Abstract
Ribosomal subunit kinases (Rsk) have been implicated in the regulation of transcription by phosphorylating and thereby activating numerous transcription factors, such as c-Fos, cAMP responsive element binding protein (CREB), and nuclear receptors. Here we describe the generation and characterization of immortalized embryonic fibroblast cell lines from mice in which the Rsk-2 gene was disrupted by homologous recombinant gene targeting. Rsk-2-deficient (knockout or KO) cell lines have no detectable Rsk-2 protein, whereas Rsk-1 expression is unaltered as compared with cell lines derived from wild-type control mice. KO cells exhibit a major reduction in platelet-derived growth factor (PDGF) and insulin-like growth factor (IGF)-1-stimulated expression of the immediate-early gene c-Fos. This results primarily from a reduced transcriptional activation of the ternary complex factor Elk-1 and reduced activation of the serum response factor. The reduced Elk-1 activation in KO cells occurs despite normal activation of the mitogen-activated protein kinase pathway and normal PDGF- and IGF-1-stimulated Elk-1 phosphorylation. By contrast, PDGF- and IGF-1-stimulated phosphorylation and transcriptional activation of CREB is unaltered in KO cells. Thus Rsk-2 is required for growth factor-stimulated expression of c-Fos and transcriptional activation of Elk-1 and the serum response factor, but not for activation of CREB or the mitogen-activated protein kinase pathway in response to PDGF and IGF-1 stimulation.
One of the main pathways potently stimulated by receptor tyrosine kinases is the mitogen-activated protein kinase (MAPK) cascade. Recruitment of Grb-2 to the activated platelet-derived growth factor (PDGF) receptor or to insulin-like growth factor (IGF)-1 receptor substrates results in the association of the guanidine nucleotide exchange factor SOS to Grb-2, thus activating the small G protein Ras (1, 2). Ras activation leads to activation of the protein kinase Raf that activates the MAPK/Erk kinase (MEK)-1. MEK-1 phosphorylates MAPK, leading to the activation of this serine/threonine kinase. Over the last several years a number of substrates for MAPK have been identified, several of which are transcription factors, including ternary complex factors Elk-1 and SAP-1 (3). Besides this direct activation of transcription factors, MAPK can also phosphorylate and activate a family of serine/threonine kinases designated p90rsk or ribosomal S6 kinase (Rsk) because they can mediate the phosphorylation of the ribosomal protein S6, in some cases, leading to cell differentiation (4). The human p90rsk family comprises three closely related isoforms Rsk-1, Rsk-2, and Rsk-3, each encoded by different genes on different chromosomes (5). These three isoforms share a highly conserved structure. Beside differential expression patterns little is known about their specific physiological role. In vitro characterized substrates of Rsk are c-Fos, serum response factor (SRF), estrogen receptor, and Nur77 (6–8). Moreover, Rsk-2 has been implicated in epidermal growth factor (EGF)-stimulated SOS phosphorylation, which leads to dissociation of the Grb-2/SOS complex and desensitization of the MAPK pathway (9). More recently, Rsk-2 has been identified as the kinase responsible for nerve growth factor (NGF) and EGF-stimulated phosphorylation of cAMP responsive element binding protein (CREB) (10). This finding revealed an important mechanism by which these growth factors can activate transcription of the immediate-early gene c-Fos.
The regulation of c-Fos transcription appears to be complex, with different stimuli targeting different cis elements in the promoter region of this immediate-early gene. NGF- and EGF-stimulated transcription of c-Fos largely depends on the activation of CREB (11). PDGF activates the sis-inducible element (sie) of the c-Fos promoter and stimulates the well-established linkage via the Ras–Raf–MAPK pathway leading to activation of the serum response element (sre) in the c-Fos promoter (12). Upon growth factor stimulation, the sre forms a ternary complex with SRF and ternary complex factors of the Elk-1 family of transcription factors. Although the exact mechanism of regulation of SRF activity remains unclear, Elk-1 has been demonstrated to be a direct target of MAPK phosphorylation for transcriptional activation (13). Besides the EGF-stimulated activation of CREB and in vitro phosphorylation of SRF, the exact action of Rsk isoforms in activation of these individual factors remains poorly understood.
The recent discovery that the Coffin Lowry syndrome results from functional null mutations of the Rsk-2 gene has added an important clinical implication to the function of Rsk isoforms (14). Coffin Lowry syndrome is characterized by the combination of complex bone malformations and mental retardation and is consistent with the linkage to mutations in the Rsk-2 gene on the X chromosome. To gain better insights into the physiological role of Rsk-2 and the pathogenesis of the Coffin Lowry syndrome, we have inactivated the murine Rsk-2 gene in mice and derived immortalized embryonic fibroblast cell lines from these animals to analyze the molecular mechanisms of Rsk-2 function in immediate-early gene regulation.
