Abstract
The TATA box-binding protein-associated factors (TAFs) are thought to play an essential role in eukaryotic RNA polymerase II transcription by mediating the expression of distinct subsets of genes. In hamster ts13 cells, a single amino acid change in TAFII250, which disrupts its acetyl-transferase activity at the restrictive temperature, alters the transcription of specific genes involved in cell cycle control. Likewise, disruption of the amino-terminal kinase domain of TAFII250 results in transcriptional defects in ts13 cells. However, it was not known whether the acetyl-transferase or kinase domains of TAFII250 modulate specific classes of genes and whether these two domains regulate distinct subsets of genes. Here we have used high-density gene-profiling to identify mammalian transcripts that require either the TAFII250 acetyl-transferase or protein kinase function for proper expression. We found that transcription of at least 18% of genes are differentially expressed at the restrictive temperature. The promoter region of one of these genes was subsequently characterized, and both upstream elements as well as the core promoter were shown to be TAFII250 responsive. We also found that expression of ≈6% of genes in ts13 cells requires a functional TAFII250 amino-terminal kinase domain, but only ≈1% of these hamster genes also require the TAFII250 acetyl-transferase activity. Our results suggest that the two TAFII250 enzymatic activities are important for regulating largely nonoverlapping sets of genes involved in a wide range of biological functions in vivo.
A central component of the transcription initiation complex is the transcription factor IID (TFIID), which is composed of the TATA box-binding protein and numerous TATA box-binding protein-associated factors (TAFs) (1). The central role that TAFs play in modulating transcription is highlighted by the high degree of conservation found in TAF sequence and function in a wide range of evolutionarily diverse organisms (2–5), and by the fact that mutations disrupting TAF function are lethal (4–8). However, deletion or mutation of any one TAF subunit is not expected to produce global defects in transcription. Indeed, the expression of only ≈30% of yeast genes is altered in cells containing a temperature-sensitive mutation in TAFII145, a homolog of human TAFII250 (9). Thus, at least in yeast, the transcription of any particular gene may not require the function of all of the TAFs; rather, each gene may only require the activity of a subset of TAFs for its appropriate expression.
The largest subunit within the TFIID complex, TAFII250, contains at least two enzymatic activities. The acetyl-transferase activity of TAFII250 can acetylate histones H2A, H3, and H4 (10) whereas the amino-terminal kinase activity of TAFII250 can autophosphorylate and transphosphorylate the large subunit of the general transcription factor TFIIF (11, 12). Interestingly, the transcriptionally active form of TFIIF is hyperphosphorylated (13), suggesting that phosphorylation of TFIIF by TAFII250 may be one mechanism for regulating gene expression. In vivo, TAFII250 function is necessary for the expression of genes involved in cell cycle control and apoptosis. The ts13 hamster cell line harbors a single point mutation in the gene encoding TAFII250 (14). When these cells are grown at the restrictive temperature, they rapidly arrest in the G1 stage of the cell cycle and undergo apoptosis (15, 16). Accordingly, expression of the cyclin A, D1, D3, and cdc2 promoters, but not the c-fos promoter, is compromised when ts13 cells are grown at the restrictive temperature (12, 17–19). It has now been shown that the acetyl-transferase activity of TAFII250 is impaired in ts13 cells at the restrictive temperature (20), suggesting that the expression of genes involved in both cell cycle control and apoptosis may depend on the acetyl-transferase activity of TAFII250 for normal expression. These results suggest the possibility that TAFII250 is required for the expression of specific sets of genes and that the majority of these genes may be involved in cell cycle regulation.
The protein kinase activity of TAFII250 also plays an important role in gene regulation in vivo. Point mutations within the amino-terminal kinase domain impair both autophosphorylation as well as transphosphorylation activities (12). These mutations reduce the ability of TAFII250 to rescue ts13 cells at the restrictive temperature (12), indicating that this kinase activity is important for TAFII250 function in vivo. Moreover, a kinase-deficient form of TAFII250 also has a reduced ability to rescue expression of the Cyclin A and cdc2 genes (12). Thus, the TAFII250 amino-terminal kinase domain is involved in transcriptional regulation of at least some genes.
Identifying genes that are modulated in part through TAFII250 should reveal novel insights into the biological basis underlying the cell cycle arrest and apoptosis observed in ts13 cells. Identifying these genes should also allow us to address the interesting question of whether two distinct functional domains of a single TAF can modulate expression of different sets of promoters. Here we use a genome-wide screen in ts13 cells to identify genes whose expression is disrupted by the temperature-sensitive form of TAFII250 (ts-TAFII250) or by mutations that inactivate the amino-terminal kinase domain of TAFII250. We found that expression of ≈18% of genes is disrupted by the ts-TAFII250 whereas expression of only ≈6% of genes is affected by mutations that inactivate the amino-terminal kinase domain. Therefore, as expected, TAFII250 is not globally required for the proper expression of all genes in animal cells. Surprisingly, however, only ≈1% of genes require the integrity of both domains, indicating that different functional domains of TAFII250 regulate distinct subsets of genes in vivo.
