Abstract
Ribonucleotide reductase (RNR) catalyzes the de novo synthesis of deoxyribonucleotides. Eukaryotes have an α2β2 form of RNR consisting of two homodimeric subunits, proteins R1 (α2) and R2 (β2). The R1 protein is the business end of the enzyme containing the active site and the binding sites for allosteric effectors. The R2 protein is a radical storage device containing an iron center-generated tyrosyl free radical. Previous work has identified an RNR protein in yeast, Rnr4p, which is homologous to other R2 proteins but lacks a number of conserved amino acid residues involved in iron binding. Using highly purified recombinant yeast RNR proteins, we demonstrate that the crucial role of Rnr4p (β′) is to fold correctly and stabilize the radical-storing Rnr2p by forming a stable 1:1 Rnr2p/Rnr4p complex. This complex sediments at 5.6 S as a ββ′ heterodimer in a sucrose gradient. In the presence of Rnr1p, both polypeptides of the Rnr2p/Rnr4p heterodimer cosediment at 9.7 S as expected for an α2ββ′ heterotetramer, where Rnr4p plays an important role in the interaction between the α2 and the ββ ′ subunits. The specific activity of the Rnr2p complexed with Rnr4p is 2,250 nmol deoxycytidine 5′-diphosphate formed per min per mg, whereas the homodimer of Rnr2p shows no activity. This difference in activity may be a consequence of the different conformations of the inactive homodimeric Rnr2p and the active Rnr4p-bound form, as shown by CD spectroscopy. Taken together, our results show that the Rnr2p/Rnr4p heterodimer is the active form of the yeast RNR small subunit.
Ribonucleotide reductases (RNRs) play a central role in the formation and control of the optimal levels of dNTPs, which are required for DNA replication and DNA repair processes; a failure to control the size of dNTP pools and/or their relative amounts leads to cell death or genetic abnormalities (1).
Three classes of RNRs have been characterized; all have an organic free radical essential for the catalytic activity (2). Most eukaryotic cells contain class Ia RNRs; this form also exists in some prokaryotes, e.g., the well studied nrdA/nrdB encoded enzyme of Escherichia coli. The functional class Ia enzyme is composed of two nonidentical homodimeric subunits, α2 (also called R1) and β2 (also called R2), which are both required for activity. Each of the larger α-polypeptides contains redox-active disulfides, an active site that binds ribonucleoside diphosphates, and two different allosteric sites binding nucleoside triphosphates. Overall enzyme activity is controlled by binding ATP (stimulation) or dATP (inhibition) to the allosteric activity site, whereas substrate specificity is controlled by binding ATP, dATP, dTTP, or dGTP to the allosteric specificity site (3). Each of the smaller β-polypeptides contributes an essential iron center-generated tyrosyl free radical. The highly flexible C-terminal tail of the β-polypeptide is essential for the formation of the α2β2 heterotetramer and becomes rigid in this complex (4).
In Saccharomyces cerevisiae, there are two genes encoding the highly homologous proteins of the large subunit, RNR1 and RNR3. RNR1 is essential for mitotic viability, whereas RNR3, whose transcription is low under normal conditions, is not (5). The small subunit is encoded by the essential RNR2 gene (6, 7). In addition, the RNR4 gene encodes an R2-like protein, which is essential for mitotic viability in some genetic backgrounds (8, 9). The lethality of rnr4 null mutations can be suppressed by overexpression of RNR1 or RNR3 (9). The transcription of all RNR genes is inducible by DNA damage or replication blocks. This effect is most pronounced for the RNR3 gene, which is induced up to 100-fold (5).