Materials and Methods
Cell Culture.
3T3-like fibroblast cell lines were established from knockout (KO) embryos as previously described (15). Experiments were performed on cells between passages 18 and 30.
Genotyping.
Genotyping was performed to detect the hemizygous male KO embryos. PCRs on DNA extracted from the embryos were performed for the Sry gene, which is located on the Y chromosome, and the neomycin-resistance gene, inserted in the Rsk-2 gene. Primers for Sry were 5′-GCACATATGTCATGAACTGGG and 3′-GTGCCATACATCACCATGTG, primers for the neo gene have been described earlier. PCR amplification was performed as previously described (16).
Cotransfection Assays.
Transient transfection assays were performed by electroporation. In brief, 1–4 × 105 cells per 2-cm2 dish were electroporated with the indicated DNA at 250 V and 400 μF. Cells were plated and allowed to reattach for 6 hr in serum-supplemented medium and thereafter changed to serum-free medium. After 36–40 hr, cells were either harvested for assay of luciferase and β-galactosidase activity (in case of MEK-1 cotransfection) or left untreated or treated with IGF-1 (10−8 M) or PDGF (10 ng/ml) for 4 hr before harvesting.
Western Blot Analysis.
Cells were starved overnight in serum-free DMEM. After stimulation with PDGF or IGF-1 for the indicated times, cells were lysed and extracts were separated on 10% polyacrylamide gels, transferred onto poly(vinylidene difluoride) membrane, and probed with the indicated antibodies, using the Boehringer enhanced chemiluminescence kit as previously described (15).
Northern Blot Analysis.
Cells were cultured overnight in serum-free medium and stimulated for the indicated time with PDGF. RNA was extracted by using Ultraspec RNA isolation reagent (Biotecx), and 15 μg of total cellular RNA was separated in formaldehyde gels according to standard procedures. After transfer to nitrocellulose, blots were probed with random prime-labeled probes for c-Fos and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as described (15).
Densitometry.
Blots were exposed to film, and autoradiographs were scanned with a Molecular Dynamics Storm PhosphorImager, and signals were quantified by using imagequant version 3.3 software.
Statistical Analysis.
All values unless otherwise indicated are expressed as mean ± SEM. Statistical analyses were carried out by using a two-tailed Student's unpaired t test, and the null hypothesis was rejected at the 0.05 level.
Results
Establishment of KO Immortalized Mouse Embryonic Fibroblasts.
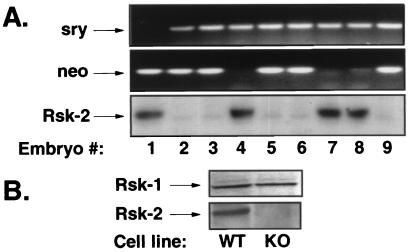
Mice deficient for Rsk-2 expression were generated by introduction of a neomycin-resistance gene into the murine Rsk-2 gene by homologous recombination in embryonic stem cells. Details of the knockout procedure and characterization of the mice will be reported elsewhere. To create immortalized embryonic fibroblast cell lines from KO mice and control littermates, homozygous Rsk-2-null females were bred with male wild-type mice. Because the murine Rsk-2 gene is located on the X chromosome, the presence of the disrupted allele in a male embryo results in complete Rsk-2 deficiency. Embryos were recovered at embryonic day 15 and genotyped as shown in Fig. 1A. The presence of the neomycin-resistance gene in the Rsk-2 locus (Fig. 1A Middle) along with the Y-chromosome marker Sry (17) (Fig. 1A Top) is characteristic of the hemizygous KO male embryos.
Figure 1.
Rsk-2 is absent in 3T3 cell lines from KO embryos. (A) PCR genotyping to identify male embryos (Top) and the presence of the neomycin-resistance gene in the targeted Rsk-2 allele (Middle). Western blot analyses using Rsk-2-specific antibody on cell extracts from primary mouse embryonic fibroblasts (Bottom). (B) Western blot analysis on extracts from immortalized 3T3 cells of control embryo no. 7 (WT) and KO embryo no. 6 (KO) with Rsk-1-specific (Upper) and Rsk-2-specific antibodies (Lower).
Western blot analysis was performed with an anti-Rsk-2 specific antibody to confirm the absence of immunologically detectable Rsk-2 protein in the primary embryonic fibroblasts. Consistent with the genotyping results, cells derived from embryos 2, 3, 5, 6, and 9 failed to express Rsk-2 protein, indicating successful targeting of the Rsk-2 locus (Fig. 1A Bottom). Immortalized embryonic fibroblast cell lines from embryo 6 (KO) and embryo number 7 (wild type; WT) were established according to a 3T3 protocol. The lack of Rsk-2 expression was also confirmed in the immortalized KO cells by Western blotting (Fig. 1B Lower), whereas expression of Rsk-1 was unaltered in these cells (Fig. 1B Upper).
