Abstract
Leptospirosis is an important zoonosis of worldwide distribution. Humans become infected via exposure to pathogenic Leptospira spp. from infected animals or contaminated water or soil. The availability of genome sequences for Leptospira interrogans, serovars Lai and Copenhageni, has opened up opportunities to examine global transcription profiles using microarray technology. Temperature is a key environmental factor known to affect leptospiral protein expression. Leptospira spp. can grow in artificial media at a range of temperatures reflecting conditions found in the environment and the mammalian host. Therefore, transcriptional changes were compared between cultures grown at 20°C, 30°C, 37°C, and 39°C to represent ambient temperatures in the environment, growth under laboratory conditions, and temperatures in healthy and febrile hosts. Data from direct pairwise comparisons of the four temperatures were consolidated to examine transcriptional changes at two generalized biological conditions representing mammalian physiological temperatures (37°C and 39°C) versus environmental temperatures (20°C and 30°C). Additionally, cultures grown at 30°C then shifted overnight to 37°C were compared with those grown long-term at 30°C and 37°C to identify genes potentially expressed in the early stages of infection. Comparison of data sets from physiological versus environmental experiments with upshift experiments provided novel insights into possible transcriptional changes at different stages of infection. Changes included differential expression of chemotaxis and motility genes, signal transduction systems, and genes encoding proteins involved in alteration of the outer membrane. These findings indicate that temperature is an important factor regulating expression of proteins that facilitate invasion and establishment of disease.
Leptospirosis is a zoonosis of global distribution caused by pathogenic spirochetes belonging to the genus Leptospira. Leptospires have been classified into more than 16 species based on DNA hybridization studies and more than 230 serovars based on their agglutinating antigens (6, 18). Leptospiral genetic and antigenic diversity reflects the broad range of mammalian species that have been found to serve as maintenance hosts (3). Maintenance hosts are carrier animals that harbor leptospires in their proximal renal tubules, in some cases, for the life of the animal. Transmission to new hosts involves either direct or indirect exposure to organisms shed in the urine of infected animals. The paradigm for direct transmission is Leptospira borgpetersenii serovar Hardjo infection of cattle. Accidental infection of humans by serovar Hardjo almost always involves a history of contact with cattle. In contrast, transmission of Leptospira interrogans serovar Copenhageni or Leptospira interrogans serovar Lai from rats to humans is usually indirect. L. interrogans is known to survive for considerable periods of time in contaminated soil or water. Invasion of the mammalian host involves an initial shift from ambient temperatures to 37°C and later to as high as 39°C during the febrile stage of infection. As in other pathogenic bacteria whose life cycle involves indirect modes of transmission, temperature is likely to be a key signal to leptospires of the transition from a free-living to an invasive stage. Although growth in laboratory media occurs optimally at 30°C, it has been unclear whether the genes expressed under in vitro conditions reflect the organism's host-associated form. For these reasons, an understanding of temperature-regulated genes is likely to provide fundamental insights into leptospiral pathogenesis.
There is some limited information about how Leptospira responds at the transcriptional or translational level to different temperatures, but no studies at a global level have been conducted. Leptospires respond to temperature upshift by the increased synthesis of standard heat shock proteins, such as GroEL, DnaK, and Hsp15 (4, 5, 25, 38). The peripheral membrane protein P31LipL45, also known as Qlp42, was shown in one study to be up-regulated by a temperature shift from 30°C to 37°C (25) and, in another study, to be up-regulated by transition from the log phase of growth to the stationary phase (23). The expression of LipL36, an outer membrane lipoprotein which is known to be down-regulated in vivo and in stationary phase (11), was also shown to be reduced at 37°C (9, 27) and under low iron concentrations (9). Other undefined outer membrane proteins have also been shown to be temperature regulated (9). On the other hand, the expression of the outer membrane lipoproteins LipL21, LipL41, LipL48, and others was unaffected by temperature (9, 27). Interestingly, the most abundant leptospiral surface protein, LipL32 (10), undergoes processing into a range of mass and pI isoforms, with the processing varying at different temperatures (9, 47). In contrast, the amount of leptospiral lipopolysaccharide produced was shown to be diminished on bacteria recovered from acutely infected guinea pig liver but not on leptospires chronically colonizing rat renal tubules (26). The Lk73.5 sphingomyelinase was also detected in leptospires grown in vivo but not in vitro (2). The environmental cue(s) for these changes are unknown.
Global analysis of gene expression on pathogenic bacteria has yielded important information on genes required for host adaptation and potential virulence-associated genes. For example, microarray analysis of the tick-borne Lyme disease spirochete Borrelia burgdorferi at 23°C and 37°C to simulate the environment encountered in unfed and fed ticks confirmed the previously known down- and up-regulation paradigm of OspA and OspC, respectively (33, 45), and was therefore able to identify a range of genes responsive to the fed tick environment and subsequently to conditions found in the mammalian host environment, although differences identified in the two studies do raise possible limitations of array studies. The majority of differentially expressed genes were those involved in cell envelope biosynthesis, energy metabolism, motility and chemotaxis, and protein synthesis and those encoding transport and binding proteins. Many genes of unknown function were both up- and down-regulated. Interestingly, the change in expression of many of these genes was transitory and returned to base levels after adaptation to the mammalian host (33). The identification of a subset of differentially expressed genes generated a priority subset for characterization with respect to their roles in pathogenesis (7). In the swine pathogen Haemophilus parasuis, microarray analysis under a range of conditions, including temperature stress, identified a total of 75 differentially regulated genes, which encoded iron and sugar transporters, metabolic enzymes, and proteins of unknown function (24). Likewise, temperature shift of Mycoplasma hyopneumoniae from 37°C to 42°C to mimic conditions encountered during febrile infection identified a range of known heat shock proteins as well as 54 genes encoding proteins with unknown function (20).
The availability of the two highly related genome sequences for serovars Lai and Copenhageni of L. interrogans (29, 32) allows, for the first time, the possibility of examining global changes in gene expression of Leptospira growing at different temperatures and also the comparison of profiles following temperature upshift with those after long-term growth.
MATERIALS AND METHODS
Culture conditions.
L. interrogans serovar Lai was grown in EMJH medium (14) at 20°C, 30°C, 37°C, and 39°C. Cultures underwent at least three passages at each of the different temperatures before being harvested for RNA isolation. For the overnight upshift to 37°C, cultures were grown to 2.5 × 108/ml at 30°C and then incubated at 37°C for 16 to 20 h before harvesting.
Microarray construction.
A revised annotation of the L. interrogans serovar Lai strain 56601 genome (which includes 3,626 coding sequences, 36 tRNA genes, and 4 rRNA genes) (http://vbc.med.monash.edu.au/genomes/) was used to design oligonucleotides of 70 bases in length using the program ArrayOligoSelector (http://arrayoligosel.sourceforge.net/). The program first calculates scores of uniqueness, sequence complexity, lack of self-binding, and GC content for every candidate oligonucleotide. The second part of the program then uses scores generated by the first program to select the optimal oligonucleotide for each gene. Since the program has a bias for the 3′ end of genes, genes were trimmed to a maximum of 750 bases, while 50 of the longest open reading frames (ORFs) and pseudogenes were represented by a 5′ and 3′ oligonucleotide. L. interrogans serovar Lai possesses 6 insertion sequences with multiple copies; each unique transposase was represented by one oligonucleotide on the microarray.
The microarrays were printed by the Australian Genomics Research Facility onto Nexterion Slide A+ substrates (Schott, Louisville, KY). Each oligonucleotide was printed in pairs in 16 subarrays, each with a 22-column by 24-row configuration. The 16 subarrays were printed in duplicate so that each oligonucleotide was present in quadruplicate.
Experimental design.