Methods
Plasmids.
The murine voltage-dependent anion channel gene 3 (mVDAC3)/CAT reporter (21) was used as a template to generate deletions by using PCR and appropriate primers, and these PCR-generated fragments were directly cloned into G5BCAT (22) digested with HindIII/SacI. Constructs containing the Gadd45 promoter fused upstream of the CAT reporter were also generated by PCR using as template the plasmid pdc45–20 (23). Hybrid mVDAC3 and Gadd45 promoters were generated by PCR and were directly cloned into G5BCAT digested either with HindIII/SacI or PstI/SacI.
Cell Culture and Transfection Assays.
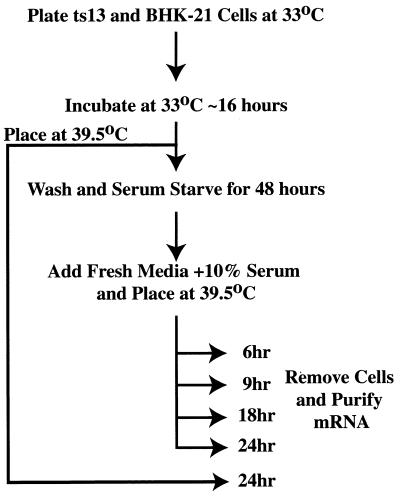
BHK-21 and ts13 cells were maintained in DMEM supplemented with 10% FBS and grown at 33°C. To examine differential gene expression profiles, ts13 or BHK-21 cells were initially seeded in 15-cm2 plates at a density of 2 × 104 cells/ml at 33°C for 12–16 hours. Cells were then treated as illustrated in Fig. 1. Stable ts13 cell lines expressing either hTAFII250N1398 or hTAFII250N1398A2 have been previously described (12) and were maintained in media supplemented with G418 (100 ng/ml).
Figure 1.
Experimental outline. See text for details.
For transient transfection assays, cells were plated in 6-well plates at a density of 2 × 104 cells/ml and were allowed to grow overnight at 33°C. DNA was transiently transfected as described (12), using 2 μg of each reporter plasmid, 2.5 μg of pUHG103–3/HA-hTAFII250, 20 ng of pUHD1-15, 25 ng of SV40-Renilla, and carrier DNA to 6 μg. Cells were placed at 39.5°C for 24–30 hours and were harvested and assayed for CAT and Renilla activity as described (12, 24).
Generation of RNA for Gene Chip Hybridizations.
After harvesting cell samples, total RNA was purified by using Sigma Tri reagent, and mRNA was subsequently purified by using a Qiagen (Chatsworth, CA) oligotex mRNA purification kit. Generation of cRNA for probing of the high-density arrays was essentially as described (25).
Biotin-labeled cRNA was used to probe Affymetrix (Santa Clara, CA) GeneChip Murine 6500K high-density oligonucleotide arrays, representing 6,500 murine genes. Probing, washing, and staining of these arrays were carried out according to standard Affymetrix protocols (25). In our analysis, ts13 and BHK-21 hybridizations were normalized to each other based on the earlier observation that the ts13 mutation does not effect the expression of the majority of genes in ts13 cells (15, 17, 19). We only scored genes as being differentially expressed in ts13 cells at the restrictive temperature if their expression levels increased or decreased two-fold or greater at more than one timepoint (i.e., in multiple independent experiments).
Primer Extension.
To examine the mVDAC3 transcription start site, the oligonucleotide 5′-TAGGCCAACTGTCCGGCTGCGCGTG-3′ was incubated with 20 μg of total RNA purified from ts13 or BHK-21 cells that had been incubated at the restrictive temperature for 24 hours. Primer extension analysis was carried out, and the products were analyzed by denaturing PAGE.
Results
Experimental Outline.
To screen for genes whose expression is selectively dependent on TAFII250 function, a high-density gene expression analysis was undertaken to identify transcripts that are differentially expressed in ts13 cells at the restrictive temperature. Because the ts13 mutation disrupts TAFII250 function (17), our experimental profiles should identify genes that are regulated, at least in part, by TAFII250. ts13 and the parental BHK-21 cells were allowed to grow for ≈16 hours at the permissive temperature (33°C) and then were switched to media containing 0.25% serum (Fig. 1). Within 48 hours, >90% of cells arrest in the G0 stage of the cell cycle. The arrested cells were then permitted to reenter the cell cycle in media supplemented with 10% serum and simultaneously were placed at the restrictive temperature of 39.5°C. We then examined gene expression profiles in ts13 versus BHK-21 cells over a range of time-points after shifting to the restrictive temperature, which is expected to reveal the temporal progression of differential gene expression as the synchronized cell population progresses through the cell cycle.
