Abstract
Mutants of Salmonella enterica serovar Typhimurium that lack the transcriptional regulator RfaH are efficient as live oral vaccines against salmonellosis in mice. We show that the attenuation of the vaccine candidate strain is associated with reduced net growth in epithelial and macrophage cells. In order to identify the relevant RfaH-dependent genes, the RfaH regulon was determined with S. enterica serovars Enteritidis and Typhimurium using whole-genome Salmonella microarrays. As well as impacting the expression of genes involved in lipopolysaccharide (LPS) core and O-antigen synthesis, the loss of RfaH results in a marked down-regulation of SPI-4 genes, the flagellum/chemotaxis system, and type III secretion system 1. However, a proportion of these effects could have been the indirect consequence of the altered expression of genes required for LPS biosynthesis. Direct and indirect effects of the rfaH mutation were dissociated by genome-wide transcriptional profiling of a structural deep-rough LPS mutant (waaG). We show that truncation of LPS itself is responsible for the decreased intracellular yield observed for ΔrfaH strains. LPS mutants do not differ in replication ability; rather, they show increased susceptibility to antimicrobial peptides in the intracellular milieu. On the other hand, evidence that deletion of rfaH, as well as some other genes involved in LPS biosynthesis, results in enhanced invasion of various mammalian cells is shown. Exposure of common minor antigens in the absence of serovar-specific antigens might be responsible for the observed cross-reactive nature of the elicited immune response upon vaccination. Increased invasiveness of the Salmonella rfaH mutant into antigen-presenting cells, combined with increased intracellular killing and the potential for raising a cross-protective immune response, renders the rfaH mutant an ideal vaccine candidate.
Salmonella enterica can cause various diseases in humans and a wide variety of farm animals. Currently available vaccines do not confer optimal protection against Salmonella infection, and so development of effective vaccines is still a high priority (for a recent review, see reference 22). Currently, over 2,500 serovariants of S. enterica have been distinguished, based on the high polymorphism of lipopolysaccharide (LPS) O antigens and flagellar antigens. Immune cross-protection between serovariants of S. enterica is limited, hindering development of an efficacious broad-spectrum Salmonella vaccine. Recently, we reported that an attenuated mutant of S. enterica serovar Typhimurium strain SL1344 that lacks the transcriptional regulator RfaH was capable of eliciting protection against subsequent challenge by wild-type Salmonella in the mouse model of typhoid fever (41). Furthermore, sera of vaccinated mice were shown to cross-react to not only the isogenic wild-type strain but also heterologous serovariants of S. enterica and even Salmonella bongori.
Transcriptional antitermination is a conserved mechanism of gene regulation based on overcoming intrinsic premature termination signals to eliminate operon polarity during transcription of long operons. RfaH, a homologue of Escherichia coli transcriptional antiterminator NusG (3), is distributed among gamma-proteobacteria. Interestingly, the regulatory role of RfaH in E. coli appears to be limited to operons encoding extracytoplasmic cell components (LPS, capsules, exotoxins, hemin uptake receptor ChuA, F pilus) involved in the virulence of E. coli pathogens. Consequently, the virulence of uropathogenic E. coli was abolished through down-regulation of several virulence factors upon deletion of RfaH (42). Unlike RfaH itself, none of the operons encoding these extracytoplasmic structures belong to the E. coli core genome; rather, they are likely to have been acquired by horizontal gene transfer (carried on plasmids or pathogenicity/genomic islands.) Moreover, RfaH-dependent operons share a short cis-acting element termed ops (for operon polarity suppressor) that is essential to allow RfaH to function (2). How all these virulence factors evolved to utilize the same core regulatory mechanism still awaits discovery. However, our knowledge of RfaH is based almost solely on observations of E. coli. Orthologues of RfaH, on the other hand, were recently shown to be able to complement an E. coli rfaH mutant, suggesting that the function of RfaH may be conserved among gamma-proteobacteria (9).
In this study, we examine the function of RfaH in Salmonella strains, and show that attenuation of Salmonella rfaH mutants is linked to impaired intracellular net growth. RfaH regulates the production of amphipathic LPS, which allow the bacteria to survive stressful environments and are required for bacterial adherence to mammalian cells (27). The genes for LPS core and O-antigen biosynthesis are clustered into long operons (see reference 47 for a review). There is a great deal of evidence that Salmonella requires full-length LPS for successful infection of mammalian hosts (18). In fact, modification of LPS chain length has a great impact upon the ability of Salmonella to kill mice (40), and it induces mammalian cell signaling via Toll-like receptor 4, which induces the inflammatory response.
Genome-wide transcriptome analysis of rfaH mutants reveals direct and, through the resulting deep-rough phenotype, indirect effects of RfaH on the expression of virulence genes, with interesting implications for vaccine development.
MATERIALS AND METHODS
Bacterial strains and mutagenesis.
Bacterial strains and plasmids used in this study are listed in Table 1. Wild-type Salmonella serovar Typhimurium strain SL1344 and Yersinia enterocolitica strain WA-314 were obtained from the IMIB strain collection (Würzburg, Germany). The recently sequenced Salmonella serovar Enteritidis PT4 strain NCTC13349 was provided by Mark Roberts (University of Glasgow, United Kingdom). All mutants used in this study were constructed using the Red recombinase method as described principally by Datsenko and Wanner (11). The cat gene encoding resistance to chloramphenicol was amplified from template plasmid pKD3 using primers that incorporated homologous sequences (40 to 50 bases) to the genes to be inactivated at their 5′ ends. The obtained PCR products were transformed to competent SL1344 cells that had been supplied with the curable helper plasmid pKD46 encoding the Red recombinase (11). Upon induction with l-arabinose (Sigma-Aldrich, Hungary) the cat cassette was introduced into the chromosome, replacing the targeted gene by Red-mediated recombination through the flanking homologies. Complementation of the rfaH mutant was performed as described by Diederich et al. (13). rfaH, together with its 5′-flanking region, was amplified with an Expand Long PCR proofreading polymerase system (Roche, Mannheim, Germany) as described elsewhere (41). The digested PCR product was cloned into the SmaI-XbaI sites of pUC18, giving rise to pGNS1. pGNS1 was digested with EcoRI and XbaI, and the resulting fragment was ligated into the corresponding sites of pLDR11, establishing pGNS11. pGNS11 was digested with NotI to eliminate the ori region of the plasmid. The recircularized fragment containing the rfaH gene located adjacent to attP and bla was electroporated into SL1344-R1 (the rfaH deletion mutant) carrying pLDR8. Induction of the λ integrase encoded on pLDR8 introduced the fragment into the attB site of the chromosome, giving rise to an ampicillin-resistant complemented strain that has been termed SL1344-R2. Appropriate insertion of the construct into attB was verified using primer set SL-att1 (GCA TTC CTG TCG CTC TCT TG) and SL-att2 (CGT AGA GCT ACA GGC GCT C).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| SL1344 | Serovar Typhimurium strain ΔhisGΔrpsL | 58 |
| SL1344-R1 | SL1344 ΔrfaH::cat | 41 |
| SL1344-R2 | SL1344 ΔrfaH::cat, attB::rfaH | 41 |
| SL1344 waaL | SL1344 ΔwaaL::cat; lacking O-antigen ligase, resulting in the loss of O antigens with intact LPS core | This study |
| SL1344 waaG | SL1344 ΔwaaG::cat; LPS is truncated at the level of inner core; deep-rough phenotype | This study |
| SL1344 waaP | SL1344 ΔwaaP::cat; lacking HepIII and phosphate residues on HepI and HepII; reduced amount of full-length LPS chains; deep-rough phenotype | This study |
| SL1344 waaY | SL1344 ΔwaaY::cat; lacking phosphate residue on HepII | This study |
| NCTC13349 | Serovar Enteritidis PT4 strain; genome sequence available at http://www.sanger.ac.uk | NCTCa |
| NCTC13349 rfaH | NCTC13349 ΔrfaH::cat | This study |
| WA-314 | Y. enterocolitica O8 prototype strain; mouse virulent | 52 |
| WA-314 rfaH | WA-314 ΔrfaH::cat | This study |
| Plasmids | ||
| pKD3 | Template plasmid for the amplification of the cat cassette; Cmr, Apr | 11 |
| pKD46 | λ Red recombinase expression vector, helper plasmid, temp-sensitive ori; Apr | 11 |
| pLDR8 | int gene expression vector, low-copy-no. helper plasmid, temp-sensitive replicon; Kmr | 13 |
| pLDR11 | Cloning vector for integration into attB; Apr, Tcr | 13 |
| pUC18 | Commercial cloning vector; Apr | 59 |
| pGNS1 | rfaH together with its promoter region cloned into pUC18; Apr | This study |
| pGNS11 | rfaH together with its promoter region cloned into pLDR11; Apr, Tcr | This study |
NCTC, National Collection of Type Cultures, Central Public Health Laboratory, Colindale, United Kingdom.