Three independent RNA samples (biological replicates) from L. interrogans serovar Lai grown at 20°C, 30°C, 37°C, and 39°C were cross-compared in a loop design (15) (Fig. 1). Three independent RNA samples were also obtained from the 37°C upshift cultures which were paired with the three 30°C and 37°C RNA samples (Fig. 1).
FIG. 1.
Experimental design for comparison of transcriptional differences at each of the four temperatures: 20°C, 30°C, 37°C, and 39°C. Each line represents a direct hybridization between the two samples, with the arrow going from the Cy3 to the Cy5 channel for biological replicate sets 1 and 3. The opposite orientation of Cy3 to Cy5 was used for biological replicate set 2. For the comparison of 30°C and 37°C samples with the overnight 37°C upshift samples, two hybridizations (dye swaps) were performed for each of the three biological replicates.
RNA purification.
Leptospires were grown to a density of 2.5 × 108 to 7.5 × 108/ml before harvesting for RNA purification. Cell count was determined as described previously (1). Cultures were treated with 1/10 volume ice-cold killing buffer (50 mM Tris-HCl, pH 7.5, 15 mg/ml sodium azide, 0.6 mg/ml chloramphenicol) and chilled on ice for 5 min before centrifugation at 9,000 × g for 10 min. RNA was isolated from bacteria by using Trizol reagent (Invitrogen, Carlsbad, CA) as described by the manufacturer and then treated with DNase (20 U for 30 min at 37°C). RNA was further purified using RNeasy minicolumns (QIAGEN, Inc., Valencia, CA) with on-column DNase treatment according to the manufacturer's protocol. Samples were quantified and checked for purity using an ND-1000 spectrophotometer (Nanodrop, Wilmington, DE) and agarose gel electrophoresis. To confirm lack of contamination with genomic DNA, 0.5 μg of RNA was used as a template in a PCR using primers for LA1647 (undecaprenyl-galactosyl transferase), BAP3315 (CACCATGAAAATTACTTACCTT), and BAP3316 (TCGTGCTCCAAAACCGGT).
Preparation of probes for microarray hybridizations.
Labeled cDNA probes were synthesized from 2.5 μg of total RNA using random primers contained in the 3DNA Array 900MPX expression array detection kit (Genisphere, Hatfield, PA) according to the instructions of the manufacturer. The cDNA hybridization mix was made using the 2× formamide-based hybridization buffer (vial 7) from the 3DNA Array 900MPX expression array detection kit (Genisphere) to a final volume of 50 μl.
Microarray hybridizations.
The microarray was prehybridized using a solution containing 25% formamide, 5× SSC (1× SSC consists of 0.15 M NaCl with 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate, 10 mg/ml bovine serum albumin, and 1 mg/ml sheared herring sperm DNA. The solution was placed on the microarray under a coverslip, and incubation was carried out for 45 min in a slide hybridization chamber (Corning, Inc., Big Flats, NY) placed in a 50°C water bath, after which the slides were washed in ultrapure H2O and dried by centrifugation at 1,000 × g for 3 min. Hybridization with the cDNA hybridization mix, 3DNA hybridization mix, and washes were carried out as per the 3DNA Array 900MPX expression array detection kit (Genisphere) protocol. The cDNA hybridization mix and 3DNA hybridization mix were both made up using 2× formamide-based hybridization buffer (vial 7) to a final volume of 50 μl and applied onto the microarray under a 22- by 40-mm LifterSlip (Erie Scientific Company, Portsmouth, NH). The cDNA hybridization was carried out at 50°C overnight, while the 3DNA hybridization was done at 50°C for 4 h. After the final set of washes, slides were dried by centrifugation at 1,000 × g for 3 min and scanned immediately.
Analysis of microarray images.
Microarray hybridizations were scanned using a GMS 418 array scanner (Genetic MicroSystems, Inc., Woburn, MA). The Cy3 and Cy5 images were aligned then overlaid with a grid using ImaGene version 5.1 (Biodiscovery, El Segundo, CA) to allow accurate gene identification and quantification of fluorescence intensity. Spots were examined manually, and poor spots were flagged for elimination from the analysis.
The four temperature conditions were examined using all six pairwise comparisons. Each comparison had three biological replicates, one being a dye-swap, resulting in 18 arrays (Fig. 1). Each array was background corrected using the “normexp” method from Limma (36), which provides smooth transformations of the intensities and results in only positive values. Each array was normalized independently using print-tip loess normalization (46). The log ratios from the arrays were then scaled such that all arrays had the same median absolute deviation. The Limma package was then used to fit a linear model to each gene, taking into account the correlation between replicate spots (37). For each gene, the coefficients from the linear model were then used to calculate the relative change for all six pairwise comparisons. The moderated t statistics and associated P values were calculated using empirical Bayes (36). The P values were then adjusted for multiple testing to control the false-discovery rate. Genes with an absolute relative ratio of greater than 1.5, and significant at a P value of <0.05, were selected as differentially expressed.
Further analysis was performed to compare the two lower-temperature conditions to the two higher-temperature conditions to represent environmental and physiological conditions, respectively. This was an analysis of a pool of the 20°C and 30°C data against the 37°C and 39°C data. The normalization and fitting of linear models were as described above. A contrast was then computed to pool the appropriate conditions. Again, the moderated t statistics were computed, and genes were selected as differentially expressed if they had a greater than 1.5-fold change and an adjusted P value of <0.05.
Raw data from the direct comparisons between 37°C upshift versus 30°C or 37°C long-term passaging were analyzed using the web-based program BioArray Software Environment (34). Raw data from replicate arrays (6 for each comparison) were combined and used for further analyses. Spot-specific median signals were corrected for local background by subtracting spot-specific median background intensities. Spots with very low intensities (<250) were omitted, and the data were normalized using the global median ratio, which scales the intensities such that the median of the ratio between the Cy3 and Cy5 channels is 1 while omitting spots within 5% of the highest or lowest intensities. Each array was then normalized independently using loess per print tip group, followed by normalization between replicate arrays such that they all had the same median absolute deviation. As before, genes were considered to be differentially expressed if they were at least 1.5-fold up- or down-regulated with a P value of <0.05 as determined by the Student t test.
Validation of microarray data by real-time RT-PCR.
Primer Express software (ABI, Foster City, CA) was used to design primers for real-time reverse transcription (RT)-PCR. RT reaction mixtures contained 2 to 10 μg of total RNA, 30 μg of random hexamers, 10 U of Superscript II (Invitrogen) reverse transcriptase, and 500 μM concentrations (each) of dATP, dCTP, dGTP, and dTTP. Samples were incubated at 42°C for 2.5 h, followed by 15 min at 70°C. The synthesized cDNA was then diluted 1/80 prior to use in real-time PCR. Reactions were done in triplicate. Each 25-μl reaction mixture contained 2.5 μl of cDNA, 50 nM concentrations of each gene-specific primer (Table 1), and 12.5 μl of SYBR green PCR master mix (ABI). Real-time PCR was carried out using an ABI PRISM model 7700 sequence detection system. Known concentrations of L. interrogans serovar Lai genomic DNA were used to construct a gene-specific standard curve so that the concentration of template in each reaction mixture could be determined. The gene encoding flagella subunit B, flaB, was used as the normalizer for all reactions. Melting curve analysis confirmed that all PCRs amplified a single product.
TABLE 1.
Oligonucleotides used for real-time quantitative RT-PCR
| Gene or ORF | Forward primer | Reverse primer |
|---|---|---|
| flaB (LA2017) | GAGAGAAACACCGAAGACGG | TGAATAGCAAGAACCCGGAT |
| LA1402 | ACAGTGGCTACCCCTGGAAA | CCCCAACCTGAATTCCAAGA |
| LA2824 | CACGGACGAGTTGGTTAGGG | CAATTGCCCCCACCATAATC |
| ligB (LA3778) | GAATATTACGGATTCGACATACATCG | CCTTGGATGGTTACAACGGATT |
| LA0594 | TTCGCGCTCTTAGAAATGGG | GCGGCAGACGTTCCTAAAAC |
| LA0980 | TGAGAAATTCACCCGTTCCAA | TCCTTTTACTTTTTCTAGGGCTTGAT |
| LA2013 | CCGACCTATATGGTCTGGGAAAGTAG | TCAATCCGCGCATCCGTCTTC |
RESULTS AND DISCUSSION
Experimental design.