Identification of Differentially Expressed Genes in ts13 Cells at 39.5°C.
Consistent with the earlier observation that there is no global defect in mRNA synthesis in ts13 cells at the restrictive temperature (15, 17, 19), we found that only ≈18% of genes with detectable expression levels were differentially expressed greater than 2-fold in ts13 cells at the restrictive temperature. It is not clear whether these effects are directly or indirectly caused by inactivating TAFII250 function. Nevertheless, our result demonstrates that expression of a subset of RNA polymerase II transcribed genes in ts13 cells is affected by a single amino acid mutation that disrupts TAFII250 function.
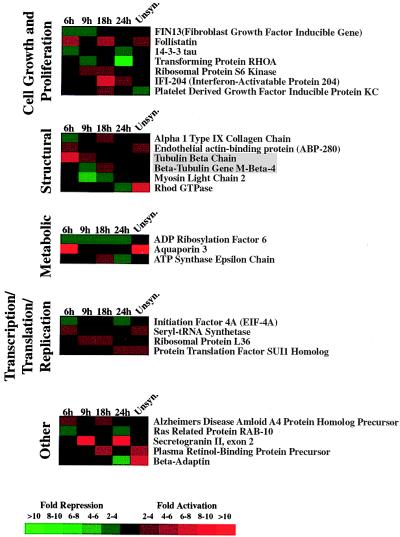
The majority of genes previously found to be differentially expressed in ts13 cells at the restrictive temperature were cell cycle-regulated genes (cyclin A, D1, D3, and cdc2) (12, 17–19), suggesting that TAFII250 may be involved predominantly in the regulation of cell cycle genes. However, our compilation of genes that are differentially expressed in ts13 cells at the restrictive temperature reveals that TAFII250 responsive genes fall into many different functional categories (Fig. 2). Thus, in ts13 cells incubated at the restrictive temperature, TAFII250 is not likely to be restricted to the regulation of a specific class of genes but, rather, functions in a more general and widespread manner.
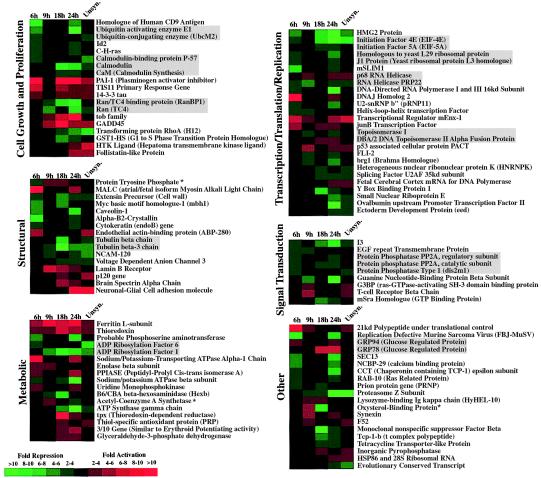
Figure 2.
Genes displaying differential expression in ts13 cells at 39.5°C. The expression pattern observed for each gene at the indicated time-points (6, 9, 18, and 24 hours) when cells were harvested after incubation at the restrictive temperature is shown. “Unsyn.” represents unsynchronized growing cells that were placed at the restrictive temperature for 24 hours. For each gene, the ratio of expression in ts13 cells compared with BHK-21 cells is represented in color according to the key. Gene identifications that are boxed in gray represent gene groups that are functionally related. An asterisk (*) after the gene identification indicates a gene whose expression is both decreased and increased at different times in ts13 cells relative to BHK-21 cells.
Transient Transfection Assays Verify Differential Expression of the mVDAC3 and Gadd45 Genes.
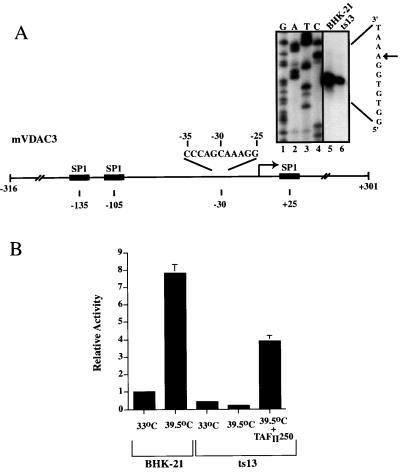
Previous studies had shown that TAFII250 functions through two independent sequence elements: the core promoter and an upstream enhancer element (18). Thus, we were interested in identifying what sequence elements were responsible for the transcriptional defect observed in ts13 cells. Unfortunately, the promoter regions of most of the genes identified in this study are not yet cloned or characterized; thus, we were limited in our selection of promoters for further study. One promoter we chose to characterize was the voltage-dependent anion channel gene 3 (VDAC3). The VDAC3 protein is a voltage-gated pore protein found in the outer membrane of the mitochondria and is involved in ATP signaling (21), and may also play an integral role in apoptosis (26). The promoter region of the murine gene (mVDAC3) had been previously sequenced (Fig. 3A) (21); however, the transcription start site was not identified. Thus, we initially used primer extension analysis to map the transcription start site in both ts13 and BHK-21 cells. Consistent with our observation from the gene-chip analysis, primer extension analysis revealed that mVDAC3 expression is significantly decreased in ts13 cells (Fig. 3A). Based on this start site, the sequence of the mVDAC3 promoter in the −30 region does not contain a canonical TATA sequence (Fig. 3A).