Bacteria were routinely grown in Luria-Bertani (LB) broth at 37°C with shaking at 250 rpm or on LB agar plates. Bacteria carrying temperature-sensitive plasmids (pLDR8 and pKD46) were grown at permissive temperatures (30°C). When appropriate, media were supplemented with chloramphenicol (20 μg/ml), kanamycin (30 μg/ml), or ampicillin (100 μg/ml).
Invasion and intracellular survival assays.
Invasion and intracellular survival tests were performed principally as described earlier (20). The human intestinal cell line INT407 was cultured in RPMI 1640 supplemented with 10% fetal calf serum, and the mouse macrophage cell line RAW264.7 was cultured using Eagle's minimal essential medium supplemented with 10% fetal calf serum and 2 mM l-glutamine. Cells were seeded 16 h prior to infection at a density of 5 × 105 cell/well (24-well plates). Bacteria were grown overnight to stationary phase, washed in phosphate-buffered saline (PBS), and diluted to 5 × 107 CFU/ml in the appropriate cell culture medium. Bacteria were added to the cells either for 30 min (RAW264.7) or 2 h (INT407) to allow invasion. Wells were washed with PBS, and the appropriate cell culture medium containing 40 μg/ml gentamicin was added for 40 min to kill extracellular bacteria. The killing medium was removed, and the cells were washed in PBS. Intracellular bacteria were liberated through disruption of eukaryotic cells by the addition of 1% Triton X-100 in PBS for 2 min. Released bacteria were diluted and plated for viable counts. For the determination of intracellular replication, cells were grown in the gentamicin-containing medium for an additional 6 or 24 h prior to lysis of cultured cells. Intracellular growth was expressed as the change (n-fold) in the bacterial number at a given time point relative to the initial invasive bacteria. For the plasmid segregation studies, a conditional suicide plasmid (pLDR8) containing a temperature-sensitive replication of origin (13) was introduced to serovar Typhimurium strains by electroporation. Plasmid-harboring bacteria were grown at 30°C (permissive temperature for plasmid replication). Invasion and intracellular survival studies were performed as described above (at 37°C; nonpermissive temperature). Replica plating onto LB agar plates containing either no antibiotic or kanamycin (30 μg/ml) allowed the determination of the extent of plasmid loss throughout the study period.
Microarray experiments.
Bacteria were grown in LB broth at 37°C (in 100-ml flasks with shaking at 250 rpm in a New Brunswick Scientific C25 shaker) until late logarithmic phase of growth (optical density at 600 nm [OD600], 0.8). Two OD units (2.4 ml) of culture was fixed by incubation on ice with a 1/5 culture volume of 5% phenol and 95% ethanol. Cultures were centrifuged at 4,000 rpm for 10 min, and the resulting pellets were frozen at −80°C. Bacteria were lysed with 100 μl of 50 μg/μl lysozyme, and RNA was isolated using an SV Total RNA system (Promega) following the protocols provided by the manufacturer. The quality of the RNA was verified using an Agilent 2100 Bioanalyzer (Agilent), and the quantity was determined with an ND-1000 spectrophotometer (Nanodrop). RNA (16 μg) from three biological replicates was labeled with Cy5-dCTP and hybridized to the SALSA microarrays with Cy3-dCTP-labeled genomic DNA as described previously (17) (for protocols, see http://www.ifr.ac.uk/safety/microarrays.html). The SALSA microarrays comprise PCR products which represent all the genes present in sequenced Salmonella serovar Typhimurium strains LT2, SL1344, and DT104, as well as the genes present in serovar Enteritidis PT4 strain NCTC13349. Details of the PCR products can be found at http://www.ifr.ac.uk/Safety/MolMicro/pubs.html. Three biological replicates for each strain were tested, and each was hybridized twice to the microarrays. Microarray image analysis was done using BlueFuse for microarrays software (BlueGnome), and data were analyzed using GeneSpring software version 6 (Silicon Genetics). Genes showing statistically significant differences (see below) and showing a minimum of twofold change in expression between the wild-type and rfaH mutant strains were considered to be differentially expressed between the mutant and wild-type strains.
Susceptibility tests.
MICs of different antimicrobial substances were determined by using 96-well tissue culture plates. Twofold serial dilutions of sodium dodecyl sulfate (SDS) (0.1 to 100 mg/ml), H2O2 (0.005 to 5 mM), and polymyxin B (0.01 to 10 μg/ml) were made across the plates. Washed bacteria (106 CFU) were inoculated and incubated for 6 h at 37°C. The optical density was determined with a conventional enzyme-linked immunosorbent assay plate reader. The threshold of inhibition was 0.1 at OD550, and assays were repeated at least three times.
Serum bactericidal test.
Bacteria grown in LB medium were washed in saline and diluted to 106 CFU/ml. Aliquots (200 μl) of bacterial suspensions were mixed with equal volumes of 20% pooled human serum and incubated at 37°C for 1 h. Viable cell counts were determined by plating aliquots onto LB plates and incubating overnight at 37°C. The assays were performed with normal and, as a control, heat-inactivated (56°C for 30 min) sera. Duplicates were used for each strain, and assays were repeated three times.
Silver staining of LPS.
LPS was purified by the procedure of Hitchcock and Brown (24). SDS-polyacrylamide gel electrophoresis was performed on 12.5% polyacrylamide gels. Gels were fixed overnight in a solution of 7% acetic acid and 25% 2-propanol and were silver stained as described by Nelson et al. (43).
Swarming.
Swarming motility was assessed with LB plates solidified with 0.6% agar and supplemented with 0.5% glucose. These swarm plates were allowed to dry at room temperature for 24 h. Bacteria collected from agar plates were washed and diluted in saline. Six microliters of bacterial suspension was spotted onto the middles of swarm plates, which were subsequently incubated at 37°C for 8 h. Radii of swarming colonies were compared. Experiments were repeated three times.
Virulence experiments.
Animal experiments were conducted according to the principles set forth in the guide for the care and use of laboratory animals in a laboratory as authorized by Hungarian decree (no. XXVII, 1998) and by the subsequent regulation (government order no. 243/1998).
Female BALB/c mice (Charles River, Budapest, Hungary) aged 6 to 8 weeks were used in all cases. For the determination of the 50% lethality dose (LD50), groups of five mice each were infected orally with inocula containing log dilutions (103 to 109 CFU) of serovar Typhimurium SL1344 or its derivatives. Bacteria were grown overnight at 37°C in LB, harvested by centrifugation, washed once, and normalized to the required inoculum density in saline by adjusting the suspension to the appropriate OD600 value justified by viable counts. Oral infections were performed using a sterile gavage without any prior neutralization of gastric acid. Animals were observed for 3 weeks postinfection, and deaths were recorded daily.
Statistical analysis.
Differences in invasion levels as well as net growth in cell cultures by strain SL1344 and its variants were analyzed by Student's t test. Probabilities (P values) of 0.05 or less were considered significant.
Genes were assessed to be statistically significantly differently expressed in the microarray experiments by an analysis of variance test with a Benjamini and Hochberg false discovery rate of 0.05 (5) and with a >2-fold change in the expression level.
Microarray data accession numbers.
Microarray data reported in this paper have been submitted to http://www.ebi.ac.uk/arrayexpress/. The accession number is E-MEXP-844 (available 1 October 2006).
RESULTS
Loss of RfaH results in decreased intracellular net growth.