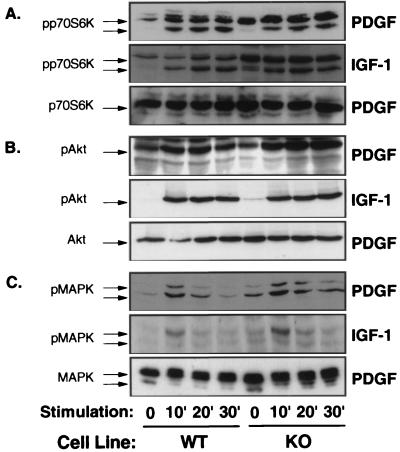
Activation of the p70S6-Kinase and MAPK Pathways Occurs Independent of Rsk-2 Expression.
The alternative ribosomal subunit kinase to p90rsk is p70S6-kinase, which is also activated by PDGF- and IGF-1 via the enzyme phosphatidylinositol 3-kinase and serine/threonine phosphorylation of the enzyme (18). Both PDGF and IGF-1 stimulated p70S6-kinase phosphorylation to similar extent in WT and KO cells as determined by Western blot analysis using phospho-specific anti-p70S6-kinase antibodies (Fig. 2A Top and Middle). Western blot analysis using anti-p70S6-kinase antibodies also revealed comparable p70S6-kinase-expression in WT and KO cells (Fig. 2A Bottom). Akt is a serine/threonine kinase involved in the regulation of gene transcription and growth factor-stimulated cell survival that is also a downstream signal of p70S6-kinase and/or phosphatidylinositol 3-kinase (19). Western blots of cell extracts from PDGF- and IGF-1-stimulated WT and KO cells using phospho-specific anti-Akt antibodies demonstrated that Akt activation and expression also were unaltered by the absence of Rsk-2 expression in KO cells (Fig. 2B).
Figure 2.
PDGF- and IGF-1-stimulated activation of p70S6-kinase, Akt, and MAPK is unaltered in KO cells. Western blot analyses were performed on cell extracts from control (WT) and KO cells, which had been stimulated with PGDF or IGF-1 for the indicated times (min). Immunoblotting was performed with antibodies against phosphorylated p70S6-kinase (A, Top and Middle), phosphorylated Akt (B, Top and Middle), and phosphorylated MAPK (C, Top and Middle) or with corresponding antibodies, detecting these proteins independent of activation (A–C, Bottom)
Because activation of MAPK has been demonstrated as an essential step in growth factor-stimulated transcription of the c-Fos gene and activation of cell proliferation and differentiation, we examined MAPK activation in response to PDGF and IGF-1 in KO cells. Activation of MAP kinases Erk-1 and -2 occurs by phosphorylation of threonine and tyrosine residues through MAPK kinase MEK-1, and this can be detected by Western blotting with an antibody that recognizes catalytically activated MAPK phosphorylated at Tyr-204. Consistent with the notion that Rsk-2 is downstream of MAPK, PDGF-, and IGF-1-stimulated phosphorylation/activation of MAPK occurred normally in the KO cells (Fig. 2C Top and Middle). Moreover, the total cellular content of MAPK was similar in Rsk-2 KO and WT cells as assessed by Western blotting with an anti-MAPK antibody (Fig. 2C Bottom).
Rsk-2 Is Required for PDGF- and IGF-1-Stimulated Transcription of c-Fos Gene.
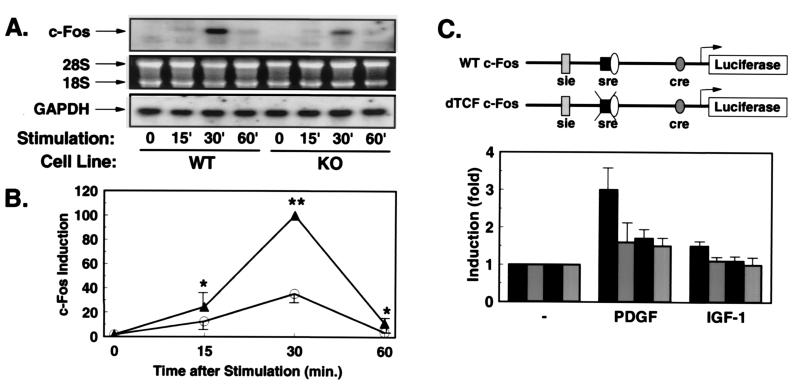
Because Rsk-2 has been implicated in growth factor-stimulated expression of the c-Fos gene, we assessed PDGF-stimulated expression of the c-Fos gene in KO and WT cells by Northern blot analysis. PDGF potently stimulated transient c-Fos expression in WT cells, peaking at 30 min after stimulation and declining by 60 min (Fig. 3A). By contrast, KO cells showed a marked reduction in PDGF-stimulated c-Fos expression (Fig. 3A). Quantitation of several experiments revealed that the ability of PDGF to induce maximal transcription of the endogenous c-Fos gene was reduced by 65% in KO cells as compared with WT cells (Fig. 3B).