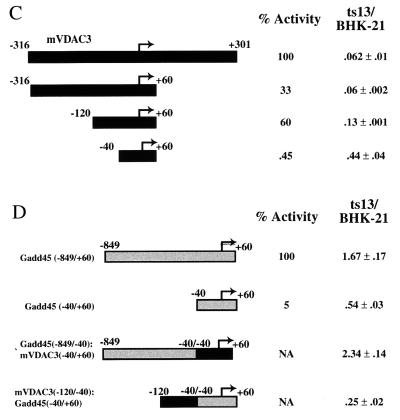
Figure 3.
The mVDAC3 and Gadd45 promoters display temperature-sensitive transcription in ts13 cells. (A) The promoter region of the mVDAC3 gene is shown, along with the location of putative Sp1 binding sites (shaded boxes) (21). Primer extension analysis was performed with total RNA purified from BHK-21 cells (lane 5) and ts13 (lane 6) cells after incubation of synchronized growing cells at the restrictive temperature for 24 hours. The corresponding DNA sequence reactions were analyzed on the same gel and are shown in lanes 1–4. The sequence surrounding the −30 region relative to the transcription start site is shown. (B) A construct containing the mVDAC3 promoter (−316 to +301) fused upstream of the CAT reporter was transiently transfected into BHK-21 or ts13 cells with an SV40-Renilla construct whose activity served as an internal standard and, where indicated, with a construct that expresses wild-type hTAFII250. Cells were incubated at the indicated temperature for 24–30 hours and were harvested, and CAT and Renilla activity was assayed. The activity of this reporter in BHK-21 cells at 33°C was standardized to 1.0, and all other values, along with their associated standard deviation, are presented relative to this. (C) Deletion mapping of the mVDAC3 promoter to identify TAFII250 responsive elements. The indicated deletions were fused upstream of the CAT reporter gene and were transiently transfected into BHK-21 and ts13 cells as described in B. The relative temperature sensitivity of each promoter (with its associated standard deviation), which is presented as a ratio of the level of activity in ts13 cells to the level of activity in BHK-21 cells, is shown. A lower number represents a greater reduction of expression in ts13 cells. The relative activity of each promoter in BHK-21 cells is also shown. (D) Transcriptional activity of hybrid mVDAC3/Gadd45 promoters in ts13 and BHK-21 cells. Constructs contained either the Gadd45 core promoter (−40 to +60), the mVDAC3 upstream enhancer region (−120 to −40) fused to the Gadd45 core promoter (−40 to +60), or the Gadd45 upstream enhancer region (−849 to −40) fused upstream of the mVDAC3 core promoter (−40 to +60). All promoters were inserted directly upstream of the CAT reporter. These constructs were transiently transfected into BHK-21 and ts13 cells, as described in B. The relative temperature sensitivity of each promoter is shown, which is presented as a ratio of the level of activity in ts13 cells relative to the level in BHK-21 cells. A number less than 1.0 represents a greater reduction of expression in ts13 cells relative to BHK-21 cells whereas a number greater than 1.0 represents a greater activity in ts13 cells relative to BHK-21 cells. The relative activity of the Gadd45 promoters is also shown. NA, not applicable because these are promoter fusions.
We used a transient transfection assay to verify that the mVDAC3 promoter displays differential expression in ts13 cells at the restrictive temperature. A construct containing mVDAC3 sequences from −316 to +301 relative to the start site fused directly upstream of the CAT reporter was co-transfected into ts13 or BHK-21 cells with an internal standard reporter (SV40-Renilla). mVDAC3 expression increased when BHK-21 cells were shifted from 33°C to 39.5°C (Fig. 3B) but was reduced ≈2-fold when ts13 cells were similarly shifted in temperature (Fig. 3B). This results in a BHK-21/ts13 differential expression level at the restrictive temperature of ≈16 fold. Importantly, co-transfection of wild-type hTAFII250 with the mVDAC3 construct substantially rescues this down-regulated expression (Fig. 3B). Thus, the reduced level of expression of the mVDAC3 promoter in ts13 cells relative to BHK-21 cells at the restrictive temperature can be attributed to a defect in TAFII250, and, furthermore, the DNA sequences necessary for this effect are contained between −316 and +301 relative to the transcription start site.
