly reduced. The strains were grown in the presence of arabinose. (Bottom) The bar graph demonstrates the distances in millimeters by which these strains migrated from the points of inoculation in soft agar plates. Data from three independent experiments were plotted with standard deviations. An asterisk denotes statistical significance by Student's t test (P = 0.0008).
FIG. 4.
Overproduction of mutated Dam decreases biological activities of T2SS-associated Act and lactone production to the level of the WT bacterium (A. hydrophila with or without the pBAD plasmid). A regulatory GidA protein is crucial in Dam-associated alteration in biological activities of Act and lactone production. (A and B) Act-associated hemolytic and cytotoxic activities in the culture supernatants of A. hydrophila strains containing pBAD plasmid alone and pBAD/damAhSSU, pBAD/damD/A, and pBAD/damY/A plasmids were measured by the release of hemoglobin from rabbit erythrocytes and LDH enzyme from macrophages, respectively (6). (C) Overproduction of native Dam (the A. hydrophila SSU strain with pBAD/damAhSSU plasmid) but not the mutated Dam (the A. hydrophila SSU strain with the pBAD/damD/A or the pBAD/damY/A plasmid) increases lactone production. The gidA mutant of A. hydrophila does not respond to overproduction of native Dam with increasing activities of T2SS-associated Act and lactone production. The strains were grown in the presence of arabinose. Three independent experiments were performed, and the arithmetic means ± standard deviations were plotted. The data were normalized to 1 × 108 CFU to account for any differences in the growth rates of the various bacterial strains used.  , statistically significant difference between the A. hydrophila control strain and the Dam-overproducing strain of WT A. hydrophila by Student's t test.
, statistically significant difference between the A. hydrophila control strain and the Dam-overproducing strain of WT A. hydrophila by Student's t test. 
 , statistically significant difference between the WT control and the gidA mutant control strain of A. hydrophila.
, statistically significant difference between the WT control and the gidA mutant control strain of A. hydrophila. 

 , statistically significant difference between the native Dam-overproducing WT strain and the gidA mutant of A. hydrophila overproducing native Dam.
, statistically significant difference between the native Dam-overproducing WT strain and the gidA mutant of A. hydrophila overproducing native Dam.
We also examined Act-associated cytotoxicity (as measured by the percentage of release of LDH enzyme) from RAW264.7 murine macrophages. As shown in Fig. 4B, compared to the control strain, a threefold increase in the release of LDH from macrophages treated with the culture filtrates of the A. hydrophila strain that overproduced native Dam was noted. However, the level of LDH release from macrophages was decreased significantly and comparable to that for the A. hydrophila (pBAD) control strain when the dam gene was mutated. These data were further confirmed by performing ELISA on the culture supernatants of the above-mentioned A. hydrophila strains and using antibodies specific to Act. As noted from Fig. 5A, the native Dam-overproducing strain produced much higher Act levels than the control strain. However, when the dam gene was mutated, the Act levels produced by the mutated A. hydrophila strains were similar to those for the control strain, clearly indicating the necessity of MTase activity in Act production. Finally, we noted a similar pattern of increased protease production when native Dam was overproduced, compared to the protease production in the culture supernatants of A. hydrophila SSU strains in which the dam gene was mutated as well as that in the control strain (Fig. 5B).
FIG. 5.
Overproduction of mutated Dam decreases Act antigen levels and protease activities to those of the WT A. hydrophila strain. (A) Production of Act based on ELISA with the culture supernatants of the A. hydrophila strain with pBAD vector only, the pBAD/damAhSSU recombinant plasmid, and the pBAD/damD/A or the pBAD/damY/A plasmid. (B) Protease activity in the culture supernatants of A. hydrophila strains with pBAD, pBAD/damAhSSU, pBAD/damD/A, or pBAD/damY/A plasmid, as measured by the hydrolysis of Hide azure powder. The strains were grown in LB medium with arabinose. Three independent experiments were performed, and the arithmetic means ± standard deviations were plotted. The data were normalized to 1 × 108 CFU to account for any differences in the growth rates of the various bacterial strains used. Asterisks denote statistically significant values by Student's t test.