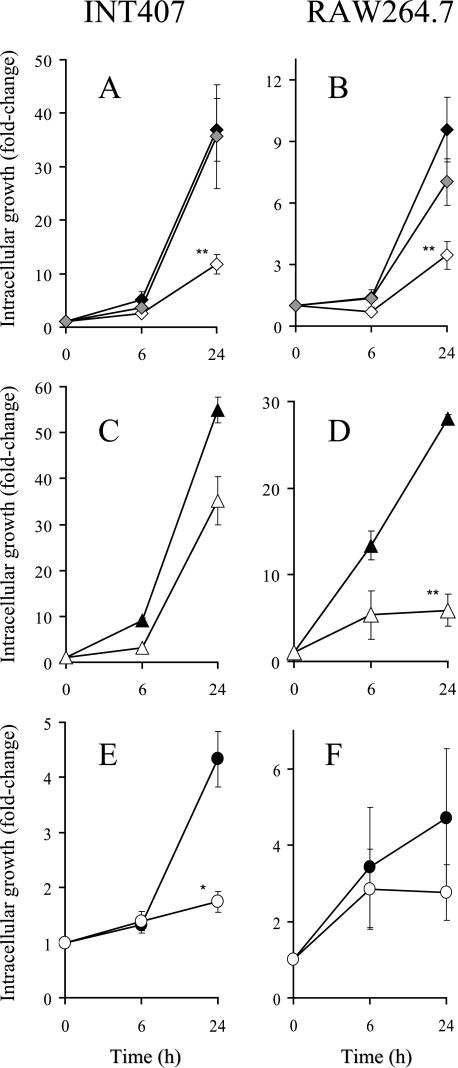
Intracellular survival and replication within epithelial cells and macrophages is an indispensable characteristic of Salmonella pathogenesis. To test whether the loss of RfaH has any influence on intracellular growth, the yield of wild-type SL1344 was compared to that of its rfaH mutant in cultured cell lines INT407 (human intestinal epithelial) and RAW264.7 (mouse macrophage). After 24 h, the numbers of wild-type bacteria recovered from epithelial cells were 35- to 40-fold higher than those of the primary invasive bacteria (Fig. 1A), while the numbers of bacteria present in macrophages had increased 10-fold (Fig. 1B). In contrast, the rfaH mutant showed impaired net growth in both cell lines, increasing 10- and 4-fold in epithelial cells and macrophages, respectively. trans-Complementation of the rfaH mutation restored the ability of serovar Typhimurium SL1344-R1 (ΔrfaH) to grow in epithelial cells and macrophages at wild-type levels. Intracellular growth of wild-type and rfaH mutant strains of Salmonella serovar Enteritidis NCTC13349 (Fig. 1C and D) and Y. enterocolitica (Fig. 1E and F) was determined for the same mammalian cell lines. The rfaH mutants exhibited reduced intracellular yield relative to their isogenic wild-type strains in all cases. These results raise the possibility that the impaired growth potential exhibited by the rfaH mutants could reflect a common mechanism in these different intracellular pathogens.
FIG. 1.
Intracellular growth of bacteria in human intestinal INT407 cells (A, C, and E) and mouse macrophage RAW264.7 cells (B, D, and F). Growth of serovar Typhimurium SL1344 (A and B), serovar Enteritidis NCTC13349 (C and D), and Y. enterocolitica WA-314 (E and F) rfaH mutants (empty symbols) were compared to that of their isogenic parental wild-type strains (filled symbols) over a 24-h period. The trans-complemented mutant of SL1344 (SL1344-R2) is shown with grey symbols. The cultured cell lines were infected as described in Materials and Methods. Intracellular bacterial growth is shown as the change (n-fold) in the number of intracellular bacteria following an additional 6-h or 24-h incubation. Means ± standard errors of the mean (SEM) for four independent experiments are shown. Asterisks indicate statistically significant differences (*, P < 0.05; **, P < 0.01).
Determination of the RfaH regulon.
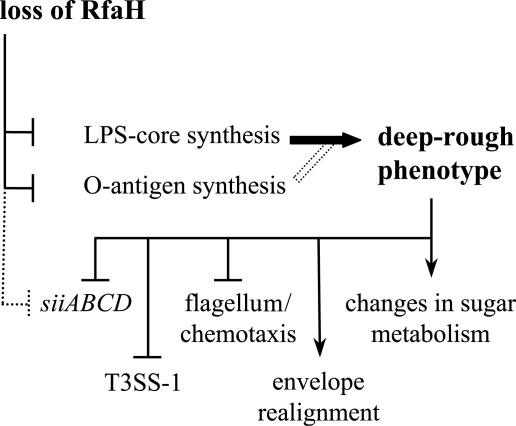
To determine how gene expression was affected by the loss of RfaH, we defined the RfaH-dependent regulon in serovar Typhimurium strain SL1344 and serovar Enteritidis PT4 strain NCTC13349 grown in LB broth culture using a transcriptomic approach. Gene expression of the rfaH mutants of these strains was compared to that of their respective parents at the late log phase of growth (OD600, 0.8). The same pan-serovar SALSA microarrays were used in all experiments (see details in Materials and Methods). The transcriptome of a structural deep-rough LPS mutant (waaG) of SL1344 known to be defective in synthesis of the LPS outer core was also determined to enable us to differentiate between genes directly affected by RfaH and those affected indirectly by the deep-rough LPS phenotype exhibited by rfaH mutants. Genes considered as being directly regulated by RfaH are therefore those which change in the rfaH mutant, not in the waaG mutant, compared to the wild-type strain. Moreover, operons directly regulated by RfaH are supposed to possess the upstream recognition site for RfaH (ops element). However, it is possible that genes directly regulated by RfaH may also be affected (independently) by the waaG mutation. In these cases, the extent of down-regulation through RfaH might not be easily evaluated (see below for siiABCD genes).
The transcriptomic data (see the supplemental material) revealed that 41 genes were differentially expressed in serovar Typhimurium SL1344-R1 (ΔrfaH), and the expression of 150 genes varied in serovar Enteritidis NCTC13349 ΔrfaH relative to the respective parental strains. A comparison of the data from the waaG and rfaH mutants of serovar Typhimurium showed that only two transcriptional units were directly affected by the loss of RfaH (i.e., changes in expression are observed in the rfaH mutant but not the waaG mutant compared to the wild-type strain), waa (formerly rfa) and wba (formerly rfb) (Fig. 2). These results are consistent with former reports showing involvement of RfaH in LPS core and O-antigen synthesis encoded by the waa and wba operons, respectively (6, 33, 53). Furthermore, both of these operons carry the short cis-acting upstream ops element that is required for RfaH-dependent regulation (2, 4).
FIG. 2.
Direct and indirect effects on the transcriptome by the loss of transcriptional regulator RfaH. Indeterminate effects are shown with dotted lines (see text).
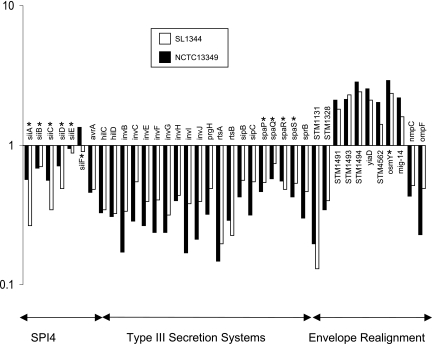
Although the six genes of SPI-4 are preceded by an ops element located just upstream of the first gene (39), results on whether there is a direct effect of RfaH on these genes have been ambiguous. The first four genes (siiABCD) appear to be one transcriptional unit, as their expression shows parallel patterns under various environmental conditions (17, 21, 29, 36, 44). In the waaG mutant of Salmonella serovar Typhimurium, the siiABCD genes were statistically significantly down-regulated compared to the wild type. Although not statistically significant, this pattern of down-regulation could also be observed with the rfaH mutants of serovar Typhimurium and serovar Enteritidis (Fig. 3). As waaG expression itself is regulated by RfaH, it is hard to estimate the WaaG-independent effect of RfaH on siiABCD. Nevertheless, the loss of RfaH elicits only a twofold down-regulation of waaG. The expressional patterns in both the waaG and the rfaH mutants therefore imply that the indirect effect through the deep-rough phenotype in rfaH mutants is likely to be more significant than the loss of direct regulatory effect through RfaH itself. The distal genes (siiEF) of SPI-4 were affected by the loss of neither RfaH nor WaaG, suggesting that they form a transcriptional unit that is detached from siiABCD.
FIG. 3.
Change (n-fold) in the expression of genes involved in SPI-4, T3SS, and envelope realignment in the rfaH mutants of serovar Enteritidis PT4 strain NCTC13349 and serovar Typhimurium strain SL1344 compared with their respective wild-type strains. *, genes which do not pass the statistical filtering.