We then mapped in greater detail the DNA sequence elements in this promoter that are responsible for the reduction in expression at the restrictive temperature. Deleting a portion of the upstream sequences to produce a construct containing sequences from −120 to +60 gives a reduction in expression level in ts13 cells compared with BHK-21 cells (Fig. 3C). By contrast, the core promoter alone (−40 to +60) displayed significantly less dependence on TAFII250 function, although a residual response to TAFII250 could still be detected (Fig. 3C). These results suggest that some upstream elements of the mVDAC3 promoter are important for modulating the TAFII250 transcriptional response, but the core promoter DNA sequences may also contribute to the TAFII250-dependent transcriptional activation, consistent with the documented biochemical functions of TAFII250 (18).
We next performed promoter swapping experiments to examine whether upstream DNA sequences could confer temperature-sensitive expression when fused with a heterologous core promoter. Our gene-chip analysis showed that expression of the hamster Growth arrest and DNA damage 45 protein (Gadd45) gene is increased in ts13 cells at the restrictive temperature (Fig. 2). We initially assayed the expression level of the hamster Gadd45 promoter in ts13 cells at the restrictive temperature and found that the promoter region spanning from −848 to +60 produced a measurable increase in the expression of the downstream CAT reporter (Fig. 3D). Interestingly, expression of the core Gadd45 promoter alone is moderately reduced in ts13 cells at the restrictive temperature (Fig. 3D). As expected, expression from a hybrid construct containing the Gadd45 upstream promoter region (−848 to −40) fused to the core mVDAC3 promoter (−40 to +60) increases in ts13 versus BHK-21 cells (Fig. 3D). Thus, the −848 to −40 region of the Gadd45 promoter contains upstream enhancer elements that directly or indirectly respond to the temperature-sensitive form of TAFII250. Fusion of the TAFII250-dependent mVDAC3 upstream promoter region immediately upstream of the Gadd45 core promoter results in decreased expression in ts13 cells (Fig. 3D). These results confirm that the upstream promoter sequences of both the mVDAC3 and Gadd45 promoters are important for conferring differential temperature sensitivity in transfected cells, suggesting that some upstream binding activators depend upon specific functions of TAFII250.
The Amino-Terminal Kinase Domain of TAFII250 Is Required for Expression of a Distinct Subset of Genes.
Expression of some genes in ts13 cells at the restrictive temperature requires a fully functional TAFII250 amino-terminal kinase domain (12). We therefore assayed gene expression profiles in ts13 cells stably transformed either with a form of TAFII250 containing mutations in the amino-terminal kinase domain (TAFII250N1398A2) or a form of TAFII250 having wild-type kinase activity (TAFII250N1398) (12). The use of these cell lines allowed us to identify genes whose expression requires the TAFII250 amino-terminal kinase domain for their correct regulation. The same experimental outline was used as shown in Fig. 1.
Of those genes whose expression was detectable, only 5.6% had an expression level significantly different (greater than 2-fold) in stable cell lines expressing the kinase deficient form of TAFII250 compared with cell lines expressing the corresponding wild-type form of TAFII250, and these genes are listed in Fig. 4. As expected, these genes encode for products with a wide range of functions in vivo, indicating that the functional requirement for the amino-terminal kinase activity of TAFII250 is not restricted to any specific functional class of genes. Importantly, most of the genes differentially expressed in the context of the kinase mutation do not overlap with the subset of genes whose expression is impaired by the ts-TAFII250. Thus, the two distinct functional domains of TAFII250 are responsible for mediating transcription of different subsets of genes in mammalian cells.
Figure 4.
Genes in ts13 cells whose expression requires the TAFII250 amino-terminal kinase domain. See Fig. 1 for details. For each gene, the ratio of expression in stable cell lines expressing the wild-type TAFII250 relative to an amino-terminal kinase deficient form of TAFII250 (12) is represented in color according to the key.