Currently, little is known about leptospiral gene transcription patterns during growth under different conditions. Leptospira spp. are able to grow under a range of temperatures, for example, at ambient temperatures in water or soil, and they are routinely cultured in laboratories at 30°C. However, most mammalian species have a body core temperature of 37°C which, during fever, can reach 39°C or higher. Therefore, we have investigated gene transcription profiles of L. interrogans grown at environmental temperatures (20°C and 30°C) and at mammalian physiological temperatures (37°C and 39°C).
Gene expression of L. interrogans grown at each of the temperatures was compared in a loop design (Fig. 1) so that differences between two different temperatures could be compared directly as well as indirectly via another sample in the loop. For example, gene expression at 20°C can be compared directly to expression at 30°C, but the two conditions can also be indirectly compared via the 37°C and 39°C samples. This method of analysis is more powerful than simply using the three biological replicates for any two given conditions, since it allows the use of the other arrays as if they were against a common reference. The loop design has been shown to be more efficient than the reference design where multiple samples are compared using a common reference sample (43).
Genes that were at least 1.5-fold up- or down-regulated at a confidence level of 95% were considered to be differentially expressed. Other studies have found that a 1.5-fold change in gene expression can be biologically relevant (13, 35). The number of genes that were differentially expressed in each pairwise comparison is shown in Table 2. The greater the differences between temperatures, the more genes were found to be differentially expressed.
TABLE 2.
Number of genes differentially expressed in L. interrogans grown at the first temperature compared to the second temperature
| Comparison | No. of genes
|
||
|---|---|---|---|
| Down-regulated | Up-regulated | Total | |
| 20°C vs 30°C | 56 | 54 | 110 |
| 20°C vs 37°C | 197 | 169 | 366 |
| 20°C vs 39°C | 218 | 173 | 391 |
| 30°C vs 37°C | 123 | 73 | 196 |
| 30°C vs 39°C | 147 | 103 | 250 |
| 37°C vs 39°C | 13 | 12 | 25 |
To determine if any groups or types of genes were overrepresented, the differentially expressed genes were sorted into functional categories based on clusters of orthologous groups (COG) (40). A summary of up- and down-regulated genes sorted by general COGs is shown in Fig. 2. Numbers of up- or down-regulated genes in each broad category is presented as a proportion of the total number of differentially expressed genes. In the pairwise comparisons, the main interest is that, in both the up- and down-regulated gene sets, the majority of genes differentially expressed are in the poorly characterized category. Additionally, in the proportion of down-regulated genes with known or predicted function (Fig. 2B), genes involved in cellular processes and signaling are the main group which are down-regulated at the lower temperatures relative to the higher temperatures, perhaps due to a slower rate of growth at the lower temperatures. The number of genes up- or down-regulated in each COG functional category for each of the comparisons is shown in Table S1 in the supplemental material.
FIG. 2.
Proportion of up-regulated (A) and down-regulated (B) genes in each general COG grouping expressed as a percentage of the total number of differentially expressed genes in each comparison. The percentage of genes up- or down-regulated in the first temperature compared to the second in three biological replicates is shown for each general COG grouping.
Genes differentially expressed at physiological temperatures compared to environmental temperatures.
Data from the four temperatures were pooled to examine two generalized biological conditions representing mammalian physiological temperatures (37°C and 39°C) versus environmental temperatures (20°C and 30°C) (Table 3). Consolidating the data in this manner also has the effect of filtering genes which are consistently differentially expressed in the two basic conditions. Given that 237 genes were significantly differentially expressed in the 37°C plus 39°C versus 20°C plus 30°C comparison but only 12 and 16 genes were significantly differentially expressed in the other pooled comparisons, we concluded that the main difference in leptospiral gene expression at different temperatures is indeed between physiological versus environmental temperatures, while there is less difference between the two environmental temperatures or between the two physiological temperatures. This is evident in Table 2, where the lowest numbers of differentially expressed genes are in the 20°C versus 30°C and 37°C versus 39°C comparisons.
TABLE 3.
Number of genes differentially expressed in L. interrogans in the first temperature pool compared to the second temperature pool
| Comparison | No. of genes
|
||
|---|---|---|---|
| Down-regulated | Up-regulated | Total | |
| 37°C + 39°C vs 20°C + 30°C | 90 | 147 | 237 |
| 20°C + 37°C vs 30°C + 39°C | 8 | 4 | 12 |
| 20°C + 39°C vs 30°C + 37°C | 11 | 5 | 16 |
Genes that were found to be significantly up- or down-regulated sorted by COG functional category are shown in Tables 4 and 5, respectively. Notably, and consistent with the high proportion of uncharacterized genes in Leptospira (50.9% of coding sequences in the L. interrogans serovar Lai genome), the majority of differentially expressed genes encoded proteins with hypothetical or poorly characterized function (52% of down-regulated and 61% of up-regulated genes). Therefore, for brevity and clarity, ORFs encoding poorly characterized or hypothetical proteins were omitted from Tables 4 and 5. The full list of differentially expressed genes is shown in Table S2 in the supplemental material. Differentially expressed genes in each broad functional category are discussed below.
TABLE 4.