Figure 3.
Growth factor-stimulated expression of c-Fos is reduced in KO cells. (A) Northern blot analyses with c-Fos and GAPDH probes were performed on RNA extracted from WT or KO cells stimulated for the indicated times (min) with PDGF. The autoradiography after incubation with the c-Fos probe (Top). The integrity of RNA on the ethidium bromide-stained gel (Middle), and the autoradiography after hybridization with the GAPDH probe (Bottom). (B) Quantification was performed by densitometric scanning of the specific c-Fos signal/intensity of the GAPDH signal. Data are the mean ± SEM of three independent experiments expressed as percentage of maximum intensity in WT cells (*, P < 0.05; **, P < 0.01). (C) Schematic representation of the c-Fos promoter-luciferase constructs. WT c-Fos contains base pairs −734 to +43 of the human c-Fos gene cloned into promoterless luciferase gene (pGL2basic). In the construct δTCF-c-Fos, the binding site for TCF (base pair −320 to −328) was mutated (12). WT cells (black bars) and KO cells (gray bars) were cotransfected with the reporter plasmid WT c-Fos (black bars with white stippling) or the mutant reporter (gray bars with white stippling). After overnight culture in serum-free medium, cells were treated with PDGF or IGF-1 for 3 hr, and cell extracts were subsequently assayed for luciferase and β-galactosidase activity. Data represent the stimulation of luciferase activity/β-galactosidase activity in PDGF- and IGF-1-stimulated vs. untreated cells. Data are the mean of four separate experiments ± SEM.
These results were confirmed in transient transfection experiments using a luciferase reporter gene regulated by bases −734 to +43 of the human c-Fos gene (Fig. 3C). When transiently transfected into WT cells, PDGF stimulated reporter gene activity 3-fold, whereas IGF-1 led to a 1.5-fold stimulation (Fig. 3C). In contrast, PDGF stimulated expression of the c-Fos reporter by only 1.6-fold in KO cells, and IGF-1 failed to significantly induce transcription of this reporter at all in KO cells (Fig. 3C).
To further characterize the Rsk-2-sensitive cis element in the promoter of the c-Fos gene, we used reporter gene constructs in which the binding site for ternary complex factor (δTCF) was mutated (Fig. 3C). As previously shown for PDGF-stimulated c-Fos expression, transcriptional activation of the c-Fos promoter by both PDGF and IGF-1 depended on an intact TCF-binding site in WT cells (Fig. 3C). Thus, in WT cells the intact c-Fos promoter was stimulated 3-fold by PDGF and 1.5-fold by IGF-1, whereas the δTCF mutant responded only 1.7-fold to PDGF stimulation and failed to respond to IGF-1 stimulation (Fig. 3C). On the other hand, in KO cells PDGF stimulated transcription of the WT promoter by 1.7-fold, and mutation of TCF-site led to no further reduction of inducibility (Fig. 3C). These data indicate that reduced transcriptional activation of c-Fos in the absence of Rsk-2 results largely from reduced TCF activation.
Rsk-2 Is Essential for PDGF- and IGF-1-Stimulated Activation of Ternary Complex Factor Elk-1.
PDGF has been demonstrated to target multiple cis elements in the c-Fos promoter, including sie, sre, and possibly cre, with possibly the most important being sre, by means of the activation of ternary complex factors (12). Similarly, IGF-1-stimulated c-Fos induction appears to depend largely on TCF activation. These transcription factors are the main targets of MAPKs in the activation of c-Fos transcription.