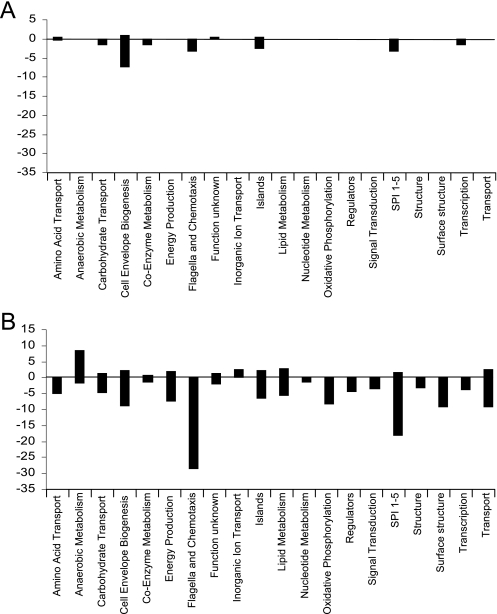
None of the remaining operons down-regulated in the serovar Typhimurium or serovar Enteritidis rfaH mutants carry the upstream ops element, and all these operons showed greater down-regulation in the structural LPS mutant strain (waaG) than the rfaH mutant, suggesting that the deep-rough phenotype exhibited by rfaH mutants is responsible for the changes in expression of these operons (Fig. 2). To determine the patterns of expression indirectly regulated by RfaH, the genes which changed in expression in the rfaH and waaG mutants were sorted into functional categories. The percentage of genes changing in each mutant in each category can be seen in Fig. 4, and many are discussed below. A full list of the genes showing differential expression can be found in Table S2 in the supplemental material. Several operons involved in modulation of the cell surface were down-regulated in the rfaH and waaG mutants. Operons down-regulated in both serovar Typhimurium and serovar Enteritidis rfaH mutants include known (aer, trg, tsr, and several che genes) and putative (STM2314, STM3138, and STM3216) chemotaxis genes as well as middle and late genes of flagellar biosynthesis (fli, flg, flh, and mot loci). Although an overall RfaH-dependent down-regulation of these genes is apparent, few of them reached the statistical threshold. However, it was clear that these genes were statistically significantly down-regulated in the waaG mutant.
FIG. 4.
Gene changes in mutant strains compared to the wild-type strain. The number of genes changing in each functional category for SL1344 ΔrfaH (A) and NCTC13349 ΔrfaH (B) compared to their respective wild types is shown.
Similarly, a general down-regulation of the type III secretion system 1 (T3SS-1) could be detected for rfaH mutants of serovar Typhimurium and serovar Enteritidis. Operons encoding the needle complex (prg), the export apparatus (inv-spa), and the secreted effectors (avrA, sip), as well as prominent regulatory genes (hilC, hilD, sprB, rtsA) of the system, were similarly down-regulated (Fig. 3). The general down-regulation of the flagellum/chemotaxis and T3SS-1 systems in the serovar Typhimurium and serovar Enteritidis rfaH mutants, as well as a more pronounced down-regulation of both systems in the waaG mutant, suggest an indirect effect of rfaH on these genes, which originates from the deep-rough phenotype (Fig. 2).
Other effects of the deep-rough phenotype were directed toward the realignment of the envelope. STM1131, STM1328, nmpC, and ompF, which encode proteins found in the outer membrane, were down-regulated, whereas the gene for the outer membrane lipoprotein, yiaD, was up-regulated (Fig. 3). These genes were significantly differently down regulated in the waaG mutant and showed the same tendency for down-regulation in the rfaH mutants. These findings correlate with previous reports that deep-rough mutants show a decrease of protein content paralleled by an increase of lipoproteins in the outer membrane (45). Such changes in gene expression may aim to compensate for a “leaky” cell wall evoked by the deep-rough phenotype. This hypothesis is supported by the up-regulation of virK and the adjacent gene, mig-14, encoding an inner membrane protein. Products of both virK and mig-14 are involved in resistance to antimicrobial peptides and may therefore play a role in the intracellular survival and replication of Salmonella (7). A similar compensatory mechanism may also account for the up-regulation of osmY (STM4561) and the possibly cotranscribed adjacent gene (STM4562), which encode a hyperosmotically inducible periplasmic protein and a putative inner membrane protein, respectively. The ABC-type transport system encoded by STM1491 to -94 was also found to be up-regulated. The first two genes of this operon show homology to proline/glycine betaine transport systems, which are known to be involved in resistance to osmotic shock. Given the observed up-regulation of osmY, the up-regulation of this operon may be due to a role played in osmotic resistance.
Numerous sugar transport systems were differentially expressed in the rfaH and waaG mutants. Down-regulation of most of these transporters may reflect the reduced demand for sugars due to altered LPS core and O-antigen synthesis.
The gene adjacent to rfaH (yigW) was highly up-regulated in both rfaH mutants, which may be a result of transcriptional read-through from the strong cat promoter inserted into rfaH. This hypothesis is supported by the unaltered expression of yigW in the waaG mutant.
The microarray data suggest that RfaH affects the transcription of genes required for outer membrane composition and LPS production. Therefore, experiments were done to determine the effect of RfaH on LPS biosynthesis, intracellular growth, susceptibility to antimicrobial peptides, and swarming motility.
Influence of genes involved in LPS synthesis on invasion and intracellular yield.
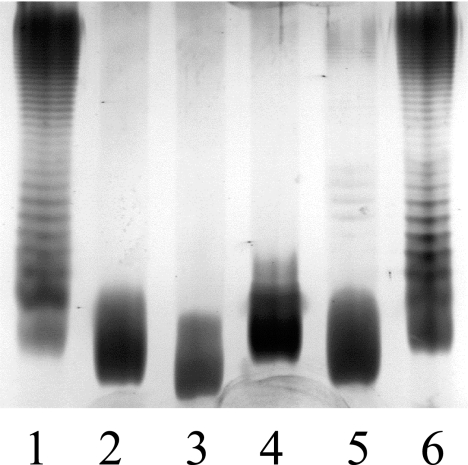
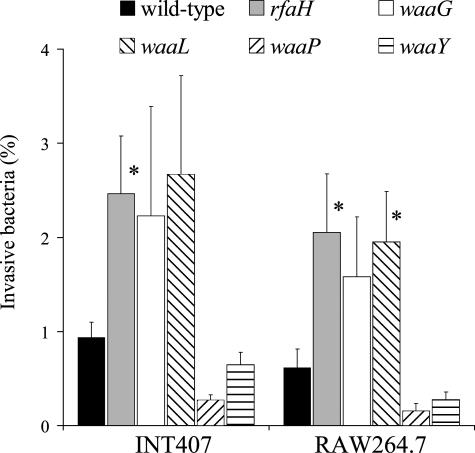
The microarray data showed that operons encoding LPS biosynthesis are directly down-regulated in the serovar Typhimurium and serovar Enteritidis rfaH mutants. To attribute a role of the waa genes in the attenuated phenotype exhibited by the rfaH mutant (41), genes involved in the LPS core biosynthesis (waaG, waaY, and waaP) or the ligation of O antigens to the core region (waaL) were deleted in SL1344. The resulting LPS phenotypes are shown in Fig. 5. The influence of mutations in rfaH or the LPS biosynthesis genes on invasiveness of epithelial cells and macrophages by Salmonella was assessed (Fig. 6). The rfaH, waaG, and waaL mutants, which have no detectable O antigens (Fig. 5), showed highly increased invasiveness of both epithelial cells (INT407) and macrophages (RAW264.7). Conversely, mutants retaining O antigens (waaY, waaP) did not considerably differ in invasiveness from the wild-type strain. Although the waaP mutant showed highly decreased amounts of O antigens, due to the requirement of WaaP-dependent phosphorylation at subsequent steps of LPS synthesis (61), its invasiveness was not increased relative to that of the wild type.
FIG. 5.
Silver staining of LPS molecules exhibited by wild-type strain SL1344 (lane 1) and its isogenic rfaH (lane 2), waaG (lane 3), waaL (lane 4), waaP (lane 5), and waaY (lane 6) mutants.
FIG. 6.
Invasion of INT407 and RAW264.7 cells by SL1344 wild-type, rfaH, and different structural LPS mutants. Data are expressed as percentages of primary inoculum recovered after a 2-h or 30-min invasion of INT407 epithelial cells or RAW264.7 macrophages, respectively. Means ± SEM for six independent experiments are shown. *, statistically significant differences between the wild-type and mutant strains.