L. interrogans genes of known or predicted function which were up-regulated at physiological temperatures compared with environmental temperatures in three sets of biological replicates
| Function and ORFa | Mean fold up-regulation | Gene | COG categoryb | Description of gene product |
|---|---|---|---|---|
| Information storage and processing | ||||
| LA1970 | 1.5 | J | poly(A) polymerase family of proteins | |
| LA0876 | 1.5 | rpoE | K | RNA polymerase sigma subunit |
| LA1282 | 2 | K | Transcriptional regulator, AcrR family | |
| LA1816 | 1.5 | K | Transcriptional regulator | |
| LA2728 | 1.9 | K | Transcriptional regulator | |
| LA3652 | 2 | K | RNA polymerase sigma subunit | |
| LA4298 | 1.8 | K | Transcriptional regulator, MarR family | |
| LB104 | 1.7 | K | AcrR family transcriptional regulator | |
| LA0984 | 1.9 | L | ATP-dependent DNA ligase | |
| LA1456 | 2 | radC | L | DNA repair protein |
| Cellular processes and signaling | ||||
| LA0145 | 1.7 | D | ATPase of the PP-loop superfamily | |
| LA1837 | 1.7 | D | ParA-related protein | |
| LA1124 | 1.5 | lnt | M | Apolipoprotein N-acyltransferase |
| LA1283 | 1.9 | M | Conserved hypothetical protein | |
| LA1284 | 1.8 | lolE | M | Lipoprotein releasing system, LolE permease component |
| LA1504 | 2 | M | Metallopeptidase | |
| LA3489 | 1.6 | flgC | N | Endoflagellar proximal basal body rod protein |
| LA1231 | 2.1 | O | Heat shock protein HtpG | |
| LA1563 | 2.4 | O | Small heat shock protein (molecular chaperone) | |
| LA1564 | 1.7 | O | Small heat shock protein (molecular chaperone) | |
| LA1879 | 3.5 | clpA | O | Endopeptidase Clp, ATP-dependent proteolytic subunit |
| LA4299 | 1.8 | btuE | O | Glutathione peroxidase |
| LB174 | 1.6 | htpX | O | Zn-dependent protease with chaperone function |
| LA0816 | 1.7 | T | Receiver component of a two-component response regulator | |
| LA1212 | 1.9 | uspA | T | Universal stress protein UspA |
| LA1743 | 3.1 | T | Chemotaxis protein, methyltransferase | |
| LA1744 | 1.8 | T | Chemotaxis protein, methylesterase | |
| LA1746 | 1.7 | T | Sensor protein of a two-component response regulator, regulator protein of a two-component response regulator | |
| LA1860 | 1.6 | T | Sensor protein of a two-component response regulator | |
| LA2117 | 1.6 | T | Anti-sigma factor antagonist | |
| LA2223 | 1.7 | T | Sensor histidine kinase of a two-component response regulator | |
| LA3657 | 1.6 | T | Receiver component of a response regulator | |
| LA3858 | 1.6 | T | Sensor histidine kinase of a two-component complex, part; response regulator of a two-component complex, part | |
| LA0802 | 2 | U | Pilus assembly protein | |
| LA3754 | 1.5 | lepB | U | Signal peptidase I |
| LA0150 | 2.1 | V | Permease of an ABC transporter complex, ATP-binding protein of an ABC transporter complex | |
| LA1285 | 1.9 | V | Lipoprotein releasing system, LolD ATPase component | |
| Metabolism | ||||
| LA2453 | 1.6 | C | 4Fe-4S binding protein | |
| LA4164 | 1.5 | E | Zinc metalloprotease (elastase) | |
| LA1372 | 1.5 | G | Permease | |
| LA1457 | 1.6 | G | Membrane protein of an ABC transporter complex | |
| LA2127 | 1.6 | G | Epimerase | |
| LB102 | 2.3 | G | Sugar phosphatase | |
| LB105 | 1.5 | ubiG | H | Transcriptional regulator, ArsR family |
| LA0502 | 1.5 | desA | I | Fatty acid desaturase |
| LA2958 | 2 | I | Hydrolase, acyltransferase | |
| LA3928 | 1.5 | caiA | I | Acyl-CoA dehydrogenase |
| LA0593 | 2.4 | copZ | P | Copper chaperone |
| LA0594 | 3.1 | P | Cation transport ATPase, possibly copper | |
| LA1859 | 2.2 | katE | P | Catalase |
| LA3110 | 1.6 | kdpC | P | Potassium-transporting ATPase C chain |
| LA3242 | 2 | P | TonB-dependent receptor | |
| LA3972 | 1.5 | trkA | P | Potassium uptake system, NAD-binding component |
| LA3712 | 1.9 | Q | Permease component of an ABC transporter complex |
Ninety-three ORFs coding for hypothetical proteins (those grouped into COG R, S, or − or which are conserved or unique hypothetical proteins) were omitted from this table. For a complete listing, see Table S2 in the supplemental material.
COG categories are as described for Fig. 3.
TABLE 5.
L. interrogans genes of known or predicted function which were down-regulated at physiological temperatures compared with environmental temperatures in three sets of biological replicates
| Function and ORFa | Mean fold down-regulation | Gene | COG categoryb | Description of gene product |
|---|---|---|---|---|
| Information storage and processing | ||||
| LA0787 | −1.6 | J | rRNA methylase | |
| LA2101 | −1.5 | K | RNA polymerase sigma subunit | |
| LA0281 | −1.9 | alkA | L | DNA-3-methyladenine glycosylase II |
| LA2347 | −1.7 | xerC | L | Tyrosine site-specific recombinase XerC |
| LA4049 | −3.7 | srmB | L | ATP-dependent RNA helicase (superfamily II) |
| Cellular processes and signaling | ||||
| LA3011 | −2.1 | D | Cell division protein with ATPase domain | |
| LA0791 | −1.5 | M | Transglycosylase | |
| LA2058 | −1.6 | ddlA | M | d-alanine-d-alanine ligase |
| LA2200 | −2 | M | Amidase | |
| LA3615 | −1.5 | M | OmpA family protein | |
| LA4078 | −1.5 | lnt | M | Apolipoprotein N-acyltransferase |
| LB323 | −1.7 | M | Transglycosylase | |
| LA2666 | −1.7 | flgA | N | Endoflagellar basal body P-ring biosynthesis protein |
| LA3778 | −1.6 | ligB | N | LigB lipoprotein |
| LA4309 | −1.6 | flgK | N | Endoflagellar hook junction protein |
| LA0036 | −1.5 | O | HSP33 family chaperone | |
| LA2278 | −1.5 | O | DnaJ-related molecular chaperone | |
| LA2949 | −1.8 | O | Trypsin-like serine protease | |
| LA0049 | −1.8 | T | Methyl-accepting chemotaxis protein | |
| LA0565 | −1.5 | T | Adenylate cyclase | |
| LA1681 | −1.5 | T | Phosphate starvation-inducible protein | |
| LA1919 | −1.6 | T | DNA-binding transcriptional activator, SARP family | |
| LA2246 | −1.5 | T | Methyl-accepting chemotaxis protein | |
| LB139 | −1.7 | T | Regulator of sigma subunit | |
| LA2014 | −1.6 | V | CreD-like protein | |
| LA3871 | −1.6 | V | Cation/multidrug efflux pump | |
| LA4287 | −1.6 | V | ATP-binding protein of an ABC transporter complex | |
| Metabolism | ||||
| LB165 | −3.3 | C | Ferredoxin related-protein | |
| LA0693 | −1.6 | lysC | E | Aspartate kinase |
| LA4200 | −1.7 | E | Amine oxidase (flavin containing) | |
| LB123 | −1.6 | aroK | E | Shikimate kinase |
| LA0786 | −1.6 | G | Glycosyltransferase | |
| LA0970 | −1.7 | G | Permease component of an ABC transporter complex | |
| LA1392 | −1.6 | G | Sugar kinase | |
| LB152 | −1.6 | cobU | H | Adenosylcobinamide kinase, adenosylcobinamide-phosphate guanylyltransferase |
| LB157 | −1.8 | cobJ | H | Precorrin-3B C(17)-methyltransferase |
| LB159 | −1.6 | cobF | H | Precorrin-6A synthase (deacetylating) |
| LB160 | −1.6 | cobL | H | Precorrin-6Y C5,15-methyltransferase (decarboxylating) |
| LB161 | −2.2 | cobH | H | Precorrin-8X methylmutase or isomerase |
| LA0438 | −1.7 | I | Acyl-CoA dehydrogenase | |
| LA4245 | −1.8 | P | Monooxygenase | |
| LA4246 | −1.9 | phoD | P | Phosphodiesterase I |
Forty-eight ORFs coding for hypothetical proteins (those grouped into COG R, S, or − or which are conserved or unique hypothetical proteins) were omitted from this table. For a complete listing, see Table S2 in the supplemental material.
COG categories are as described for Fig. 3.
Information storage and processing.
Several genes involved in translation, transcription, and DNA replication, recombination, and repair were differentially expressed. Of particular interest was the increased expression of several genes encoding transcriptional regulators, including two with similarity to the AcrR family of transcriptional regulators and another with similarity to the MarR family. Proteins in both families are transcriptional repressors which, in Escherichia coli, respond to stress conditions and are part of a global regulatory pathway involved in antibiotic resistance (19, 39). AcrR represses acrA and acrB, which encode membrane-associated polypeptide components of a multidrug efflux pump (19). The up-regulation of similar transcriptional factors in L. interrogans at physiological temperatures may thus play a role in modulating the expression of membrane-associated proteins.
Two ORFs (LA0872 and LA3652) encoding the alternative sigma subunit σE were found to be 1.5- and 2-fold up-regulated, respectively. Sigma factors are directly involved in transcription of sets of genes, and their expression is induced in response to stress. In E. coli, the rpoE gene was found to be essential for bacterial growth at high temperatures (12), while in Salmonella enterica serovar Typhimurium, σE facilitates survival under nutritional deprivation and oxidative stress and is strongly induced upon reaching stationary phase (41). In other bacteria, σE has been found to play a role in virulence. For example, σE mutants of Vibrio cholerae were highly attenuated, with a marked decrease in their ability to colonize the intestine (16), while nontypeable Haemophilus influenzae requires σE expression to persist in mammalian cells and survive in macrophages (8). Since L. interrogans rpoE was up-regulated at physiological temperatures relative to environmental temperatures, it is possible that leptospiral σE may regulate genes required for pathogenesis and/or virulence.