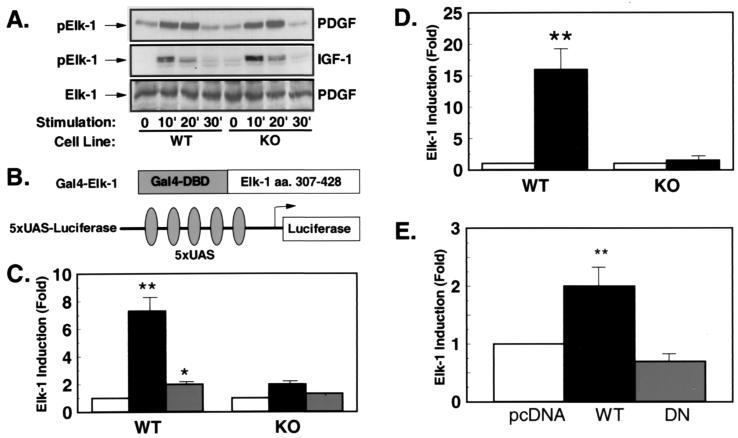
Elk-1 has been shown to be directly phosphorylated by MAPKs on Ser-383 in the C-terminal domain. This phosphorylation is thought to be essential and sufficient to activate transcriptional activity of Elk-1. To analyze PDGF- and IGF-1-stimulated Elk-1 phosphorylation, we performed Western blot experiments on extracts from KO and WT cells, using an antibody that recognizes Elk-1 only in the phosphorylated state. Consistent with an unaltered activation of MAPKs in KO cells, PDGF- and IGF-1-stimulated phosphorylation of Elk-1 occurred normally in the absence of Rsk-2 (Fig. 4A Top and Middle). Elk-1 phosphorylation occurred transiently, peaking 10 min after growth factor stimulation and declining to basal levels by 30 min in cells from both WT and KO embryos (Fig. 4A Top and Middle). Moreover, the total cellular content of Elk-1 was comparable in WT and KO cells as assessed by Western blotting with an anti-Elk-1 antibody (Fig. 4A Bottom).
Figure 4.
PDGF- and IGF-1-stimulated transcriptional activity of Elk-1 is reduced in the presence of unaltered Elk-1 phosphorylation. (A) Western blot analyses were performed with an anti-Elk-1 antibody (Bottom) or an phospho-specific anti-Elk-1 antibody (Top and Middle) on cell extracts of PDGF- (Top and Bottom) or IGF-1-treated (Middle) cells. (B) Schematic representation of the heterologous Elk-1 reporter system. (C) WT and KO cells were cotransfected with the described reporter plasmid, Gal-4–Elk-1 and the plasmid PEF–β-galactosidase (PEF–β-gal) expressing β-galactosidase under control of the elongation factor promoter. After culture in serum-free medium for 36 hr, cells were left untreated (white bars) or treated with PDGF (black bars) or IGF-1 (gray bars) for 4 hr before lysis. Data represent the stimulation of luciferase activity/β-galactosidase activity in stimulated vs. untreated cells. Data are the mean of four independent experiments ± SEM. (*, P < 0.05; **, P < 0.01). (D) Cotransfections were performed with the described Elk-1-reporter in the presence of pcDNA or a MEK-1-expression plasmid (black bars) (**, P < 0.01). (E) KO cells were transfected with 5XUAS-luciferase and Gal-4–ElkC together with the expression plasmid pcDNA (white bar), RSV-WTRsk (black bar), or RSV-DNRsk (gray bar) expressing wild-type Rsk (WTRsk) or a dominant–negative mutant of Rsk-2 (DNRsk). Data presented are the mean of four experiments ± SEM expressed as stimulation compared with pcDNA-transfected cells (**, P < 0.01).
To determine transcriptional activation of Elk-1 in KO cells we used a heterologous reporter system cotransfecting an expression plasmid coding for a fusion protein of the Gal-4–DNA-binding domain with the C-terminal transcriptional activation domain encompassing amino acids 307–421 of human Elk-1 with a Gal-4-responsive luciferase reporter gene [(UAS)5-tk-luc] (Fig. 4B). When WT cells were transfected with this reporter system, PDGF stimulated transcription through the Elk-1 activation domain by 7-fold, whereas IGF-1 led to a 2-fold induction (Fig. 4C). In contrast, in KO cells PDGF-stimulated transcriptional activation of the same reporter was reduced to 2-fold and IGF-1-stimulated activation was reduced to 1.3-fold (Fig. 4C). To determine whether this effect was specific to PDGF and IGF-1 stimulation or a general mechanism after transcriptional activation through MAPKs, the same reporter plasmids were transfected in the absence or presence of an expression plasmid for a constitutively active MEK-1 mutant. MEK-1 stimulated transcriptional activation through the Elk-1 activation domain 16-fold in WT cells, whereas MEK-1 failed to activate Elk-1 KO cells (Fig. 4D).
To further analyze the role of Rsk-2 in Elk-1 activation, we cotransfected KO cells with the heterologous Elk-1-reporter system and expression plasmids for WT Rsk-2 or a dominant-negative, kinase-deficient mutant of the enzyme. In this case, cotransfection with Rsk-2 resulted in a 2-fold activation of transcription through the Elk-1 activation domain, whereas cotransfection with the dominant-negative mutant resulted in a 30% reduced activation as compared to mock-transfected cells (Fig. 4E). These data indicate that Rsk-2 is required for full transcriptional activation through Elk-1 even in the presence of unaltered growth factor-stimulated Elk-1 phosphorylation.