We have previously demonstrated that RNR4 is directly involved in yeast ribonucleotide reduction: the addition of bacterially expressed Rnr4p to yeast extracts lacking endogenous RNR4 dramatically increases RNR activity (8). Although Rnr4p is necessary for the normal function of the yeast RNR and is rather similar to Rnr2p in overall sequence, 6 of the 16 essential amino acid residues completely conserved among other R2 proteins through evolution are replaced by other residues in Rnr4p. Notably, substitution of two histidines and one glutamate, which are ligands of the iron center, by two tyrosines and an arginine makes it highly unlikely that Rnr4p could form a functional iron-radical center. Moreover, it has been shown that the cellular function of the Rnr4p does not depend on its ability to form a radical (8). The conserved radical-bearing tyrosine residue can be replaced by a phenylalanine, which cannot form a radical, without impairing the in vitro RNR4 activity. This situation is unprecedented among RNRs, and the role of the Rnr4p in yeast ribonucleotide reduction is not understood.
The genes encoding yeast RNR are among the best studied transcriptional targets of the DNA replication and DNA damage checkpoint pathways (10). Mec1p and Rad53p are two essential proteins involved in these pathways (11, 12); in the presence of DNA damage or a DNA replication block, these proteins are required to arrest cell-cycle progression. At the same time, they induce transcription of the RNR genes (10) and inhibit the Sml1 (13), a recently discovered protein, which normally inhibits yeast RNR by binding to the Rnr1p (14). Interestingly, overexpression of RNR1 or RNR3 suppresses the lethality of mec1 and rad53 null mutations (15), as does deletion of the SML1 gene (13). These findings suggest that the essential function of the MEC1 and RAD53 genes is the regulation of dNTP levels via RNR and Sml1p (13, 15).
Although the transcriptional regulation of the yeast RNR genes has been studied in great detail, little is known about the structure and biochemical properties of the yeast RNR enzyme (16, 17). Recently, Stubbe and coworkers described the expression and purification of recombinant yeast RNR subunits and implied that Rnr4p plays a key role in the assembly of the Rnr2p diiron cluster (18).
Herein, we report the purification and characterization of recombinant yeast RNR produced in E. coli. We demonstrate that the yeast Rnr2p is unable to fold correctly by itself and therefore is unable to form an iron-radical center. A tyrosyl radical is detectable only in the presence of the Rnr4p, which together with Rnr2p forms a stable heterodimer, the only active form of the small subunit in the RNR assay.
Materials and Methods
Protein Expression and Purification.
Rnr1p and Rnr3p were expressed in E. coli BL21(DE3) by using the pET21a expression vector (Novagen). The bacterial culture was grown in LB medium with 50 μg/ml carbenicillin at 37°C to an OD of 0.6 at 600 nm, chilled on ice, and, after the addition of isopropyl-1-thio-β-d-galactopyranoside to a final concentration 0.5 mM, further incubated at 13–15°C for 13–20 h. All subsequent procedures were carried out at 4°C. Bacteria were harvested by centrifugation, washed once in 50 mM Tris⋅HCl buffer (pH 7.6; buffer A), resuspended in buffer A containing 20 mM DTT and protease inhibitors (2 μg/ml chymostatin/0.3 μg/ml leupeptin/1.4 μg/ml pepstatin A/1 mM phenylmethylsulfonyl fluoride/2.75 mM benzamidine), and disrupted in a Bead-Beater (Biospec Products, Bartlesville, OK). The rest of the purification followed the procedure described for the mouse R1 protein (19).
The Rnr2p was overexpressed in the yeast strain GRF18 (MATα leu2-3,112 his3-11,15) by using the backbone of the pJDBA12 plasmid, where the RNR2 coding sequence was cloned between the PHO5 promoter and terminator sequences (20). The Rnr2p expression was achieved by incubating yeast in a low phosphate medium. Yeast cells were disrupted in a Bead-Beater, and the rest of the purification was carried out as described below for the bacterially expressed Rnr2p.