Cellular processes and signaling.
Export of membrane proteins is mediated by the Sec pathway in conjunction with signal peptidase I (LepB), the lipoprotein synthesis pathway (Lgt, LspA, and Lnt), and proteins involved in transport and incorporation of lipoproteins into the outer membrane (LolA, LolC, and LolD). At physiological versus environmental temperatures, lnt (LA1124), lolD (LA1285), lolE (LA1284), and lepB were up-regulated, while another homolog of lnt (LA4078) was down-regulated. Genes encoding LolA (LA0410 and LA1136) and other copies of LolD (LA0274 and LA2982) and LolE (LA0273 and LA2983) were not significantly differentially expressed. It is likely that bacteria would need to express a different repertoire of outer membrane proteins to adapt to different conditions. Differences in expression of lnt, lolD, and lolE homologs suggest variation in the spectrum of lipoproteins expressed under the different temperature conditions. The lipoproteins LipL36 and Qlp42 were previously shown to be differentially expressed at the protein level at different temperatures (9, 11, 25, 27). However, these lipoproteins were not found to be differentially expressed in our experiments, suggesting regulation at the translational level.
In B. burgdorferi, changes in morphology, motility, and growth rate were observed in cultures grown at 35°C versus 23°C (42). The changes coincided with up-regulation of chemotaxis and sensing regulons as well as proteins involved in protein processing. Pathogens need to be able to sense and respond rapidly to different environmental signals. Therefore, it was unsurprising that many differentially expressed genes with known or predicted function encoded proteins involved in two-component signal transduction systems, chemotaxis, and motility. L. interrogans is a highly invasive pathogen and, therefore, motility is likely to play a major role in the disease process. Motility and chemotaxis are essential for the organism to target and migrate to specific organs and initiate infection. Three genes involved in endoflagellar synthesis or encoding components of endoflagella were differentially expressed in L. interrogans grown at physiological versus environmental temperatures. However, while flgC was up-regulated, flgA and flgK were down-regulated. Since these genes are found on different parts of the genome and do not constitute an operon, it is conceivable that different endoflagellar-related genes would undergo different mechanisms of regulation to modulate flagellar structure and/or rotary speed in response to environmental cues.
Several genes encoding chaperones or proteins involved in the stress response were differentially expressed, such as clpA, htpG, and htpX. The uspA gene, which is commonly found across several bacterial species to be up-regulated under stress conditions (17), was also 1.9-fold up-regulated at physiological temperatures. However, genes encoding proteins commonly expressed in response to heat shock, such as GroEL and DnaK, were not found to be differentially expressed in our experiments despite being shown to be up-regulated at the protein level (5, 38). In B. burgdorferi, genes encoding typical heat shock proteins, such as GroEL and DnaK, were also not up-regulated by temperature shift (30). Expression of ClpA, which performs the ATP-dependent chaperone function of DnaK and DnaJ (44), was up-regulated 3.5-fold at physiological temperatures, while a DnaJ-related molecular chaperone was down-regulated 1.5-fold. Therefore, it appears that in spirochetes which are able to survive in a range of temperatures either in the environment or in a host, transition and subsequent adaptation to higher temperatures does not induce a substantial heat shock or stress response.
Metabolism.
A broad range of metabolic genes was differentially expressed. It is unclear why genes involved in cobalamin synthesis would be expressed at lower levels at physiological versus environmental temperatures, or conversely, expressed more highly at the lower temperatures. Although vitamin B12 is a component of EMJH in vitro growth medium, it appears that metabolic requirements differ at physiological versus environment temperatures, and for reasons unknown, de novo synthesis at lower temperatures is necessary. Genes encoding proteins involved in defense against oxidative stress were also up-regulated at physiological temperatures, namely katE (which encodes a catalase) and btuE (encoding a glutathione peroxidase). Several genes involved in transport of nutrients and ions across the outer membrane were differentially expressed. Those that were up-regulated at physiological temperatures include genes involved in potassium and copper transport, trkA and copZ, respectively, as well as LA3242, which encodes a homolog of a TonB-dependent receptor. Genes involved in amino acid transport and metabolism, lysC and aroK, were down-regulated at physiological versus environmental temperatures. Lipid metabolism and, therefore, lipid composition of the membrane and lipopolysaccharide may also be altered at physiological temperatures due to up-regulation of caiA (acyl coenzyme A [acyl-CoA] dehydrogenase involved in β-oxidation of fatty acids), desA (fatty acid desaturase), and an epimerase homolog. A glycosyltransferase homolog and another acyl-CoA dehydrogenase homolog were down-regulated at physiological temperatures compared to environmental temperatures.
Effects of temperature upshift on gene expression patterns.
To identify genes which are potentially involved in establishment of disease upon entry into the host from the environment, early- to mid-log-phase cultures of L. interrogans were subjected to overnight (16 to 20 h) upshift from 30°C to 37°C. Gene expression levels from these upshifted cultures were compared to those from cultures grown at 30°C long term and also with cultures grown at 37°C long term to identify genes which may be involved in long-term adaptation to the host environment. Table 6 shows the number of genes which were found to be differentially expressed in each upshift comparison. The number of genes that were up- or down-regulated in each COG category for 37°C upshift versus 30°C long term and 37°C upshift versus 37°C long term are shown in Fig. 3 and 4, respectively. For full lists of differentially expressed genes in each upshift comparison, refer to Tables S3 and S4 in the supplemental material. The 37°C upshift versus 30°C long term comparison showed the greatest number of differentially expressed genes of all comparisons in this study, a finding consistent with the notion that many changes in gene expression are indeed required for the immediate and/or short-term transition from environmental temperatures to physiologically relevant temperatures. Again, many of the genes found to be differentially expressed in both of the upshift comparisons are those encoding hypothetical or poorly characterized proteins (53% of those at 37°C upshift versus 30°C long term and 61% at 37°C upshift versus 37°C long term).
TABLE 6.
Number of genes differentially expressed in L. interrogans during 37°C overnight upshift compared to L. interrogans grown at 30°C and 37°C long term
| Comparison | No. of genes
|
||
|---|---|---|---|
| Down-regulated | Up-regulated | Total | |
| 37°C upshift vs 30°C | 208 | 299 | 507 |
| 37°C upshift vs 37°C | 49 | 74 | 123 |
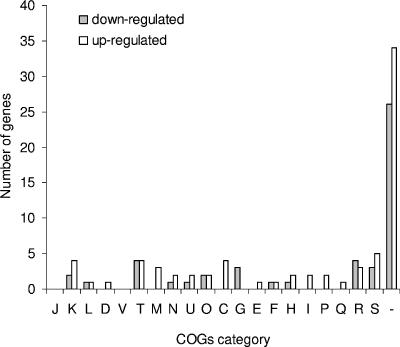
FIG. 3.
Number of differentially expressed genes by COG category during overnight upshift to 37°C compared with long-term growth of L. interrogans at 30°C across three sets of biological replicates. The COG functional categories are as follows: information storage and processing (11.2% of coding sequences in L. interrogans serovar Lai genome) (includes J, translation; K, transcription; L, replication, recombination, and repair); cellular processes and signaling (19% of coding sequences in the serovar Lai genome) (includes D, cell cycle control, cell division, chromosome partitioning; V, defense mechanisms; T, signal transduction mechanisms; M, cell wall, membrane, or envelope biogenesis; N, cell motility; U, intracellular trafficking, secretion, and vesicular transport; O, posttranslational modification, protein turnover, chaperones); metabolism (18.9% of coding sequences in the serovar Lai genome) (includes C, energy production and conversion; G, carbohydrate transport and metabolism; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolite biosynthesis, transport, and catabolism); poorly characterized (50.9% of coding sequences in the serovar Lai genome) (includes R, general function prediction only; S, function unknown; and −, not in COGs).