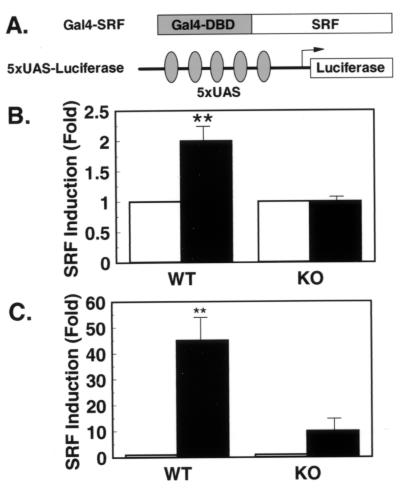
Rsk-2 Is Required for Transcriptional Activation of SRF.
There is evidence from in vitro experiments that SRF can be phosphorylated by Rsk-family members on Ser-103. In contrast to Elk-1, growth factor-stimulated phosphorylation of SRF seems not to correlate with its transcriptional activation but appears to modulate DNA binding (20). To assess transcriptional activation of SRF in KO cells, we performed transient transfection experiments with SRF fused to the heterologous DNA-binding domain of the yeast transcription factor Gal-4 and a Gal-4-responsive luciferase reporter gene (Fig. 5A). In WT cells transiently transfected with this reporter system, PDGF stimulated transcriptional activation by 1.9-fold (Fig. 5B). On the other hand, PDGF failed to induce transcriptional activation through the heterologous SRF-transcription factor in the absence of Rsk-2 (Fig. 5B). This difference was even more pronounced when activated MEK-1 was cotransfected. Thus, in this case, MEK-1 stimulated transcription through SRF 45-fold in WT cells, whereas this response was blunted to a 10-fold stimulation in KO cells (Fig. 5C). These data argue for the existence of a Rsk-2-dependent pathway in Ras-mediated SRF activation required for full transcriptional activation of the ternary complex at the sre.
Figure 5.
Transcriptional activation of a heterologous SRF in response to PDGF and MEK-1 stimulation is reduced in KO cells. (A) Schematic representation of the heterologous SRF-reporter system with a fusion protein of the Gal-4–DNA-binding domain (amino acids 1–147 of Gal-4) and human SRF (amino acids 10–301 of SRF). (B) WT and KO cells were cotransfected with the described reporter plasmid. After overnight culture in serum-free medium, cells were left untreated or treated with PDGF for 3 hr before lysis. Data represent the stimulation of luciferase activity/β-galactosidase activity in stimulated vs. untreated cells. Data are the mean of four independent experiments ± SEM. (C) Cotransfections were performed as described for B with the exception that PDGF stimulation was replaced by cotransfection of an MEK-1 expression plasmid (**, P < 0.01).
Rsk-2 Is Not Required for CREB Phosphorylation and Activation in Response to PDGF and IGF-1.
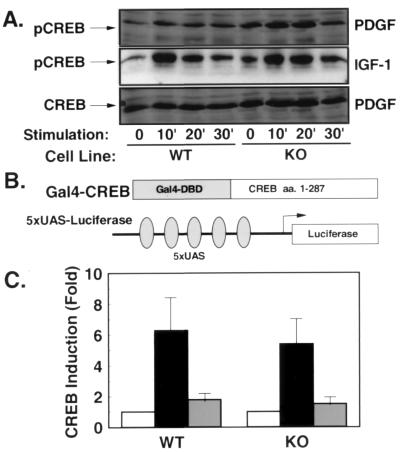
Previous studies have revealed an essential role for Rsk-2 in EGF- and NGF-stimulated c-Fos transcription by means of phosphorylation and activation of CREB (10). To test the role of Rsk-2-mediated CREB phosphorylation in PDGF- and IGF-1-stimulated c-Fos transcription, we performed Western blot analyses using antibodies that recognize CREB only in its activated form—i.e., phosphorylated on Ser-133. In these experiments, PDGF and IGF-1 stimulated CREB-phosphorylation in WT cells 2- to 3-fold after 10 min of stimulation (Fig. 6A Top and Middle). Surprisingly, PDGF and IGF-1 stimulated CREB phosphorylation in KO with similar kinetics and to the same magnitude (Fig. 6A Top and Middle). Moreover, the expression of CREB as assessed by Western blot with an anti-CREB antibody was unaltered in KO cells (Fig. 6A Bottom). These data indicate that CREB phosphorylation in response to PDGF and IGF-1 stimulation occurs independent from functional Rsk-2 expression.
Figure 6.