Quorum sensing and Dam activity.
QS, the ability of bacteria to communicate and coordinate behavior via signaling molecules, has been shown in several pathogenic microorganisms (15, 17, 35). We described a correlation between the T3SS and Act production in A. hydrophila with lactones (34) and provided evidence that mutations in the act and aopB genes resulted in reduced lactone production. To determine whether Dam overproduction altered lactone production, which is a signaling molecule of QS, we noted that the Dam-overproducing strain with the native dam gene produced significantly more lactones than the control strain (Fig. 4C). However, when the mutated dam gene (in the pBAD/damD/A or the pBAD/damY/A plasmid) was overexpressed, the lactone production level was similar to that of the A. hydrophila (pBAD) control strain (Fig. 4C). To eliminate any possible nonspecific contribution from protein overproduction, using an arabinose-inducible pBAD vector, we included the original A. hydrophila SSU strain (without any plasmid) as an additional control, whose lactone production level was similar to that of the A. hydrophila SSU strain with the pBAD vector alone (data not shown). These control cultures were the most appropriate for these studies, as they also ruled out any influence on lactone production by other virulence factors produced by A. hydrophila SSU that are present in the culture filtrates (8).
Regulation of Act and lactone production in A. hydrophila SSU by Dam occurs via GidA.
We demonstrated previously that Act production was significantly reduced when the gidA gene was deleted from A. hydrophila, an effect which could be complemented (32). Based on the upstream sequence of the gidA structural gene, multiple GATC sequences (a total of 15) that potentially could be methylated by Dam, resulting in increased Act production, were detected within the putative promoter region by using a software program from Softberry, Inc. Indeed, we noted that the hemolytic activity in the culture filtrate of the WT A. hydrophila strain harboring the pBAD/damAhSSU plasmid was considerably greater than that of the A. hydrophila control strain harboring the pBAD vector alone (Fig. 4A). As expected, truncation of the gidA gene resulted in a reduced level of hemoglobin release from erythrocytes (Fig. 4A). More interestingly, when the native dam gene was overexpressed in the gidA mutant, no further increase in the hemolytic activity (compared to that of the gidA mutant with pBAD vector alone) was noted (Fig. 4A). Similar findings were observed when we examined cytotoxic activity (as a percentage of LDH release) associated with Act in the culture filtrates of the native Dam-overproducing WT and the gidA mutant of A. hydrophila (Fig. 4B). As noted from Fig. 4B, overproduction of Dam in WT A. hydrophila increased Act-associated cell toxicity compared to that in the control strain. The cytotoxicity was decreased in the gidA mutant with the pBAD vector compared to that in the WT A. hydrophila control strain. Overproduction of native Dam in the gidA mutant exhibited no further changes in cytotoxicity (Fig. 4B).
Finally, overproduction of native Dam in the gidA isogenic mutant failed to increase lactone production compared to that in WT A. hydrophila expressing the native dam gene (Fig. 4C). As expected, a significant increase in lactone production in WT A. hydrophila overexpressing the native dam gene, compared to that in the A. hydrophila control strain, was noted. Taken together, these data indicate the involvement of GidA in Dam-associated biological functions. We confirmed that Dam activity was indeed increased in the gidA mutant harboring the pBAD/damAhSSU plasmid after arabinose induction, as measured by the transfer of methyl groups from [methyl-3H]AdoMet to N6-methyladenine-free lambda DNA (data not shown).
Protection of mice immunized with the Dam-overproducing strain against WT A. hydrophila challenge.
As indicated in our previous studies, the native Dam-overproducing strain of A. hydrophila SSU was less virulent and attenuated in a mouse infection model (6). To further examine whether this strain might immunize mice to subsequent Aeromonas infection, animals were infected i.p. with the native-Dam-overproducing bacteria at a dose of 1 × 107 CFU. Mice immunized with Dam-overproducing bacteria were completely protected when they were subsequently challenged at 1 to 6 weeks postimmunization with WT A. hydrophila containing pBAD vector alone, indicating possibly the role of both cell-mediated and humoral immune responses in animal protection (Table 2). Since immune responses to this pathogen are largely unknown, our future studies will examine in detail the immunological basis of protection by Dam overproduction in a mouse model of infection. The nonimmunized group of mice, given only DPBS, died within 2 days after subsequent challenge with the lethal dose of the A. hydrophila control strain.