FIG. 4.
Number of differentially expressed genes by COG category during overnight upshift to 37°C compared with long-term growth of L. interrogans at 37°C across three sets of biological replicates. The COG functional categories are as described in the legend to Fig. 3.
A substantial heat shock response was not seen in the upshift experiments, although there was down-regulation of some ribosomal proteins after 37°C upshift versus 30°C. Using [35S]methionine labeling, previous studies have shown that the expression of GroEL and DnaK homologs in L. interrogans is increased following temperature upshifts from 30°C to 37°C or to 42°C, but cells were incubated at the higher temperatures for only 1 h 15 min (38). However, since the leptospiral cultures had been upshifted to 37°C overnight, our experiments were not an evaluation of gene expression in response to immediate heat stress but rather an assessment of gene expression patterns likely to take place during the early stages of infection and/or adaptation to the host. However, a universal stress protein, UspA, was up-regulated 1.6-fold, implying that growth at higher temperatures may be stressful to the organism.
As observed in the physiological versus environmental comparison, many genes that were differentially expressed upon upshift from 30°C to 37°C are involved in signal transduction mechanisms, chemotaxis, or processing of membrane proteins (COG functional categories T and M), a not unexpected finding, given that pathogens need the ability to respond rapidly to changes upon invasion of a host.
Validation of microarray data by real-time RT-PCR.
Real-time quantitative RT-PCR was used to validate the microarray data. Six genes which appear in at least 2 of the 3 gene lists (physiological versus environmental and the two temperature upshift lists) were selected at random. The flaB gene did not show any variation in expression under the different temperature conditions and, therefore, was used for normalization. There was a high correlation between the expression values obtained by microarray analysis and those measured by quantitative real-time RT-PCR for all three comparisons (r > 0.95) (Fig. 5).
FIG. 5.
Comparison of real-time RT-PCR and microarray data. Six genes which appear in at least 2 of the 3 gene lists (physiological versus environmental [A], 37°C upshift versus 30°C [B], and 37°C upshift versus 37°C [C]) were selected at random for real-time RT-PCR analysis. The correlation coefficient (r) in each case was >0.95.
Comparison of common genes between the physiological versus environmental and 37°C upshift gene lists.
To gain more insight into long- and short-term gene expression changes in response to different temperatures, the expression profile of genes with known or predicted function which appear in at least 2 of the 3 gene lists were examined (Table 7). Of the differentially expressed hypothetical genes, 44% which were up- or down-regulated at physiological versus environmental temperatures were likewise up- or down-regulated at 37°C upshift versus 30°C long term. In contrast, only 3.5% of the differentially expressed hypothetical genes at physiological versus environmental temperatures were also differentially expressed at 37°C upshift versus 37°C long term. There was no correlation in gene transcription patterns according to location on chromosome 1 or 2.
TABLE 7.
Comparison of differentially expressed genes with known or predicted function that are common between the physiological versus environmental and 37°C upshift gene lists
| Response to temp change and ORF | Gene | COG categorya | Description of gene product | Differential expression atb:
|
||
|---|---|---|---|---|---|---|
| 37°C + 39°C vs 20°C + 30°C | 37°C upshift vs 30°C | 37°C upshift vs 37°C | ||||
| No change upon temp adaptation | ||||||
| LA2453 | C | 4Fe-4S binding protein | + | + | 0 | |
| LA3011 | D | Cell division protein with ATPase domain | − | − | 0 | |
| LA0693 | lysC | E | Aspartate kinase | − | − | 0 |
| LB123 | aroK | E | Shikimate kinase | − | − | 0 |
| LA1372 | G | Permease | + | + | 0 | |
| LB102 | G | Sugar phosphatase | + | + | 0 | |
| LA0786 | G | Glycosyltransferase | − | − | 0 | |
| LA0970 | G | Permease component of an ABC transporter complex | − | − | 0 | |
| LA1392 | G | Sugar kinase | − | − | 0 | |
| LA2958 | I | Hydrolase, acyltransferase | + | + | 0 | |
| LA3928 | caiA | I | Acyl-CoA dehydrogenase | + | + | 0 |
| LA0438 | I | Acyl-CoA dehydrogenase | − | − | 0 | |
| LA1970 | J | poly(A) polymerase family of proteins | + | + | 0 | |
| LA1282 | K | Transcriptional regulator, AcrR family | + | + | 0 | |
| LA1816 | K | Transcriptional regulator | + | + | 0 | |
| LA2728 | K | Transcriptional regulator | + | + | 0 | |
| LA3652 | K | RNA polymerase sigma subunit | + | + | 0 | |
| LA4298 | K | Transcriptional regulator, MarR family | + | + | 0 | |
| LB104 | K | AcrR family transcriptional regulator | + | + | 0 | |
| LA0984 | L | ATP-dependent DNA ligase | + | + | 0 | |
| LA1456 | radC | L | DNA repair protein | + | + | 0 |
| LA0281 | alkA | L | DNA-3-methyladenine glycosylase II | − | − | 0 |
| LA2347 | xerC | L | Tyrosine site-specific recombinase XerC | − | − | 0 |
| LA4049 | srmB | L | ATP-dependent RNA helicase (superfamily II) | − | − | 0 |
| LA1284 | lolE | M | Lipoprotein releasing system, LolE permease component | + | + | 0 |
| LA2058 | ddlA | M | d-alanined-alanine ligase | − | − | 0 |
| LB323 | M | Transglycosylase | − | − | 0 | |
| LA3489 | flgC | N | Endoflagellar proximal basal body rod protein | + | + | 0 |
| LA2666 | flgA | N | Endoflagellar basal body P-ring biosynthesis protein | − | − | 0 |
| LA1563 | O | Small heat shock protein (molecular chaperone) | + | + | 0 | |
| LA1879 | clpA | O | Endopeptidase Clp, ATP-dependent proteolytic subunit | + | + | 0 |
| LA4299 | btuE | O | Glutathione peroxidase | + | + | 0 |
| LB174 | htpX | O | Zn-dependent protease with chaperone function | + | + | 0 |
| LA2949 | O | Trypsin-like serine protease | − | − | 0 | |
| LA0593 | copZ | P | Copper chaperone | + | + | 0 |
| LA0594 | P | Cation transport ATPase, possibly copper | + | + | 0 | |
| LA1859 | katE | P | Catalase | + | + | 0 |
| LA3110 | kdpC | P | Potassium-transporting ATPase C chain | + | + | 0 |
| LA4245 | P | Monooxygenase | − | − | 0 | |
| LA4246 | phoD | P | Phosphodiesterase I | − | − | 0 |
| LA0816 | T | Receiver component of a two-component response regulator | + | + | 0 | |
| LA1743 | T | Chemotaxis protein, methyltransferase | + | + | 0 | |
| LA1744 | T | Chemotaxis protein, methylesterase | + | + | 0 | |
| LA1746 | T | Sensor and regulator protein of a two-component response regulator | + | + | 0 | |
| LA1860 | T | Sensor protein of a two-component response regulator | + | + | 0 | |
| LA2117 | T | Anti-sigma factor antagonist | + | + | 0 | |
| LA2223 | T | Sensor histidine kinase of a two-component response regulator | + | + | 0 | |
| LA3858 | T | Sensor histidine kinase and response regulator of a two-component complex, part (C terminus) | + | + | 0 | |
| LA0049 | T | Methyl-accepting chemotaxis protein | − | − | 0 | |
| LA0565 | T | adenylate cyclase | − | − | 0 | |
| LA0802 | U | Pilus assembly protein | + | + | 0 | |
| LA1285 | V | Lipoprotein releasing system, LolD ATPase component | + | + | 0 | |
| LA2014 | V | CreD-like protein | − | − | 0 | |
| LA3871 | V | Cation, multidrug efflux pump | − | − | 0 | |
| Transiently expressed | ||||||
| LA2598 | gmk | F | Guanylate kinase | 0 | + | + |
| LA0980 | thiC | H | Thiamine biosynthesis protein ThiC | 0 | − | − |
| LA2335 | ubiX | H | 3-polyprenyl-4-hydroxybenzoate decarboxylase | 0 | + | + |
| LA0001 | dnaA | L | Chromosomal replication initiator protein | 0 | + | + |
| LA1964 | M | AcrA-related membrane protein, part (N terminus) | 0 | + | + | |
| LA2417 | flgL | N | Endoflagellar hook-filament protein | 0 | + | + |
| LA3778 | ligB | N | LigB lipoprotein | − | + | + |
| LA0666 | mauG | P | Cytochrome c peroxidase | 0 | + | + |
| LA0280 | T | Cyclic AMP-binding protein | 0 | − | − | |
| LA2427 | cheW | T | Chemotaxis signal transduction protein | 0 | + | + |
| LA2473 | T | Cyclic AMP-binding protein, regulatory protein | 0 | + | + | |
| LA3699 | T | Protein-tyrosine-phosphatase | 0 | + | + | |
| LA1142 | secD | U | Preprotein translocase, SecD subunit | 0 | − | − |
| LA1457 | G | DNA repair protein | + | 0 | − | |
| Up-regulated across all 3 comparisons | ||||||
| LA0145 | D | ATPase of the PP-loop superfamily | + | + | + | |
| LA1212 | uspA | T | Universal stress protein UspA | + | + | + |
| LA1504 | M | Metallopeptidase | + | + | + | |
COG categories are as described for Fig. 3.