PDGF- and IGF-1-stimulated phosphorylation and activation of CREB are unaltered in the absence of Rsk-2. (A) Western blot analysis using an anti-CREB antibody (Bottom) shows comparable cellular content of CREB in WT and KO cells. Western blot with an Ser-133 phospho-specific anti-CREB antibody reveals normal phosphorylation of CREB in KO cells in response to PDGF (Top) and IGF-1 stimulation (Middle). (B) Schematic representation of the heterologous CREB reporter system. (C) WT and KO cells were cotransfected with the described reporter plasmid. After serum depletion for 36 hr, cells were left untreated (open bars) or treated with PDGF (black bars) or IGF-1 (gray bars) for 4 hr before lysis. Data represent the stimulation of luciferase activity/β-galactosidase activity in stimulated vs. untreated cells.
To assess transcriptional activation of CREB in KO cells, we performed transient transfection experiments with the CREB-transcriptional activation domain fused to the heterologous DNA-binding domain of the yeast transcription factor Gal-4 and a Gal-4-responsive luciferase reporter gene (Fig. 6B). When transiently transfected with this reporter system, PDGF stimulated transcriptional activation in WT cells by 6-fold and, not significantly different, by 5.3-fold in KO cells (Fig. 6C). Similarly, transcriptional activation of the same reporter system by IGF-1 did not differ in WT and KO cells (Fig. 6C), indicating that functional Rsk-2-expression is not required for PDGF- and IGF-1-stimulated CREB activation.
Discussion
The ribosomal subunit kinases are a family of molecules that play major roles in the regulation of cell proliferation induced by serum and growth factors. The function of Rsk-2 has been studied most extensively with respect to EGF- and NGF-stimulated c-Fos transcription. Rsk-2 appears to be the predominant CREB-kinase in response to EGF stimulation (10). In cell lines from patients suffering from Coffin Lowry syndrome in whom the Rsk-2 gene is mutant, EGF fails to induce c-Fos transcription due to an apparent failure to phosphorylate CREB, whereas serum and UV-induced CREB phosphorylation are preserved (21). Our data indicate that PDGF- and IGF-1-stimulated CREB phosphorylation and transcriptional activation also occur independent of Rsk-2. At least three possibilities exist regarding alternative kinases responsible for PDGF- and IGF-1-stimulated CREB phosphorylation. First, it is possible that a yet-unidentified kinase is responsible for PDGF- and IGF-1-stimulated CREB phosphorylation. Alternatively, Rsk-1 and Rsk-3 compensate for the lack of Rsk-2 in the knockout cells, but fail to compensate in cells from patients with Coffin Lowry syndrome, where the truncated, catalytically inactive Rsk-2 might act as a dominant-negative enzyme on other Rsk isoforms. Finally, it possible that the PDGF- and IGF-1-stimulated CREB kinase is p70S6-kinase. Activation of p70S6-kinase and Akt is unaltered in the absence of Rsk-2, and it has been shown that serum-stimulated p70S6-kinase can result in phosphorylation of the cAMP-responsive element modulator (CREM) (22). Whatever the kinase responsible for PDGF- and IGF-1-stimulated CREB phosphorylation, it is interesting that PDGF and IGF-1, as opposed to EGF, exhibit profoundly different effects on gene transcription even though these growth factors engage similar postreceptor signaling cascades including the activation of MAPK.
Another intriguing finding of the present study is the involvement of Rsk-2 in sre-mediated transcription. Although growth factor-stimulated phosphorylation of Elk-1 has been thought to be sufficient for transcriptional activation (13), we have shown an additional level of control of this transcription factor. Apparently, Rsk-2 is required for Elk-1 activation in response to PDGF and IGF-1 stimulation independent from MAPK-mediated Elk-1 phosphorylation. Different mechanistic explanations for this phenomenon exist. It has been shown that Rsk expression is required for growth factor-stimulated recruitment of the transcriptional coactivators p300/CBP (23). Because p300/CBP also act as a coactivator for both Elk-1 and SRF, reduced activation of these transcription factors might result from the failure of p300/CBP recruitment (24). Rsk-kinase activity also appears to be required for Elk-1 activation, and Rsk-2 is required for EGF-stimulated histone H3 phosphorylation resulting in unfolding of chromatin (25). Therefore, it is likely that growth factor-stimulated Rsk activity not only targets individual transcription factors directly but also results in general transcriptional activity because of chromatin remodeling. Alternatively, Rsk-2 might directly phosphorylate Elk-1 on a site different from the well-characterized MAPK-phosphorylation site, and this phosphorylation might be essential for Elk-1 activation. Clearly, further studies are required to determine the exact mechanism by which Rsk-2 stimulates sre-mediated transcription.
In summary, we have provided evidence for an essential role of Rsk-2 in PDGF- and IGF-1-stimulated transcription of c-Fos gene targeting the activity of both SRF and Elk-1. The cell lines described here provide an excellent tool to further characterize the molecular mechanisms through which Rsk-2 participates in the growth factor-stimulated activation of these transcription factors.