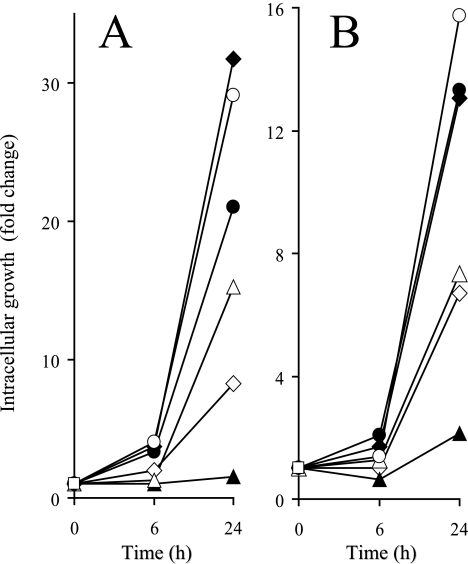
As the effect of the LPS core mutations on the intracellular yield of serovar Typhimurium has not been defined, the intracellular net growth of the waaG, waaY, waaP, and waaL mutants was compared to that of the parental wild-type strain and of the rfaH mutant in two eukaryotic cell lines. The LPS structure appeared to have a major influence on the intracellular yield of Salmonella, both in epithelial cells (Fig. 7A) and macrophages (Fig. 7B). Loss of O antigens with a retained intact core structure (waaL), as well as lack of modifications of the heptose residues located within the main branch (waaP and waaY), had much less effect on intracellular yield than truncation of the main branch at the level of the inner core (waaG). None of the mutants showed an altered MIC to gentamicin or resistance to Triton X-100 at the conditions used during cell culture assays (data not shown), indicating that these factors could not explain the differences in intracellular net growth exhibited by the five mutants.
FIG. 7.
Intracellular growth of SL1344 (filled diamond) and its isogenic rfaH (empty diamond), waaG (filled triangle), waaL (empty triangle), waaP (filled circle), and waaY (empty circle) mutants in human intestinal INT407 cells (A) and mouse macrophage RAW264.7 cells (B). Cells were infected as described in Materials and Methods. Intracellular bacterial growth is denoted as the change (n-fold) in the number of intracellular bacteria following an additional 6-h or 24-h incubation. Means for six independent experiments are shown.
Impaired intracellular yield of waa and rfaH mutants of serovar Typhimurium is a consequence of increased susceptibility to antimicrobial peptides.
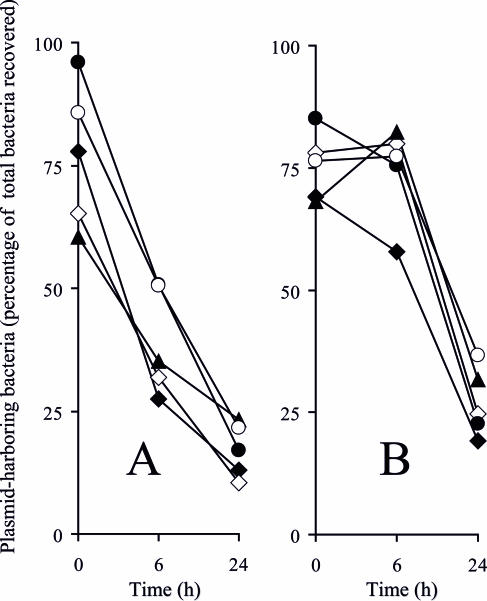
The observed decreased intracellular net growth of the mutated strains could reflect either reduced intracellular replication or the increased vulnerability of Salmonella to intracellular antimicrobial products. The up-regulation of mig-14 and virK indicated by the microarray data suggests that changes in susceptibility to antimicrobial peptides may be the reason for the decreased numbers of bacteria recovered from the eukaryotic cells. To monitor the replication rate of the bacteria in the mammalian cells, the segregation levels of pLDR8, a low-copy-number plasmid unable to replicate under the conditions used, were determined for the LPS mutants and the wild-type strain. Regardless of the total bacterial number recovered (Fig. 7), the levels of plasmid segregation were not significantly different for any mutant strains throughout the study period (Fig. 8), indicating that rates of intracellular replication were similar for these strains. Moreover, in the case of the deep-rough mutants, there was a significant decrease in the numbers of plasmid-harboring bacteria following the 24-h intracellular incubation (data not shown). These data imply that the reduction of the total amount of bacteria recovered at 24 h postinfection was likely due to an increased vulnerability of the mutants to intracellular antimicrobial products.
FIG. 8.
Segregation of pLDR8 during intracellular replication within human intestinal INT407 cells (A) and mouse macrophage RAW264.7 cells (B). SL1344 (filled diamond) and its isogenic rfaH (empty diamond), waaG (filled triangle), waaP (filled circle), and waaY (empty circle) mutants were transformed with pLDR8 and grown at a permissive temperature (30°C). Cell lines were infected and incubated at 37°C, a nonpermissive temperature for plasmid replication. The percentage of plasmid-harboring intracellular bacteria was determined at 6 h and 24 h by replica plating onto selective plates.
The LPS mutations shown to have the most pronounced effect on intracellular survival had formerly been described to cause the deep-rough phenotype (60, 62). To test how intracellular yield of the LPS mutants correlates with susceptibilities to various environmental stressors, MICs of a detergent (SDS), an oxidative agent (H2O2), and a small cationic antimicrobial peptide (polymyxin B) were determined. The results are summarized in Table 2. In agreement with previous reports on E. coli (62), LPS core mutants (waaG, waaP) exhibited high susceptibilities to SDS compared to the smooth wild-type strain. Loss of the O-antigen chains alone (waaL), on the other hand, resulted in a relatively minor change in resistance to SDS. No impaired resistance to oxidative stress elicited by H2O2 could be detected for any of the five mutants. In fact, resistance was slightly increased in the waaP mutant. Most notably, a marked difference was seen in resistance to polymyxin B, a small cationic peptide used to mimic effects of similar antimicrobial substances produced by mammalian cells. The MIC of this substance was approximately eightfold lower for the rfaH mutant than for the wild-type strain. The susceptibilities of the LPS core mutants exhibiting the deep-rough phenotype were increased even more (up to 32-fold), whereas those of the waaL and waaY mutants were only slightly increased (2-fold).
TABLE 2.
Susceptibility to different antimicrobial substances, serum resistance, swarming motility, and virulence of SL1344 and its derivatives
| Strain | MICa
|
Serum resistanceb | Swarming motilityc | Virulence (LD50)d | ||
|---|---|---|---|---|---|---|
| SDS (mg/ml) | H2O2 (mM) | Polymyxin B (μg/ml) | ||||
| SL1344 | >100 | 0.64 | 5 | 100.3 ± 17 | 29.6 ± 1.8 | 2 × 104 |
| SL1344-R1 (rfaH) | 3.2 | 0.64 | 0.64 | 0.06 ± 0.02 | 1.7 ± 0.3 | >109 |
| SL1344 waaG | <0.1 | 0.64 | 0.16 | 0.55 ± 0.4 | 0 | >109 |
| SL1344 waaL | 25 | 0.64 | 2.5 | 16.9 ± 0.75 | 0.9 ± 0.1 | >109 |
| SL1344 waaP | <0.1 | 1.25 | 0.64 | 0.16 ± 0.11 | 7.3 ± 2.0 | >109 |
| SL1344 waaY | 12.5 | 0.64 | 2.5 | 96.2 ± 11.9 | 28 ± 5.6 | >109 |
MICs were determined as described in Materials and Methods.
Percent survival following a 1-h incubation in 10% serum.
Average diameter (mm) ± SEM of swarming colony following an 8-h incubation.
Virulence is expressed as the LD50 value (CFU) for the murine model of typhoid.
Swarming motility.
Lipopolysaccharide structure has been reported to play a crucial role in the swarming motility of Salmonella (51), and the microarray data showing down-regulation of genes involved in chemotaxis and flagellum production suggest that the rfaH and LPS mutants may vary in motility. A plate assay was used to determine whether the down-regulation of genes encoding LPS synthesis and flagellum/chemotaxis correlates with swarming motility of the rfaH as well as various structural LPS mutants (Table 2). The swarming motility of the mutants seemed to be proportional to the level of truncation of LPS molecules. The waaY mutant that retained all O antigens showed swarming motility similar to that of the wild type, while loss of WaaP resulted in reduced amounts of O antigens and reduced swarming ability (Fig. 5 and Table 2). Truncation of the LPS branch at the level of the inner core (waaG) or the core-O-antigen junction (waaL) resulted in a complete or almost complete inability of the bacteria to swarm. Deletion of rfaH elicited a highly decreased yet detectable swarming phenotype.