TABLE 2.
Immunization of mice with the native Dam-overproducing strain of A. hydrophila provides protection against subsequent challenge with the lethal dose of WT A. hydrophila SSU
| Period of immunization with the native Dam overproducera (no. of wks) | No. of survivals/total no. of animals infected
|
||
|---|---|---|---|
| Over the period of immunization | After immunization and challenge with two LD50s of WT A. hydrophilab | For control animals given DPBS and then challenged with two LD50s of WT A. hydrophilab | |
| 1 | 10/10 | 10/10 | 0/10 |
| 3 | 10/10 | 10/10 | 0/10 |
| 5 | 10/10 | 10/10 | 0/10 |
| 6 | 10/10 | 10/10 | 0/10 |
Animals were immunized with 1 × 107 CFU of the Dam-overproducing strain.
Animals were challenged with 1 × 107 CFU of WT A. hydrophila, which represented approximately two LD50s.
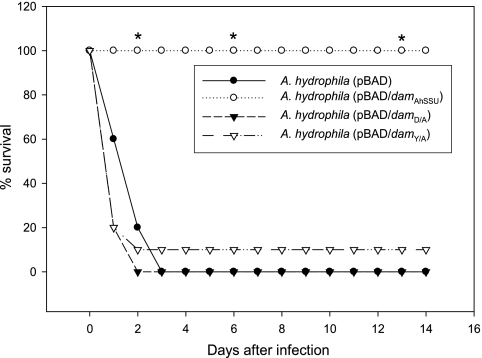
On the other hand, inoculation of mice with the mutated-Dam-overproducing strains indicated that such mutants (D/A and Y/A) induced lethality in mice similar to that of the control A. hydrophila (i.e., WT bacterium with pBAD vector alone), while the A. hydrophila strain with the native overproduced Dam did not cause any mortality at the same dose (Fig. 6). Our data indicate that mutations in the conserved catalytic DPPY motif of M.AhySSUDam indeed caused the phenotype of the Dam-overproducing strain to revert to that of the WT bacterium.
FIG. 6.
A. hydrophila with mutated Dam is highly virulent in a mouse model. Mutants of A. hydrophila with pBAD/damD/A or pBAD/damY/A plasmid led to mouse mortality within 48 h, similar to what was found for the WT A. hydrophila strain harboring only the pBAD vector. The Dam-overproducing A. hydrophila strain with pBAD/damAhSSU plasmid (with native Dam) fully protected mice. Asterisks denote statistically significant values by Fisher's exact test.
DISCUSSION
We demonstrated previously that Dam is crucial for the viability of A. hydrophila and that it affects bacterial virulence by altering the biological activities associated with type 2- and type 3-secreted proteins. In addition, Dam overproduction in A. hydrophila attenuated bacterial virulence in a mouse infection model (6).
Functional roles for strongly or moderately conserved amino acid residues were investigated for some MTases which can modify DNA by two classes of these enzymes, C-MTases modifying cytosines at the C-5 position, and N-MTases transferring a methyl group to adenine-N6 or cytosine-N4 positions (24, 42). The notion that region IV, which contains the DPPY motif, is possibly involved in enzymatic activity was described with regard to the phage T4 Dam, E. coli DNA N6-adenine MTases (e.g., EcoRV Dam), and the BcgI restriction modification system (9, 14, 20, 21, 29, 42).
In this study, we generated two M.AhySSUDam mutants in the DPPY motif (D/A and Y/A), tested them for the ability to methylate gDNA of the dam-deficient E. coli GM33 strain, and evaluated the virulence of the A. hydrophila SSU strain with overproduced native and mutated Dam. Our data indicate that mutations in the DPPY motif of Dam resulted in no detectable MTase activity (Fig. 1 and 2).