+, up-regulated genes; −, down-regulated genes; 0, genes which were not significantly differentially expressed.
A subset of genes was found to be differentially expressed at physiological versus environmental temperatures and 37°C upshift versus 30°C long term but not at 37°C upshift versus 37°C long term. The lack of differential expression at 37°C upshift versus 37°C long term appears to indicate that, once up- or down-regulated, the level of expression remains the same, possibly as a consequence of adaptation to temperature or the host environment. These are possibly genes which are required to establish and maintain infection. Interestingly, genes found in this subset but not the others are grouped into COG categories C, D, E, I, J, K, and O. Rapid detection of changes in growth conditions followed by subsequent changes in metabolism and alteration of membrane proteins would facilitate invasion of the host. Therefore, it is unsurprising that genes in these COG groups are primarily involved in transcriptional regulation, posttranslational modification, protein turnover and chaperones, carbohydrate and amino acid transport, and metabolism (Table 7). Genes encoding LolD (LA1285) and LolE (LA1284) were also found in this subset.
A subset of genes appears to be transiently expressed or switched off. These genes are differentially expressed in the upshift comparisons but not at physiological versus environmental temperatures. The exception to this was ligB, which was 1.6-fold down-regulated at physiological versus environmental temperatures but 1.7-fold and 2.2-fold up-regulated during 37°C upshift versus 30°C long term and 37°C long term, respectively. This supports data from previous studies which found that ligA and ligB are expressed by low-passage virulent strains of L. interrogans serovar Copenhageni, but expression is lost upon repeated subculturing, which is concurrent with loss in virulence (21, 31). L. interrogans serovar Lai lacks a copy of ligA but has ligC as well as ligB (28, 32). At 37°C upshift versus 30°C long term, ligC was also up-regulated by 1.6-fold (see Table S3 in the supplemental material), while there was no significant difference in expression at 37°C versus 37°C long term. Taken together, our microarray data indicate that ligB and ligC are expressed immediately upon exposure to stress or sudden changes in environmental cues and may assist in initial colonization and establishment of disease in the host. Expression of ligB and other genes in the transiently expressed subset may continue during infection of the host, but in our experiments, in the absence of other environmental cues apart from upshift in temperature, ligB expression decreased with prolonged culturing at 37°C. Osmolarity has also been found to induce expression of LigB (22), and due to its similarity to other bacterial host cell adherence and invasion proteins, such as invasin in Yersinia spp. and intimin in enteropathogenic E. coli, it is thought that LigB is also likely to play a similar role in pathogenesis (21).
Another interesting, transiently up-regulated gene was cheW, which encodes a chemotaxis-related protein. In E. coli, CheW is a coupling protein which stabilizes the complex between CheA, a histidine kinase, and its receptor. CheW expression was also found to be up-regulated in B. burgdorferi in response to temperature upshift and the presence of blood (42). A motility-associated gene, flgL, was also transiently up-regulated, but flgC and flgA were found to exhibit no change in expression upon temperature adaptation. It can be surmised that flagellar activity and structure may need to be modulated upon host entry and during the course of infection.
Of the three genes consistently expressed across all three gene lists, up-regulation of uspA, which encodes a universal stress protein, implies that growth at and transition to higher temperatures is stressful to the organism.
Concluding remarks.
We have identified genes which are differentially expressed under different temperature conditions. Furthermore, we have conducted a novel investigation into differences in long- and short-term changes in gene expression in response to temperature. However, proteins which were shown in previous studies to be differentially expressed at different temperatures, namely LipL36 (9, 27) and Qlp42 (25), were not found to be differentially expressed in our experiments. It is thus possible that regulation of these proteins occurs at the translational level rather than during transcription.
Genes that were found to be up-regulated may potentially be involved in colonization of the host and/or the early stages of pathogenesis. Differentially expressed genes of unknown function may hold clues to processes involved in establishment of disease and persistence in host species.
Supplementary Material
Acknowledgments
We gratefully acknowledge the excellent technical assistance of Vicki Vallance and Chen Ai Khoo.
This work was supported by a grant from the National Health and Medical Research Council Program in Medical Genomics, Canberra, Australia (B.A.), by NIH grant AI-34431 (D.A.H.), and by VA Medical Research funds (D.A.H.).