Virulence.
The mouse model of typhoid was used to assess the virulence of SL1344 and its LPS mutant derivatives (structural and regulatory). The LD50 values were calculated from lethality rates elicited by different infectious doses (see Materials and Methods). While the wild-type strain was highly virulent when administered by the oral route, the rfaH mutant and all structural LPS mutants were attenuated (Table 2). Apart from those mice infected with the wild-type strain, only one of five mice infected with the highest dose (109 CFU) of SL1344 waaY died. All other mice infected with any mutant at any dose survived.
DISCUSSION
Due to the serious medical and veterinary problems caused by S. enterica worldwide, we need to be able to specifically protect humans and animals against S. enterica infection. The recent emergence of multiresistant Salmonella strains has emphasized this necessity (46). Vaccination has long been used against the human pathogen S. enterica serovar Typhi. In some countries, vaccination strategies against other serovars are being developed for use in animal husbandry. Vaccines currently available against Salmonella infection, however, do not confer optimal protection. Therefore, several rationally attenuated live vaccine candidates have been investigated for the past two decades. These include auxotrophic mutants (aroA, aroCD, purA) and strains deficient in stress responses (htrA) or adenylate cyclase functions (cya, crp) (reviewed in reference 22). Mutants lacking known virulence genes (31) or affected in virulence gene regulation have also been tested as promising novel vaccine candidates. The latter group includes mutants lacking regulation of virulence by PhoP-Q (37), Dam (14), SlyA (28), and SirA/HilA (1).
An ideal live vaccine strain combines efficient immunogenicity with minimal reactogenicity. Consequently, strains with mutations of genes which regulated virulence factors could make ideal vaccine candidates, as they may down-regulate the expression of virulence factors to a level resulting in avirulence of the strain but still retain immunogenicity of key antigens. We have previously shown that an rfaH mutant of the S. enterica serovar Typhimurium prototype strain fulfils both of these criteria; the mutant becomes attenuated and, at the same time, elicits protective immunity against a subsequent challenge with the wild-type strain (41). Here, we have used a transcriptomic approach to determine the manner of virulence attenuation caused by the loss of RfaH.
We have shown that the absence of RfaH results in a decreased intracellular net growth of serovar Typhimurium in epithelial and macrophage cells (Fig. 1). Furthermore, this phenomenon was observed for other intracellular pathogens, namely, serovar Enteritidis and Y. enterocolitica, showing that the impaired intracellular yield of rfaH mutants is common to other intracellular pathogens (Fig. 1). As intracellular growth is a prominent virulence characteristic of Salmonella, this phenotype may contribute to virulence attenuation of rfaH mutants.
Transcriptomic analysis of rfaH mutants showed that, in contrast to results with global regulatory mutants, expression of relatively few genes is altered by the loss of RfaH. Moreover, change in expression of most of the affected genes is an indirect effect of the deep-rough phenotype exhibited by the rfaH mutants (Fig. 2). These compensatory changes were identified by determining the transcriptome of a structural deep-rough mutant of SL1344 lacking WaaG. The direct regulatory influence of RfaH could be shown for operons encoding LPS core and O-antigen synthesis, in agreement with former reports (6, 33, 53). The direct effect of RfaH on the siiABCD genes located on SPI-4 is ambiguous. SPI-4 comprises six genes (siiABCDEF) which show homology to components of a type 1/ABC transporter system. The SPI-4 cluster possesses the upstream cis-acting ops element (39) required for RfaH activity (2) and shows slight down-regulation in both rfaH mutants investigated. The expression of siiABCD, however, is down-regulated to a greater extent in the waaG mutant than in the rfaH mutant, indicating an indirect effect of the deep-rough phenotype. The expression of the remaining two genes in the island (siiEF) is unaltered by the rfaH or waaG mutations. Our data suggest, therefore, that siiABCD forms one transcriptional unit, which is transcriptionally uncoupled from siiEF. This idea is supported by analysis of the DNA sequence of siiABCD, which suggests that stop codons overlap start codons, and previous reports showing that the siiABCD genes exhibit parallel changes under different culture conditions. These changes include growth in the presence of butyrate (21), inside macrophages (17), and in Fis (29) and IHF (36) mutants. SPI-4 has been implicated in intestinal colonization of calves (39), and so the impaired colonization of LPS mutants may be due to the subsequent down-regulation of the siiABCD genes (57).
The remaining functional groups of genes affected by the absence of rfaH are clearly down-regulated through the deep-rough phenotype (Fig. 2). These include sugar transport, envelope realignment, T3SS-1, and the flagellum/chemotaxis system. Due to decreased LPS synthesis, the bacterial demand for sugars is much reduced, which could account for the down-regulation of sugar transporters. Moreover, the (deep-)rough phenotype has a huge impact on the stability of the bacterial envelope (19), rearrangement of which can be considered a clear compensatory change. Interestingly, virK and mig-14 have been shown to be involved in resistance to antimicrobial peptides (7). Up-regulation of these genes in the rfaH and waaG mutants suggests that these genes may be involved in the preservation of the envelope barrier.
Parallel down-regulation of the SPI-1 and flagellar systems was observed for deep-rough mutants. These two secretion systems show structural homology to each other (23). Moreover, they are transcribed under similar conditions and affected by several global regulators, including CsrA, Fis, and H-NS (recently reviewed in reference 50). In the waaG mutant, rtsA and rtsB are both down-regulated. These genes are regulated in a manner similar to that of the SPI-1 regulators HilC and HilD and the master regulator HilA (15). RtsA and RtsB have also been shown to regulate SPI-1 genes and flhDC (which direct flagellum biosynthesis), respectively (16). Furthermore, the flagellar regulator FliZ was shown to affect hilA and, hence, SPI-1 expression (26). It is uncertain why a mutation in rfaH or waaG affects transcription of the SPI-1 and flagellum/chemotaxis systems, but there is a clear regulatory link between these type III secretion systems and the deep-rough phenotype of LPS mutants.
SPI-1, SPI-4, and LPS genes have been shown to be up-regulated during swarming motility (54). Rough mutants of Salmonella are unable to swarm, as the lack of LPS results in insufficient surface wetness (51). The transcriptomic analysis of the waaG mutant showed down-regulation of SPI-1, SPI-4, and the flagellar system, as would be expected in a nonswarming strain. Recently, Wang et al. showed that the flagellar system itself senses surface wetness, blocks expression of class 3 flagellar genes (55), and down-regulates SPI-1 and SPI-4 genes. The definite regulators connecting expression of these systems still need to be identified. A recent report showed that the SPI-1 master regulator HilA has a binding site on SPI-4 (12), and so HilA may be an important regulator co-utilized by these systems.
Reduced intracellular yield could be the mechanism whereby rfaH mutants are attenuated, but what is responsible for this phenotype? Neither SPI-1 nor SPI-4 affect intracellular growth of Salmonella (10, 39). Moreover, nonflagellated mutants were shown to be fully virulent in the mouse model of salmonellosis (34), suggesting that intracellular net growth is unaffected by these mutations. Therefore, the rough LPS phenotype might be directly responsible for the reduced intracellular yield. Mutations (including rfaH) provoking the loss of O antigens and/or part of the LPS core resulted in highly elevated invasiveness (Fig. 6). This corroborates observations of Shigella LPS mutants (56), in which increased T3SS at the host cell membrane was proposed to be responsible for higher invasiveness. Although LPS mutants seem to be defective in net intracellular growth (Fig. 7), no impaired growth in LB cultures can be detected (data not shown). The decreased intracellular yield seems to be related to the level of truncation of LPS molecules. Reduced intracellular recovery could, theoretically, originate from a decreased replication or a higher intracellular death rate. With LPS mutants carrying a plasmid which is unable to replicate at the conditions used, we were able to show that intracellular replication of the LPS mutants did not differ significantly from that of the wild-type strain. Furthermore, we confirmed former reports for E. coli (62) showing that deep-rough LPS mutants exhibit enhanced susceptibilities to antimicrobial peptides (Table 2), such as polymyxin B. However, resistance to oxidative stress is affected in neither the rfaH nor the waaG mutants, supporting the hypothesis that killing by naturally occurring antimicrobial peptides impedes intracellular net growth of rough mutants.