We recently investigated the role of overproduced Dam in the pathogenesis of A. hydrophila (6). In this study, we examined the effect of overproduced but mutated Dam in the DPPY motif (region IV), which is responsible for the catalytic and substrate (AdoMet and DNA) binding activities of MTase relative to the virulence of the bacterium. Our data clearly indicate that the motility of the A. hydrophila SSU strain, as well as Act-associated biological activities, was directly linked to Dam activity. We also provided evidence that those Dam mutants with an altered catalytic site affected MTase activity, as mutations in two other tested amino acid residues outside the catalytic domain exhibited MTase activities similar to that of the native Dam. Further, the levels of overproduced native and mutated Dam in both E. coli and A. hydrophila strains were essentially similar.
Bacteria that use QS produce and secrete certain signaling compounds, autoinducers (AIs), or pheromones, such as AHLs. It is known that culture supernatants from both A. hydrophila and Aeromonas salmonicida activate a range of biosensors responsive to AHLs, and the genes for a QS signal generator and a response regulator were cloned from each of the above-mentioned Aeromonas species and termed ahyRI and asaRI, respectively (36). Recently, we revealed that lactone production levels were decreased in the act (32%) and aopB (64%) mutants compared to that of the WT A. hydrophila SSU strain (34). The effect of Dam activity on lactone production is demonstrated in Fig. 4C, showing that the levels increased when the native dam gene was overexpressed and returned to that of the WT bacterium when the mutated dam genes were overexpressed. Since we previously demonstrated that the act mutant produced fewer lactones (34), our data indicate that when Act production was increased due to native Dam overproduction, lactone production was also increased, and this was indeed the case (Fig. 4A to C). However, one should not overlook the possible effect of AdoMet-dependent Dam on QS.
Interestingly, AdoMet (a source of methyl groups for MTases) is a direct precursor of the Vibrio fischeri AI (12). Hanzelka and Greenberg (12) showed that AdoMet is required for AI synthesis by LuxI in amino acid-starved E. coli mutants. Investigations of the in vivo source of the acyl chain and homoserine lactone components of AI synthesized by the LuxI homolog, TraI, showed that decreased levels of intracellular AdoMet caused by expression of bacteriophage T3 AdoMet hydrolase resulted in a marked reduction in AI synthesis. These data provided direct in vivo evidence that the homoserine lactone ring of LuxI family autoinducers is derived from AdoMet (39). In our future studies, this interesting correlation between Dam activity and lactone production as it relates to QS will be pursued.
The role that Dam plays in the regulation of bacterial virulence is only beginning to be understood (23), and studies have shown that adenine methylation can either directly or indirectly alter the interaction of regulatory proteins with DNA (4). Dam can act as a de novo methylase by methylating both nonmethylated and hemimethylated GATC sites (38) and plays a pivotal role in the control of gene expression by the formation of DNA methylation patterns (40). A typical example is the pyelonephritis-associated pilus (pap) operon of uropathogenic E. coli (16). The DNA methylation patterns influence the binding of the regulatory proteins Lrp and PapI to the papBA pilin promoter, which correlates with the ON and OFF stages of pilus expression in this bacterium.
In our previous studies, we demonstrated a direct role for GidA in the regulation of Act production (32). GidA was initially thought to be involved in chromosome replication and cell division; however, recent studies implicated GidA in a number of biological and pathogenic processes, suggesting a global regulatory role for GidA (41). It has been reported that GATC methylation sites are unevenly distributed in the genome of E. coli, and the oriC (chromosome origin of replication) has a GATC-rich region (2). These observations are in agreement with the role of Dam in chromosomal DNA replication. GidA is a FAD (flavin adenine dinucleotide) binding protein and might act as a sensor for the redox state of cells in a manner similar to that of the flavoproteins, e.g., aerotaxis signal transducer in E. coli (41). Importantly, we noted multiple DNA methylation sites within the putative promoter region of the gidA gene.