Acknowledgments
We thank J. Flier for generous support during creation of KO mice at the Beth Israel Deaconess Medical Center provided by National Institutes of Health Grant support (to J. F.). We thank A. Bonni and M. E. Greenberg for providing the Gal-4–SRF plasmid, R. Treisman for providing plasmids δTCF, and M. Montminy for the generous gift of Gal-4–ElkC and Gal-4–CREB plasmids. This study was funded by grants from the Deutsche Forschungsgemeinschaft (DFG 1492–1, to J.C.B.), the Center of Molecular Medicine (ZMMK 2, to D.M.-W., J.C.B., and W.K.), and National Institutes of Health Grant DK33201 (to C.R.K.).
Abbreviations
- CREB
cAMP responsive element binding protein
- IGF
insulin-like growth factor
- SRF
serum response factor
- EGF
epidermal growth factor
- NGF
nerve growth factor
- TCF
ternary complex factor
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- MAPK
mitogen-activated protein kinase
- MEK
MAPK/Erk kinase
- KO
Rsk-2-deficient (knockout)
- WT
wild type
References
- 1.Lowenstein E J, Daly R J, Batzer A G, Li W, Margolis B, Lammers R, Ullrich A, Skolnik E Y, Bar-Sagi D, Schlessinger J. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 2.Baltensperger K, Kozma L M, Cherniack A D, Klarlund J K, Chawla A, Banerjee U, Czech M P. Science. 1993;260:1950–1952. doi: 10.1126/science.8391166. [DOI] [PubMed] [Google Scholar]
- 3.Marais R, Wynne J, Treisman R. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 4.Chen R H, Sarnecki C, Blenis J. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, Bjorbaek C, Weremowicz S, Morton C C, Moller D E. Mol Cell Biol. 1995;15:4353–4363. doi: 10.1128/mcb.15.8.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorbaek C, Zhao Y, Moller D E. J Biol Chem. 1995;270:18848–18852. doi: 10.1074/jbc.270.32.18848. [DOI] [PubMed] [Google Scholar]
- 7.Chen R H, Abate C, Blenis J. Proc Natl Acad Sci USA. 1993;90:10952–10956. doi: 10.1073/pnas.90.23.10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joel P B, Smith J, Sturgill T W, Fisher T L, Blenis J, Lannigan D A. Mol Cell Biol. 1998;18:1978–1984. doi: 10.1128/mcb.18.4.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douville E, Downward J. Oncogene. 1997;15:373–383. doi: 10.1038/sj.onc.1201214. [DOI] [PubMed] [Google Scholar]
- 10.Xing J, Ginty D D, Greenberg M E. Science. 1996;273:959–963. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 11.Ginty D D, Bonni A, Greenberg M E. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 12.Hill C S, Treisman R. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb M H, Shaw P E. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trivier E, De Cesare D, Jacquot S, Pannetier S, Zackai E, Young I, Mandel J L, Sassone-Corsi P, Hanauer A. Nature (London) 1996;384:567–570. doi: 10.1038/384567a0. [DOI] [PubMed] [Google Scholar]
- 15.Bruning J C, Winnay J, Cheatham B, Kahn C R. Mol Cell Biol. 1997;17:1513–1521. doi: 10.1128/mcb.17.3.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruning J C, Winnay J, Bonner-Weir S, Taylor S I, Accili D, Kahn C R. Cell. 1997;88:561–572. doi: 10.1016/s0092-8674(00)81896-6. [DOI] [PubMed] [Google Scholar]
- 17.Sinclair A H, Berta P, Palmer M S, Hawkins J R, Griffiths B L, Smith M J, Foster J W, Frischauf A M, Lovell-Badge R, Goodfellow P N. Nature (London) 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 18.Virkamaki A, Ueki K, Kahn C R. J Clin Invest. 1999;103:931–943. doi: 10.1172/JCI6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohn A D, Takeuchi F, Roth R A. J Biol Chem. 1996;271:21920–21926. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]
- 20.Janknecht R, Hipskind R A, Houthaeve T, Nordheim A, Stunnenberg H G. EMBO J. 1992;11:1045–1054. doi: 10.1002/j.1460-2075.1992.tb05143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Cesare D, Jacquot S, Hanauer A, Sassone-Corsi P. Proc Natl Acad Sci USA. 1998;95:12202–12207. doi: 10.1073/pnas.95.21.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Groot R P, Ballou L M, Sassone-Corsi P. Cell. 1994;79:81–91. doi: 10.1016/0092-8674(94)90402-2. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 24.Janknecht R, Nordheim A. Biochem Biophys Res Commun. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- 25.Sassone-Corsi P, Mizzen C A, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis C D. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]