The rfaH mutant strains may be good candidates for the generation of a cross-immunogenic vaccine. The major immunogenic molecules on the surface of S. enterica are the lipopolysaccharide and flagella, antigenic variants of which serve as the basis for classification into serovariants. Indeed, humoral protection against Salmonella is characterized by the bulk of antibodies directed against the O determinant (8, 18), and cross-immunity among strains belonging to different serovariants is normally not elicited at high levels (25, 49). We propose that down-regulation of both flagellar and O antigens in rfaH mutants will allow a more efficient immune response to those antigens that otherwise possess minor immunogenicity; this theory is supported by previously published data which show cross-reactivity of immune sera obtained from mice that had been vaccinated by SL1344-R1 (rfaH) to heterologous serovariants (41). Developing an immunization strategy against conserved antigens of salmonellae that allows protection against multiple serovariants is of high importance due to the huge number of serovars capable of colonizing farm animals, with a consequent danger of infecting humans (30).
Generating a Salmonella strain which is safe (virulence attenuated) and also retains its immunogenicity is the biggest challenge in the development of live vaccine candidates. The role of LPS in the virulence of enteric bacteria is well documented (18, 27, 32, 40, 48, 51). Usually, rough mutants are avirulent to a degree that makes them inappropriate as live oral vaccine candidates (18, 35), as impaired intestinal colonization, susceptibility to bile and antimicrobial peptides, and serum sensitivity might all contribute to the inefficient presentation of LPS mutants to the immune system. On the other hand, smooth virulent strains that possess complete LPS structures overwhelm the immune system with enormous amounts of O antigens, and so no adequate immune response against minor antigens is elicited. Although silver staining could not detect any O antigens produced by the rfaH mutant, phenotypic tests, including swarming, susceptibility to antimicrobial compounds, and intracellular growth, suggest that the truncation of the LPS molecules is partial. The mechanism of transcriptional antitermination might justify this observation—the absence of the RfaH regulator results in operon polarity; however, a complete lack of any gene products is not expected (53). The highly reduced amount of O antigen in rfaH mutants allows a higher invasiveness of antigen-presenting cells (Fig. 6), but net intracellular growth, which is affected mainly by the LPS core structure, is impaired, resulting in attenuation of the strain. The waaG mutant, which has a discrete truncation of all LPS molecules at the level of the inner core, shows the same phenotypes as the rfaH mutant but to a much greater extent. Therefore, whereas rfaH mutants may survive for as long as 2 weeks in vivo (41), allowing development of an efficacious immune response, the extent of the attenuation of a waaG mutant is so great that the bacteria are quickly cleared and no effective immune response can be mounted (unpublished observation). This setting is similar to that of the “sword and shield” theory, according to which bacteria try to fine-tune the length of the LPS chains in order to provide enough protection and, at the same time, allow the action of surface virulence factors (56). In this case, the loss of RfaH appears to elicit a balanced situation in which the dramatically reduced amount of full-length LPS attenuates virulence, but the remaining LPS molecules allow sufficient in vivo survival for antigen presentation. Furthermore, down-regulation of major immunogenic molecules on the surface of the rfaH mutant bacteria may improve immunogenicity of conserved protein antigens located on the bacterial surface of other serovars, thus improving cross protection.
Further experiments are needed to identify the antigens that are responsible for the observed cross-reactivity upon vaccination and to assess whether they, in fact, provide in vivo cross-protection against a heterologous challenge. As Salmonella strains have been shown to be very effective in delivering heterologous antigens (22, 38), cross-immunity may be further enhanced by the overexpression of protective antigens. Lack of cross-protective immunity between different serotypes is a major problem in enteric pathogens other than Salmonella; therefore, it is worth testing rfaH mutants as live vaccines against other pathogens belonging to the Enterobacteriaceae.
Supplementary Material
Acknowledgments
The cell line RAW264.7 was kindly provided by Michael Hensel. We are grateful to Sacha Lucchini and Yvette Wormstone for help during the microarray experiments and Rózsa Lajkó for technical assistance.
This work was performed in frame of the EuroPathoGenomics network of excellence.
This work was supported by grants OTKA F048526 (to G.N.), OTKA T037833 (to L.E.), SFB479, and TP A1 (to the Wüzburg group) and a BBSRC core strategic grant (to J.C.H.). G.N. was supported by Bolyai and Humboldt fellowships.
Editor: A. D. O'Brien
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Ahmer, B. M., J. van Reeuwijk, P. R. Watson, T. S. Wallis, and F. Heffron. 1999. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol. Microbiol. 31:971-982. [DOI] [PubMed] [Google Scholar]
- 2.Artsimovitch, I., and R. Landick. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109:193-203. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, M. J., C. Hughes, and V. Koronakis. 1996. Increased distal gene transcription by the elongation factor RfaH, a specialized homologue of NusG. Mol. Microbiol. 22:729-737. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, M. J., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. [DOI] [PubMed] [Google Scholar]
- 5.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 6.Brazas, R., E. Davie, A. Farewell, and L. I. Rothfield. 1991. Transcriptional organization of the rfaGBIJ locus of Salmonella typhimurium. J. Bacteriol. 173:6168-6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodsky, I. E., N. Ghori, S. Falkow, and D. Monack. 2005. Mig-14 is an inner membrane-associated protein that promotes Salmonella typhimurium resistance to CRAMP, survival within activated macrophages and persistent infection. Mol. Microbiol. 55:954-972. [DOI] [PubMed] [Google Scholar]
- 8.Brown, A., and C. E. Hormaeche. 1989. The antibody response to salmonellae in mice and humans studied by immunoblots and ELISA. Microb. Pathog. 6:445-454. [DOI] [PubMed] [Google Scholar]
- 9.Carter, H. D., V. Svetlov, and I. Artsimovitch. 2004. Highly divergent RfaH orthologs from pathogenic proteobacteria can substitute for Escherichia coli RfaH both in vivo and in vitro. J. Bacteriol. 186:2829-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, K., C. C. Kim, and S. Falkow. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect. Immun. 73:5438-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Keersmaecker, S. C., K. Marchal, T. L. Verhoeven, K. Engelen, J. Vanderleyden, and C. S. Detweiler. 2005. Microarray analysis and motif detection reveal new targets of the Salmonella enterica serovar Typhimurium HilA regulatory protein, including hilA itself. J. Bacteriol. 187:4381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diederich, L., L. J. Rasmussen, and W. Messer. 1992. New cloning vectors for integration in the lambda attachment site attB of the Escherichia coli chromosome. Plasmid 28:14-24. [DOI] [PubMed] [Google Scholar]
- 14.Dueger, E. L., J. K. House, D. M. Heithoff, and M. J. Mahan. 2001. Salmonella DNA adenine methylase mutants elicit protective immune responses to homologous and heterologous serovars in chickens. Infect. Immun. 69:7950-7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellermeier, C. D., J. R. Ellermeier, and J. M. Slauch. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57:691-705. [DOI] [PubMed] [Google Scholar]
- 16.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 18.Fierer, J., and D. G. Guiney. 2001. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 107:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frirdich, E., and C. Whitfield. 2005. Lipopolysaccharide inner core oligosaccharide structure and outer membrane stability in human pathogens belonging to the Enterobacteriaceae. J. Endotoxin Res. 11:133-144. [DOI] [PubMed] [Google Scholar]