In a recent study using E. coli gene arrays, it was noted that those genes which were located proximal to the oriC gene were transcribed with enhanced frequency in a Dam-overproducing E. coli strain (22). The position of the gidA gene in A. hydrophila is immediately downstream of the oriC gene, indicating that the expression of the gidA gene could possibly be upregulated in the Dam-overproducing strain. Therefore, it is plausible that Dam might regulate gene expression through a global regulator, GidA. This scenario matches with our data indicating that the expression of the act gene was downregulated in the gidA isogenic mutant but upregulated in the native Dam-overproducing strain of A. hydrophila.
To test this possibility, we overproduced native Dam in a gidA isogenic mutant and demonstrated that increased MTase activity was unable to induce increased production of Act and lactones in the absence of GidA (Fig. 4A to C and 5A). These studies provided the first evidence of an intermediate protein that could be involved in the mechanism for the influence of Dam on bacterial virulence. These studies also suggested that different virulence genes may share a common regulatory mechanism whose activity is dependent on Dam methylation. Examination of the putative promoter region of the act gene indicated two potential GATC sequences which could be methylated by Dam. However, since Act production remained unaltered in the gidA mutant control strain versus that in the gidA mutant that overproduced native Dam, our future studies will focus on the potential role of GATC sequences of gidA in the alteration of virulence factor production by A. hydrophila.
Overproduction of Dam significantly attenuated the virulence of Y. pseudotuberculosis and provided a fully protective immune response in the vaccinated hosts (19). We provided evidence that the Dam-overproducing strain with pBAD/damAhSSU plasmid was avirulent in a mouse model compared to the WT bacterium with pBAD vector alone (6). Dysregulation of Dam activity can disable the ability of a pathogen to cause disease via aberrant virulence gene expression and contribute to heightened immunity in vaccinated hosts through the ectopic production of an expanded repertoire of potential antigens (19). While a concern with this approach is that virulence in Dam-overproducing strains can revert to WT levels by mutation, the insertion of multiple, nontandem copies of Dam-overproducing cassettes into the chromosome should reduce the likelihood of this undesired scenario (19). In this study, we showed that mutated Dam without MTase activity exhibited a virulence phenotype similar to that of the WT A. hydrophila SSU strain in both the in vitro and in vivo models. Further, immunization of mice with the Dam-overproducing strain provided protection to animals challenged with the lethal dose of WT A. hydrophila. Dam represents a possible target for vaccine development because the dam gene is highly conserved among a large majority of pathogenic bacteria and across lower and higher eukaryotes (13, 28). Thus, Dam is a global control factor for gene expression and overproduction of Dam results in altered virulence in bacterial pathogens.
.
Acknowledgments
This work was supported by grants from the NIH/NIAID (AI41611), the American Water Works Association Research Foundation, and the Environmental Protection Agency. A. A. Fadl was supported by a McLaughlin Postdoctoral Fellowship. L. Pillai, a Predoctoral Fellow, obtained funding from the NIH T32 training grant in Emerging and Tropical Infectious Diseases. B. K. Khajanchi, from the International Centre for Diarrheal Diseases Research, Dhaka, Bangladesh, was a recipient of the UNESCO-American Society for Microbiology travel award.
The C. violaceum CV026 strain was provided by J. Graf, University of Connecticut, Storrs, CT. We thank M. J. Susman for editing the manuscript.