Discussion
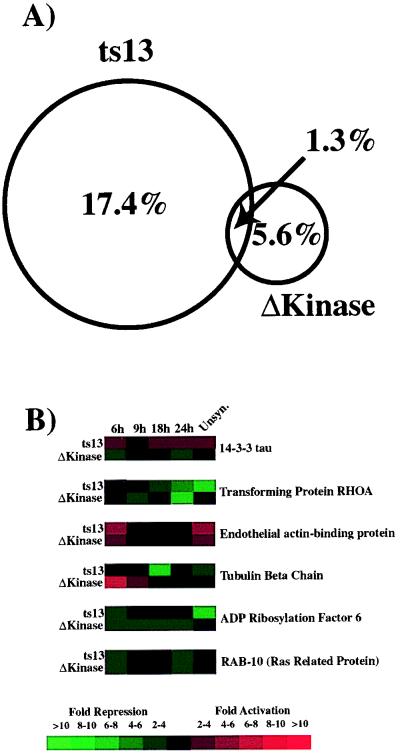
TAFII250 contains at least two distinct enzymatic activities, an acetyl-transferase and a kinase activity (10, 11). Our results indicate that transcription of ≈18% of the genes in hamster cells are differentially expressed when these two distinct TAFII250 functions are disrupted. Moreover, the proper expression of only a small percentage of genes (≈1.3%) requires the function of both of these activities (Fig. 5 A and B). In contrast, the majority of genes whose expression is affected by the kinase-deficient form of TAFII250 does not overlap with those genes whose expression requires the acetyl-transferase function. Thus, in vivo, the regulated expression of distinct sets of genes likely requires different enzymatic activities of TAFII250.
Figure 5.
Comparison of genes whose expression depends on the TAFII250 acetyl-transferase activity versus those whose expression depends on the TAFII250 protein kinase activity. (A) Venn diagram showing the percentage of genes in ts13 cells whose expression requires the TAFII250 acetyl-transferase and/or the amino-terminal kinase activities. (B) This lists all of the genes in ts13 cells whose expression was found to require the activities of both the TAFII250 acetyl-transferase and kinase domains. See Fig. 2 for details.
TAFII250 Kinase and Acetyl-Transferase Activities Modulate Expression of Distinct Subsets of Genes in Vivo.
It was recently demonstrated that the temperature-sensitive form of TAFII250 has significantly decreased acetyl-transferase activity when assayed at the restrictive temperature (20). This suggests that transcriptional modulation of genes involved in cell cycle control and apoptosis requires the acetyl-transferase activity of TAFII250. However, expression of these genes may also require other functional activities of TAFII250 because this single point mutation may also disrupt the functional activity of other domains. TAFII250 can acetylate histones H2A, H3, and H4 (10) and also the general transcription factor TFIIE (27). However, the functional relevance of histone or TFIIE acetylation by TAFII250 has not been demonstrated, nor is it known whether other substrates may be biologically relevant targets of the TAFII250 acetyl-transferase activity.
Because TAFII250 is a component of the TFIID protein complex, it is likely that the activity of the core promoter, and possibly upstream enhancer regions, would be responsive to TAFII250. In vitro experiments have demonstrated that TAFII250 can directly interact with the core promoter DNA and that a TATA box-binding protein/TAF complex containing TAFII250 has a different DNA-binding specificity than TATA box-binding protein alone (28). In support of the core promoter being critical for TAF function, promoter mapping experiments of the cyclin A gene in ts13 cells have revealed that its core promoter is responsive to TAFII250 (18). Similarly, in yeast, the core promoter can render a gene sensitive to mutations that affect the function of TAFII145(29). Because TAFs are also known to function as transcriptional co-activators (30), it would seem likely that upstream enhancer elements may also be TAFII250 responsive. Again, promoter mapping studies of the cyclin A gene in ts13 cells revealed that the upstream binding element for the ATF enhancer protein is TAFII250-responsive (18), suggesting that, in mammalian cells, TAFs play a general role in gene regulation. In contrast, at least some yeast enhancer elements appear to function independently of TAFII145 (29). Here, promoter mapping studies were performed with two genes identified in our gene-profiling analysis. Consistent with observations made with the cyclin A promoter, we find that both the upstream enhancer regions and the core promoters of the mVDAC3 and Gadd45 genes display temperature sensitivity in ts13 cells. Thus, it appears that the acetyl-transferase activity of TAFII250 may contribute to both its core promoter and upstream enhancer-dependent functions.
The amino-terminal kinase activity of TAFII250 was previously shown to be involved in modulating gene expression in vivo (12) and, consistent with this role, we found here that ≈6% of hamster genes require a functionally intact TAFII250 kinase domain for their proper regulation. Genes regulated by the kinase activity of TAFII250 are represented by a diverse group of functional categories. Thus, the kinase activity of TAFII250 appears important for the regulation of genes involved in a wide range of biological processes. Interestingly, expression of fewer genes require the kinase activity compared with the acetyl-transferase activity of TAFII250, suggesting that the TAFII250 kinase may play a more specialized role in gene regulation.
Biological Basis Underlying Cell Cycle Arrest and Apoptosis in ts13 Cells.