- 20.Gahring, L. C., F. Heffron, B. B. Finlay, and S. Falkow. 1990. Invasion and replication of Salmonella typhimurium in animal cells. Infect. Immun. 58:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gantois, I., R. Ducatelle, F. Pasmans, F. Haesebrouck, I. Hautefort, A. Thompson, J. C. Hinton, and F. Van Immerseel. 2006. Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl. Environ. Microbiol. 72:946-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garmory, H. S., K. A. Brown, and R. W. Titball. 2002. Salmonella vaccines for use in humans: present and future perspectives. FEMS Microbiol. Rev. 26:339-353. [DOI] [PubMed] [Google Scholar]
- 23.Gophna, U., E. Z. Ron, and D. Graur. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151-163. [DOI] [PubMed] [Google Scholar]
- 24.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hormaeche, C. E., P. Mastroeni, J. A. Harrison, R. Demarco de Hormaeche, S. Svenson, and B. A. D. Stocker. 1996. Protection against oral challenge three months after i.v. immunization of BALB/c mice with live Aro Salmonella typhimurium and Salmonella enteritidis vaccines is serotype (species)-dependent and only partially determined by the main LPS O antigen. Vaccine 14:251-259. [DOI] [PubMed] [Google Scholar]
- 26.Iyoda, S., T. Kamidoi, K. Hirose, K. Kutsukake, and H. Watanabe. 2001. A flagellar gene fliZ regulates the expression of invasion genes and virulence phenotype in Salmonella enterica serovar Typhimurium. Microb. Pathog. 30:81-90. [DOI] [PubMed] [Google Scholar]
- 27.Jacques, M. 1996. Role of lipo-oligosaccharides and lipopolysaccharides in bacterial adherence. Trends Microbiol. 4:408-409. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko, A., M. Mita, K. Sekiya, H. Matsui, K. Kawahara, and H. Danbara. 2002. Association of a regulatory gene, slyA with a mouse virulence of Salmonella serovar Choleraesuis. Microbiol. Immunol. 46:109-113. [DOI] [PubMed] [Google Scholar]
- 29.Kelly, A., M. D. Goldberg, R. K. Carroll, V. Danino, J. C. Hinton, and C. J. Dorman. 2004. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150:2037-2053. [DOI] [PubMed] [Google Scholar]
- 30.Khakhria, R., D. Woodward, W. M. Johnson, and C. Poppe. 1997. Salmonella isolated from humans, animals and other sources in Canada, 1983-92. Epidemiol. Infect. 119:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirkpatrick, B. D., R. McKenzie, J. P. O'Neill, C. J. Larsson, A. L. Bourgeois, J. Shimko, M. Bentley, J. Makin, S. Chatfield, Z. Hindle, C. Fidler, B. E. Robinson, C. H. Ventrone, N. Bansal, C. M. Carpenter, D. Kutzko, S. Hamlet, C. LaPointe, and D. N. Taylor. 2006. Evaluation of Salmonella enterica serovar Typhi (Ty2 aroC-ssaV-) M01ZH09, with a defined mutation in the Salmonella pathogenicity island 2, as a live, oral typhoid vaccine in human volunteers. Vaccine 24:116-123. [DOI] [PubMed] [Google Scholar]
- 32.Licht, T. R., K. A. Krogfelt, P. S. Cohen, L. K. Poulsen, J. Urbance, and S. Molin. 1996. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect. Immun. 64:3811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindberg, A. A., and C. G. Hellerqvist. 1980. Rough mutants of Salmonella typhimurium: immunochemical and structural analysis of lipopolysaccharides from rfaH mutants. J. Gen. Microbiol. 116:25-32. [DOI] [PubMed] [Google Scholar]
- 34.Lockman, H. A., and R. Curtiss III. 1990. Salmonella typhimurium mutants lacking flagella or motility remain virulent in BALB/c mice. Infect. Immun. 58:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyman, M. B., J. P. Steward, and R. J. Roantree. 1976. Characterization of the virulence and antigenic structure of Salmonella typhimurium strains with lipopolysaccharide core defects. Infect. Immun. 13:1539-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangan, M. W., S. Lucchini, V. Danino, T. O. Croinin, J. C. Hinton, and C. J. Dorman. 2006. The integration host factor (IHF) integrates stationary-phase and virulence gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 59:1831-1847. [DOI] [PubMed] [Google Scholar]
- 37.Miller, S. I., J. J. Mekalanos, and W. S. Pulkkinen. 1990. Salmonella vaccines with mutations in the phoP virulence regulon. Res. Microbiol. 141:817-821. [DOI] [PubMed] [Google Scholar]
- 38.Mollenkopf, H., G. Dietrich, and S. H. Kaufmann. 2001. Intracellular bacteria as targets and carriers for vaccination. Biol. Chem. 382:521-532. [DOI] [PubMed] [Google Scholar]
- 39.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 40.Murray, G. L., S. R. Attridge, and R. Morona. 2003. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol. Microbiol. 47:1395-1406. [DOI] [PubMed] [Google Scholar]
- 41.Nagy, G., U. Dobrindt, J. Hacker, and L. Emody. 2004. Oral immunization with an RfaH mutant elicits protection against salmonellosis in mice. Infect. Immun. 72:4297-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy, G., U. Dobrindt, G. Schneider, A. S. Khan, J. Hacker, and L. Emody. 2002. Loss of regulatory protein RfaH attenuates virulence of uropathogenic Escherichia coli. Infect. Immun. 70:4406-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson, D., W. Neill, and I. R. Poxton. 1990. A comparison of immunoblotting, flow cytometry and ELISA to monitor the binding of anti-lipopolysaccharide monoclonal antibodies. J. Immunol. Methods. 133:227-233. [DOI] [PubMed] [Google Scholar]
- 44.Ono, S., M. D. Goldberg, T. Olsson, D. Esposito, J. C. Hinton, and J. E. Ladbury. 2005. H-NS is a part of a thermally controlled mechanism for bacterial gene regulation. Biochem. J. 391:203-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabsch, W., H. Tschape, and A. J. Baumler. 2001. Non-typhoidal salmonellosis: emerging problems. Microbes Infect. 3:237-247. [DOI] [PubMed] [Google Scholar]
- 47.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 48.Shaio, M. F., and H. Rowland. 1985. Bactericidal and opsonizing effects of normal serum on mutant strains of Salmonella typhimurium. Infect. Immun. 49:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, B. P., G. W. Dilling, J. K. House, H. Konrad, and N. Moore. 1995. Enzyme-linked immunosorbent assay for Salmonella serology using lipopolysaccharide antigen. J. Vet. Diagn. Investig. 7:481-487. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, A., G. Rowley, M. Alston, V. Danino, and J. C. Hinton. 2006. Salmonella transcriptomics: relating regulons, stimulons and regulatory networks to the process of infection. Curr. Opin. Microbiol. 9:109-116. [DOI] [PubMed] [Google Scholar]
- 51.Toguchi, A., M. Siano, M. Burkart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trulzsch, K., T. Sporleder, E. I. Igwe, H. Russmann, and J. Heesemann. 2004. Contribution of the major secreted Yops of Yersinia enterocolitica O:8 to pathogenicity in the mouse infection model. Infect. Immun. 72:5227-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, L., S. Jensen, R. Hallman, and P. R. Reeves. 1998. Expression of the O antigen gene cluster is regulated by RfaH through the JUMPstart sequence. FEMS Microbiol. Lett. 165:201-206. [DOI] [PubMed] [Google Scholar]
- 54.Wang, Q., J. G. Frye, M. McClelland, and R. M. Harshey. 2004. Gene expression patterns during swarming in Salmonella typhimurium: genes specific to surface growth and putative new motility and pathogenicity genes. Mol. Microbiol. 52:169-187. [DOI] [PubMed] [Google Scholar]
- 55.Wang, Q., A. Suzuki, S. Mariconda, S. Porwollik, and R. M. Harshey. 2005. Sensing wetness: a new role for the bacterial flagellum. EMBO J. 24:2034-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.West, N. P., P. Sansonetti, J. Mounier, R. M. Exley, C. Parsot, S. Guadagnini, M. C. Prevost, A. Prochnicka-Chalufour, M. Delepierre, M. Tanguy, and C. M. Tang. 2005. Optimization of virulence functions through glucosylation of Shigella LPS. Science 307:1313-1317. [DOI] [PubMed] [Google Scholar]
- 57.West, N. P., P. J. Sansonetti, G. Frankel, and C. M. Tang. 2003. Finding your niche: what has been learnt from STM studies on GI colonization. Trends Microbiol. 11:338-344. [DOI] [PubMed] [Google Scholar]
- 58.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]
- 59.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 60.Yethon, J. A., J. S. Gunn, R. K. Ernst, S. I. Miller, L. Laroche, D. Malo, and C. Whitfield. 2000. Salmonella enterica serovar Typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo. Infect. Immun. 68:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yethon, J. A., D. E. Heinrichs, M. A. Monteiro, M. B. Perry, and C. Whitfield. 1998. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 273:26310-26316. [DOI] [PubMed] [Google Scholar]
- 62.Yethon, J. A., E. Vinogradov, M. B. Perry, and C. Whitfield. 2000. Mutation of the lipopolysaccharide core glycosyltransferase encoded by waaG destabilizes the outer membrane of Escherichia coli by interfering with core phosphorylation. J. Bacteriol. 182:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.