Editor: J. T. Barbieri
REFERENCES
- 1.Albert, M. J., M. Ansaruzzaman, K. A. Talukder, A. K. Chopra, I. Kuhn, M. Rahman, A. S. Faruque, M. S. Islam, R. B. Sack, and R. Mollby. 2000. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J. Clin. Microbiol. 38:3785-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barras, F., and M. G. Marinus. 1988. Arrangement of Dam methylation sites (GATC) in the Escherichia coli chromosome. Nucleic Acids Res. 16:9821-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chopra, A. K., and C. W. Houston. 1999. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1:1129-1137. [DOI] [PubMed] [Google Scholar]
- 4.Diekmann, S. 1987. DNA methylation can enhance or induce DNA curvature. EMBO J. 6:4213-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dueger, E. L., J. K. House, D. M. Heithoff, and M. J. Mahan. 2003. Salmonella DNA adenine methylase mutants elicit early and late onset protective immune responses in calves. Vaccine 21:3249-3258. [DOI] [PubMed] [Google Scholar]
- 6.Erova, T. E., L. Pillai, A. A. Fadl, J. Sha, S. Wang, C. L. Galindo, and A. K. Chopra. 2006. DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect. Immun. 74:410-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galindo, C. L., C. Gutierrez, Jr., and A. K. Chopra. 2006. Potential involvement of galectin-3 and SNAP23 in Aeromonas hydrophila cytotoxic enterotoxin-induced host cell apoptosis. Microb. Pathog. 40:56-68. [DOI] [PubMed] [Google Scholar]
- 8.Galindo, C. L., J. Sha, A. A. Fadl, L. Pillai, and A. K. Chopra. 2006. Host immune responses to Aeromonas virulence factors. Curr. Immunol. Rev. 2:13-26. [Google Scholar]
- 9.Guyot, J. B., J. Grassi, U. Hahn, and W. Guschlbauer. 1993. The role of the preserved sequences of Dam methylase. Nucleic Acids Res. 21:3183-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanninen, M. L., S. Salmi, L. Mattila, R. Taipalinen, and A. Siitonen. 1995. Association of Aeromonas spp. with travellers' diarrhoea in Finland. J. Med. Microbiol. 42:26-31. [DOI] [PubMed] [Google Scholar]
- 12.Hanzelka, B. L., and E. P. Greenberg. 1996. Quorum sensing in Vibrio fischeri: evidence that S-adenosylmethionine is the amino acid substrate for autoinducer synthesis. J. Bacteriol. 178:5291-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hattman, S. 2005. DNA-[adenine] methylation in lower eukaryotes. Biochemistry (Moscow) 70:550-558. [DOI] [PubMed] [Google Scholar]
- 14.Hattman, S., J. Wilkinson, D. Swinton, S. Schlagman, P. M. Macdonald, and G. Mosig. 1985. Common evolutionary origin of the phage T4 dam and host Escherichia coli dam DNA-adenine methyltransferase genes. J. Bacteriol. 164:932-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henke, J. M., and B. L. Bassler. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186:3794-3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernday, A., M. Krabbe, B. Braaten, and D. Low. 2002. Self-perpetuating epigenetic pili switches in bacteria. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16470-16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogardt, M., M. Roeder, A. M. Schreff, L. Eberl, and J. Heesemann. 2004. Expression of Pseudomonas aeruginosa exoS is controlled by quorum sensing and RpoS. Microbiology 150:843-851. [DOI] [PubMed] [Google Scholar]
- 18.Horton, J. R., K. Liebert, M. Bekes, A. Jeltsch, and X. Cheng. 2006. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J. Mol. Biol. 358:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julio, S. M., D. M. Heithoff, D. Provenzano, K. E. Klose, R. L. Sinsheimer, D. A. Low, and M. J. Mahan. 2001. DNA adenine methylase is essential for viability and plays a role in the pathogenesis of Yersinia pseudotuberculosis and Vibrio cholerae. Infect. Immun. 69:7610-7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong, H., and C. L. Smith. 1997. Substrate DNA and cofactor regulate the activities of a multi-functional restriction-modification enzyme, BcgI. Nucleic Acids Res. 25:3687-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kossykh, V. G., S. L. Schlagman, and S. Hattman. 1993. Conserved sequence motif DPPY in region IV of the phage T4 Dam DNA-[N6-adenine]-methyltransferase is important for S-adenosyl-L-methionine binding. Nucleic Acids Res. 21:4659-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lobner-Olesen, A., M. G. Marinus, and F. G. Hansen. 2003. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc. Natl. Acad. Sci. USA 100:4672-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Low, D. A., N. J. Weyand, and M. J. Mahan. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69:7197-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malone, T., R. M. Blumenthal, and X. Cheng. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253:618-632. [DOI] [PubMed] [Google Scholar]