One reason for identifying genes that are differentially expressed in ts13 cells at the restrictive temperature is to better understand the biological basis underlying the cell cycle arrest and apoptosis observed previously for TAFII250 mutant cells. At least ≈18% of genes in ts13 cells are differentially expressed at the restrictive temperature, and it was gratifying to find that many of these genes are components of signaling pathways that are directly or indirectly involved in cell cycle control and DNA synthesis. Similarly, in yeast cells bearing a ts-mutation in TAFII145, a homolog of TAFII250, the expression of ≈30% of the genes is disrupted at the restrictive temperature (9). These cells also undergo cell cycle arrest (31), and many of the genes showing differential expression are involved in cell cycle regulation and DNA synthesis (9). Thus, both in yeast and mammalian cells, cycle arrest induced by TAF mutations is likely caused by the misexpression of not just one gene but, rather, to the accumulative misexpression of many genes resulting in the inappropriate regulation of multiple distinct pathways.
These microarray experiments, however, failed to detect a significant reduction in the expression of previously identified cyclin A, D1, D3, or cdc2 genes (12, 17–19). Our inability to measure a difference in the steady state mRNA levels of these genes could be attributable to a number of factors. For instance, we may not be examining mRNA levels at the optimum time, or we may be losing a significant proportion of the hybridization signal due to species differences because we are hybridizing RNA derived from hamster cells with oligonucleotide arrays generated by using mouse sequence information. Therefore, it is likely that the proportion of hamster genes that we find here affected by the temperature-sensitive allele of TAFII250 is an underestimate.
Although there were numerous classes of genes whose expression profiles were affected in ts13 cells at the restrictive temperature, it is clear that many of these can be grouped into functionally related categories. For instance, expression levels of both the Ran/TC4 and the Ran/TC4 binding protein (RanBP1) are coordinately reduced. Both of these gene products are implicated in choreographing progression through the cell cycle, and overexpression of Ran/TC4 is sufficient to reduce cell viability (32). We also observed that members of the protein phosphatase PP2A family are down-regulated at the restrictive temperature. It has previously been suggested that the regulatory subunit of protein phosphatase PP2A is necessary for cell growth (33). More recently, it has been shown that down-regulation of PP2A activity affects the expression of genes involved in regulating entry into S-phase and DNA synthesis (34). A similar defect in expression of the PP2A regulatory subunit was also noted in yeast cells containing a ts-mutation in TAFII145(9). Interestingly, we observe a down-regulation in ts13 cells of both the regulatory and catalytic subunits of PP2A.
ts13 cells incubated at the restrictive temperature also undergo apoptosis (16), suggesting that TAFII250 may be necessary for the regulated expression of genes whose protein products are components of the apoptotic pathway. Thus, in our analysis, we should observe the differential expression of genes whose misexpression may be implicated in the initiation of apoptosis. Two of the genes identified in this study are good candidates for genes involved in the apoptotic pathway. Recent results have suggested that, at least in yeast, the mitochondrial channel VDAC protein regulates the release of cytochrome c into the cytoplasm, which then activates the caspase family of proteins (26). Thus, misexpression of the VDAC3 gene in ts13 cells may result in the inappropriate regulation of the cellular caspases, which are apoptosis-driving proteolytic enzymes. The growth arrest and DNA damage protein 45 (Gadd45) gene is rapidly induced in cells that have been exposed to UV irradiation (35) and is implicated as being an important component of the JNK/SAPK signaling pathways leading to apoptosis (36). It is likely that apoptosis in ts13 cells is not initiated primarily by a single pathway but, rather, by multiple pathways, some of which we may not detect by the micro-array assay.
Conclusions
Our results indicate that the acetyl-transferase and kinase activities of TAFII250 are involved in the regulation of a wide range of genes in vivo. It is interesting to note that many of these genes require either the TAFII250 acetyl-transferase or kinase activities, but not both. Thus, each of these functional domains regulates only a subset of gene in vivo, and, importantly, different domains regulate predominantly different subsets of genes in vivo. It is also likely that other functional domains of TAFII250 not characterized here may regulate distinct groups of genes. Our study further emphasizes the complex and diverse range of functions that TAFII250 performs in the regulation of cellular gene expression.
Acknowledgments
We thank Al Fornace and William Craigen for providing us with the Gadd45 and mVDAC3 promoter constructs, respectively. We are grateful to Rich Freiman, Shane Albright, Andreas Ladurner, and Edith Wang for critical reading of this manuscript. T.O. is a Leukemia Society of America Special Fellow. This work was supported in part by the Howard Hughes Medical Institute and National Institutes of Health grants to R.T.