Editor: J. T. Barbieri
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Adler, B., and S. Faine. 1976. Susceptibility of mice treated with cyclophosphamide to lethal infection with Leptospira interrogans serovar Pomona. Infect. Immun. 14:703-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artiushin, S., J. F. Timoney, J. Nally, and A. Verma. 2004. Host-inducible immunogenic sphingomyelinase-like protein, Lk73.5, of Leptospira interrogans. Infect. Immun. 72:742-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babudieri, B. 1958. Animal reservoirs of leptospires. Ann. N. Y. Acad. Sci. 70:393-413. [DOI] [PubMed] [Google Scholar]
- 4.Ballard, S. A., M. Go, R. P. Segers, and B. Adler. 1998. Molecular analysis of the dnaK locus of Leptospira interrogans serovar Copenhageni. Gene 216:21-29. [DOI] [PubMed] [Google Scholar]
- 5.Ballard, S. A., R. P. Segers, N. Bleumink-Pluym, J. Fyfe, S. Faine, and B. Adler. 1993. Molecular analysis of the hsp (groE) operon of Leptospira interrogans serovar Copenhageni. Mol. Microbiol. 8:739-751. [DOI] [PubMed] [Google Scholar]
- 6.Brenner, D. J., A. F. Kaufmann, K. R. Sulzer, A. G. Steigerwalt, F. C. Rogers, and R. S. Weyant. 1999. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int. J. Syst. Bacteriol. 49:839-858. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig, J. E., A. Nobbs, and N. J. High. 2002. The extracytoplasmic sigma factor, σE, is required for intracellular survival of nontypeable Haemophilus influenzae in J774 macrophages. Infect. Immun. 70:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen, P. A., S. J. Cordwell, D. M. Bulach, D. A. Haake, and B. Adler. 2002. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 70:2311-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen, P. A., X. Xu, J. Matsunaga, Y. Sanchez, A. I. Ko, D. A. Haake, and B. Adler. 2005. Surfaceome of Leptospira spp. Infect. Immun. 73:4853-4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haake, D. A., C. Martinich, T. A. Summers, E. S. Shang, J. D. Pruetz, A. M. McCoy, M. K. Mazel, and C. A. Bolin. 1998. Characterization of leptospiral outer membrane lipoprotein LipL36: downregulation associated with late-log-phase growth and mammalian infection. Infect. Immun. 66:1579-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiratsu, K., M. Amemura, H. Nashimoto, H. Shinagawa, and K. Makino. 1995. The rpoE gene of Escherichia coli, which encodes σE, is essential for bacterial growth at high temperature. J. Bacteriol. 177:2918-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, R. C., J. Walby, R. A. Henry, and N. E. Auran. 1973. Cultivation of parasitic leptospires: effect of pyruvate. Appl. Microbiol. 26:118-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerr, M. K., and G. A. Churchill. 2001. Experimental design for gene expression microarrays. Biostatistics 2:183-201. [DOI] [PubMed] [Google Scholar]
- 16.Kovacikova, G., and K. Skorupski. 2002. The alternative sigma factor σE plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 70:5355-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kvint, K., L. Nachin, A. Diez, and T. Nystrom. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6:140-145. [DOI] [PubMed] [Google Scholar]
- 18.Levett, P. N., R. E. Morey, R. L. Galloway, and A. G. Steigerwalt. 2006. Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int. J. Syst. Evol. Microbiol. 56:671-673. [DOI] [PubMed] [Google Scholar]
- 19.Ma, D., M. Alberti, C. Lynch, H. Nikaido, and J. E. Hearst. 1996. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19:101-112. [DOI] [PubMed] [Google Scholar]
- 20.Madsen, M. L., D. Nettleton, E. L. Thacker, R. Edwards, and F. C. Minion. 2006. Transcriptional profiling of Mycoplasma hyopneumoniae during heat shock using microarrays. Infect. Immun. 74:160-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsunaga, J., M. A. Barocchi, J. Croda, T. A. Young, Y. Sanchez, I. Siqueira, C. A. Bolin, M. G. Reis, L. W. Riley, D. A. Haake, and A. I. Ko. 2003. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49:929-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsunaga, J., Y. Sanchez, X. Xu, and D. A. Haake. 2005. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect. Immun. 73:70-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsunaga, J., T. A. Young, J. K. Barnett, D. Barnett, C. A. Bolin, and D. A. Haake. 2002. Novel 45-kilodalton leptospiral protein that is processed to a 31-kilodalton growth-phase-regulated peripheral membrane protein. Infect. Immun. 70:323-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melnikow, E., S. Dornan, C. Sargent, M. Duszenko, G. Evans, N. Gunkel, P. M. Selzer, and H. J. Ullrich. 2005. Microarray analysis of Haemophilus parasuis gene expression under in vitro growth conditions mimicking the in vivo environment. Vet. Microbiol. 110:255-263. [DOI] [PubMed] [Google Scholar]
- 25.Nally, J. E., S. Artiushin, and J. F. Timoney. 2001. Molecular characterization of thermoinduced immunogenic proteins Q1p42 and Hsp15 of Leptospira interrogans. Infect. Immun. 69:7616-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nally, J. E., E. Chow, M. C. Fishbein, D. R. Blanco, and M. A. Lovett. 2005. Changes in lipopolysaccharide O antigen distinguish acute versus chronic Leptospira interrogans infections. Infect. Immun. 73:3251-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nally, J. E., J. F. Timoney, and B. Stevenson. 2001. Temperature-regulated protein synthesis by Leptospira interrogans. Infect. Immun. 69:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nascimento, A. L., A. I. Ko, E. A. Martins, C. B. Monteiro-Vitorello, P. L. Ho, D. A. Haake, S. Verjovski-Almeida, R. A. Hartskeerl, M. V. Marques, M. C. Oliveira, C. F. Menck, L. C. Leite, H. Carrer, L. L. Coutinho, W. M. Degrave, O. A. Dellagostin, H. El-Dorry, E. S. Ferro, M. I. Ferro, L. R. Furlan, M. Gamberini, E. A. Giglioti, A. Goes-Neto, G. H. Goldman, M. H. Goldman, R. Harakava, S. M. Jeronimo, I. L. Junqueira-de-Azevedo, E. T. Kimura, E. E. Kuramae, E. G. Lemos, M. V. Lemos, C. L. Marino, L. R. Nunes, R. C. de Oliveira, G. G. Pereira, M. S. Reis, A. Schriefer, W. J. Siqueira, P. Sommer, S. M. Tsai, A. J. Simpson, J. A. Ferro, L. E. Camargo, J. P. Kitajima, J. C. Setubal, and M. A. Van Sluys. 2004. Comparative genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. J. Bacteriol. 186:2164-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nascimento, A. L., S. Verjovski-Almeida, M. A. Van Sluys, C. B. Monteiro-Vitorello, L. E. Camargo, L. A. Digiampietri, R. A. Harstkeerl, P. L. Ho, M. V. Marques, M. C. Oliveira, J. C. Setubal, D. A. Haake, and E. A. Martins. 2004. Genome features of Leptospira interrogans serovar Copenhageni. Braz. J. Med. Biol. Res. 37:459-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palaniappan, R. U., Y. F. Chang, S. S. Jusuf, S. Artiushin, J. F. Timoney, S. P. McDonough, S. C. Barr, T. J. Divers, K. W. Simpson, P. L. McDonough, and H. O. Mohammed. 2002. Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect. Immun. 70:5924-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 33.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saal, L. H., C. Troein, J. Vallon-Christersson, S. Gruvberger, A. Borg, and C. Peterson. 2002. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 3:software0003.1-0003.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [Online.] http://www.bepress.com/sagmb/vol3/iss1/art3/. [DOI] [PubMed]
- 37.Smyth, G. K., J. Michaud, and H. S. Scott. 2005. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21:2067-2075. [DOI] [PubMed] [Google Scholar]
- 38.Stamm, L. V., F. C. Gherardini, E. A. Parrish, and C. R. Moomaw. 1991. Heat shock response of spirochetes. Infect. Immun. 59:1572-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulavik, M. C., L. F. Gambino, and P. F. Miller. 1995. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol. Med. 1:436-446. [PMC free article] [PubMed] [Google Scholar]
- 40.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 41.Testerman, T. L., A. Vazquez-Torres, Y. Xu, J. Jones-Carson, S. J. Libby, and F. C. Fang. 2002. The alternative sigma factor sE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol. Microbiol. 43:771-782. [DOI] [PubMed] [Google Scholar]
- 42.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 72:5419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vinciotti, V., R. Khanin, D. D'Alimonte, X. Liu, N. Cattini, G. Hotchkiss, G. Bucca, O. de Jesus, J. Rasaiyaah, C. P. Smith, P. Kellam, and E. Wit. 2005. An experimental evaluation of a loop versus a reference design for two-channel microarrays. Bioinformatics 21:492-501. [DOI] [PubMed] [Google Scholar]
- 44.Wickner, S., S. Gottesman, D. Skowyra, J. Hoskins, K. McKenney, and M. R. Maurizi. 1994. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc. Natl. Acad. Sci. USA 91:12218-12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470-1479. [DOI] [PubMed] [Google Scholar]
- 46.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuerner, R. L., W. Knudtson, C. A. Bolin, and G. Trueba. 1991. Characterization of outer membrane and secreted proteins of Leptospira interrogans serovar pomona. Microb. Pathog. 10:311-322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.