- 25.Marinus, M. G., and N. R. Morris. 1973. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J. Bacteriol. 114:1143-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McClean, K. H., M. K. Winson, L. Fish, A. Taylor, S. R. Chhabra, M. Camara, M. Daykin, J. H. Lamb, S. Swift, B. W. Bycroft, G. S. Stewart, and P. Williams. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703-3711. [DOI] [PubMed] [Google Scholar]
- 27.Mohler, V. L., D. M. Heithoff, M. J. Mahan, K. H. Walker, M. A. Hornitzky, C. S. McConnell, L. W. Shum, and J. K. House. 2006. Cross-protective immunity in calves conferred by a DNA adenine methylase deficient Salmonella enterica serovar Typhimurium vaccine. Vaccine 24:1339-1345. [DOI] [PubMed] [Google Scholar]
- 28.Robertson, K. D., and A. P. Wolffe. 2000. DNA methylation in health and disease. Nat. Rev. Genet. 1:11-19. [DOI] [PubMed] [Google Scholar]
- 29.Roth, M., S. Helm-Kruse, T. Friedrich, and A. Jeltsch. 1998. Functional roles of conserved amino acid residues in DNA methyltransferases investigated by site-directed mutagenesis of the EcoRV adenine-N6-methyltransferase. J. Biol. Chem. 273:17333-17342. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Wiley & Co., Cold Spring Harbor, N.Y.
- 31.Sha, J., E. V. Kozlova, and A. K. Chopra. 2002. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sha, J., E. V. Kozlova, A. A. Fadl, J. P. Olano, C. W. Houston, J. W. Peterson, and A. K. Chopra. 2004. Molecular characterization of a glucose-inhibited division gene, gidA, that regulates cytotoxic enterotoxin of Aeromonas hydrophila. Infect. Immun. 72:1084-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sha, J., M. Lu, and A. K. Chopra. 2001. Regulation of the cytotoxic enterotoxin gene in Aeromonas hydrophila: characterization of an iron uptake regulator. Infect. Immun. 69:6370-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sha, J., L. Pillai, A. A. Fadl, C. L. Galindo, T. E. Erova, and A. K. Chopra. 2005. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect. Immun. 73:6446-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swift, S., A. V. Karlyshev, L. Fish, E. L. Durant, M. K. Winson, S. R. Chhabra, P. Williams, S. Macintyre, and G. S. Stewart. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179:5271-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor, V. L., R. W. Titball, and P. C. Oyston. 2005. Oral immunization with a dam mutant of Yersinia pseudotuberculosis protects against plague. Microbiology 151:1919-1926. [DOI] [PubMed] [Google Scholar]
- 38.Urieli-Shoval, S., Y. Gruenbaum, and A. Razin. 1983. Sequence and substrate specificity of isolated DNA methylases from Escherichia coli C. J. Bacteriol. 153:274-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Val, D. L., and J. E. Cronan, Jr. 1998. In vivo evidence that S-adenosylmethionine and fatty acid synthesis intermediates are the substrates for the LuxI family of autoinducer synthases. J. Bacteriol. 180:2644-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Woude, M., W. B. Hale, and D. A. Low. 1998. Formation of DNA methylation patterns: nonmethylated GATC sequences in gut and pap operons. J. Bacteriol. 180:5913-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White, D. J., R. Merod, B. Thomasson, and P. L. Hartzell. 2001. GidA is an FAD-binding protein involved in development of Myxococcus xanthus. Mol. Microbiol. 42:503-517. [DOI] [PubMed] [Google Scholar]
- 42.Willcock, D. F., D. T. Dryden, and N. E. Murray. 1994. A mutational analysis of the two motifs common to adenine methyltransferases. EMBO J. 13:3902-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, Z., J. R. Horton, L. Zhou, X. J. Zhang, A. Dong, X. Zhang, S. L. Schlagman, V. Kossykh, S. Hattman, and X. Cheng. 2003. Structure of the bacteriophage T4 DNA adenine methyltransferase. Nat. Struct. Biol. 10:849-855. [DOI] [PMC free article] [PubMed] [Google Scholar]