Abbreviations
- TFIID
transcription factor IID
- TAF
TATA box-binding protein-associated factor
- ts
temperature-sensitive
- mVDAC3
murine voltage-dependent anion channel gene 3
References
- 1.Goodrich J, Tjian R. Curr Opin Cell Biol. 1994;6:403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 2.Tanese N, Pugh B F, Tjian R. Genes Dev. 1991;5:2212–2224. doi: 10.1101/gad.5.12a.2212. [DOI] [PubMed] [Google Scholar]
- 3.Dynlacht B D, Hoey T, Tjian R. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 4.Poon D, Bai Y, Cambell A M, Bjorklund S, Kim Y J, Zhou S, Kornberg R D, Weil P A. Proc Natl Acad Sci USA. 1995;92:8224–8228. doi: 10.1073/pnas.92.18.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Nature (London) 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 6.Sauer F, Wasserman D, Rubin G, Tjian R. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 7.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. Nature (London) 1996;383:188–191. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 8.Michel B, Komarnitsky P, Buratowski S. Mol Cell. 1998;2:663–673. doi: 10.1016/s1097-2765(00)80164-1. [DOI] [PubMed] [Google Scholar]
- 9.Holstege F C P, Jennings E G, Wyrich J J, Lee T I, Hengartner C J, Green M R, Goluc T R, Lander E S, Young R A. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 10.Mizzen C, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, et al. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 11.Dikstein R, Ruppert S, Tjian R. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 12.O' Brien T, Tjian R. Mol Cell. 1998;1:905–911. doi: 10.1016/s1097-2765(00)80089-1. [DOI] [PubMed] [Google Scholar]
- 13.Kitajima S, Chibazakura T, Yohana M, Yasukochi Y. J Biol Chem. 1994;269:29970–29977. [PubMed] [Google Scholar]
- 14.Hayashida T, Sekiguchi T, Noguchi E, Sunamoto H, Ohba T, Nishimoto T. Gene. 1994;141:267–270. doi: 10.1016/0378-1119(94)90583-5. [DOI] [PubMed] [Google Scholar]
- 15.Talavera A, Basilico C. J Cell Physiol. 1977;92:425–436. doi: 10.1002/jcp.1040920310. [DOI] [PubMed] [Google Scholar]
- 16.Sekiguchi T, Nakashima T, Hayashida T, Kuraoka A, Hashimoto S, Tsuchida N, Shibata Y, Hunter T, Nishimoto T. Exp Cell Res. 1995;218:490–498. doi: 10.1006/excr.1995.1183. [DOI] [PubMed] [Google Scholar]
- 17.Wang E H, Tjian R. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 18.Wang E, Zhou S, Tjian R. Genes Dev. 1997;11:2658–2669. doi: 10.1101/gad.11.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki-Yagawa Y, Guermah M, Roeder R G. Mol Cell Biol. 1997;17:3284–3294. doi: 10.1128/mcb.17.6.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunphy E L, Johnson T, Auerbach S S, Wang E H. Mol Cell Biol. 2000;20:1134–1139. doi: 10.1128/mcb.20.4.1134-1139.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampson M J, Lovell R S, Craigen W J. J Biol Chem. 1997;272:18966–18973. doi: 10.1074/jbc.272.30.18966. [DOI] [PubMed] [Google Scholar]
- 22.Lillie J W, Green M R. Nature (London) 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 23.Hollander M C, Alamo I, Jackman J, Wang M, McBride O W, Fornace A J. J Biol Chem. 1993;268:24385–24393. [PubMed] [Google Scholar]
- 24.Cutler G, Perry K, Tjian R. Mol Cell Biol. 1998;18:2252–2261. doi: 10.1128/mcb.18.4.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu S, Narita M, Tsujimoto Y. Nature (London) 1999;399:483–487. doi: 10.1038/20959. [DOI] [PubMed] [Google Scholar]
- 27.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 28.Verrijzer C P, Chen J-L, Yokomori K, Tjian R. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- 29.Shen W C, Green M R. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- 30.Chen J-L, Attardi L, Verrijzer C P, Yokomori K, Tjian R. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 31.Walker S S, Reece J C, Apone L M, Green M R. Nature (London) 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 32.Milano J, Strayer D S. Exp Cell Res. 1998;239:31–39. doi: 10.1006/excr.1997.3869. [DOI] [PubMed] [Google Scholar]
- 33.Mayer-Jaekel R E, Ohkura H, Gones R, Sunkel E, Baumgartner S, Hemmings B A, Glover D M. Cell. 1993;72:621–633. doi: 10.1016/0092-8674(93)90080-a. [DOI] [PubMed] [Google Scholar]
- 34.Altiok S, Xu M, Spiegelman B M. Genes Dev. 1997;11:1987–1998. doi: 10.1101/gad.11.15.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fornace A J, Alamo I, Hollander M C. Proc Natl Acad Sci USA. 1988;85:8800–8804. doi: 10.1073/pnas.85.23.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takwkawa M, Saito H. Cell. 1998;9:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]