The Rnr2p, H6Rnr2p (Rnr2p containing 6 extra histidine residues after the first methionine residue), Rnr4p, and ΔRnr4p (Rnr4p with a deletion of the eight C-terminal amino acid residues) were expressed in E. coli BL21(DE3) bacteria by using the pET3a expression vector (Novagen). The H6Rnr2p was created by overlap extension PCR by using the RNR2-pET3a vector and oligonucleotides encoding six histidine residues. The ΔRnr4p was created by replacing the Glu-338 (GAA) by a stop-codon (TAA) by using PCR modification of the RNR4-pET3a vector. To coexpress the H6Rnr2p and Rnr4p or ΔRnr4p in the same bacterium, a kanamycin-resistance gene was inserted into the PstI restriction site of the β-lactamase gene of the H6Rnr2p-pET3a vector. The Rnr4p was expressed by growing a bacterial culture at 37°C to an OD of 0.6 at 600 nm, followed by a 3-h induction with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside at 37°C. Expression and coexpression of the other proteins and preparation of the bacterial crude extracts were carried out as described above for the Rnr1p, except that 50 μg/ml kanamycin was used when appropriate. All crude extracts were centrifuged at 150,000 × g at 4°C for 1 h. Proteins without a His tail (Rnr2p and Rnr4p) were precipitated from the supernatant by adding solid ammonium sulfate (AS) at 0°C. The Rnr2p was precipitated in the fraction from 0 to 50% AS saturation and the Rnr4p was precipitated in the fraction from 60 to 85% AS saturation. The pellets were dissolved in buffer A, and the AS was removed by chromatography on a Sephadex G-25 column equilibrated with the same buffer. Finally, the proteins were purified by ion-exchange chromatography on a Bioscale Q2 column by using the BioLogic Chromatography System (Bio-Rad; Rnr2p) or a DE52 column (Whatman; Rnr4p) by using gradients of increasing NaCl concentrations in buffer A. The proteins with a His tail (H6Rnr2p alone, coexpressed H6Rnr2p/Rnr4p, and coexpressed H6Rnr2p/ΔRnr4p) were purified without AS fractionation on Ni-NTA agarose columns (Qiagen, Chatsworth, CA). Elution was carried out stepwise with imidazole in buffer A. Unspecifically bound proteins were eluted by 20 mM imidazole, and then proteins of interest were eluted by 100 mM imidazole. After elution, the imidazole was immediately removed by chromatography on a Sephadex G-25 column equilibrated with buffer A. The final purification of the H6Rnr2p/Rnr4p and H6Rnr2p/ΔRnr4p was made on a Bioscale Q10 column as described for the Bioscale Q2 column.
Protein Concentrations.
Molar extinction coefficients ɛ280–310 of pure Rnr4p, H6Rnr2p, and H6Rnr2p/Rnr4p were calculated by using data from quantitative amino acid analyses as described earlier (21). The ɛ280–310 values were 57,000 M−1⋅cm−1 for Rnr2p and H6Rnr2p, 45,000 M−1⋅cm−1 for Rnr4p, and 51,000 M−1⋅cm−1 for H6Rnr2p/Rnr4p (expressing concentration as molar polypeptide). The concentration of Rnr1p was determined by the A280–310 absorbance by using a value of 12 for the absorbance of a 1% solution determined for the calf thymus R1 protein (22).
Western Blotting.
To detect Rnr2p and Rnr4p, we used the monoclonal rat YL 1/2 antibodies (23) in combination with rabbit anti-rat IgG, conjugated with alkaline phosphatase (Sigma).
RNR Assay.
Assay mixtures containing 20 mM Hepes-KOH (pH 7.4), 200 mM KOAc, 5 mM ATP, 20 mM MgOAc, 1 mM [3H]cytidine 5′-diphosphate (CDP; specific activity 27,000 cpm/nmol), 20 μM FeCl3, 20 mM DTT, and yeast RNR proteins in different combinations were incubated at 30°C for 10–20 min in a final volume of 50 μl. After incubation, the samples were processed as described earlier to obtain the amount of dCDP formed (24).
Regeneration of the Iron Center and Iron Assay.
The regeneration of the iron center was carried out by using argon-purged 59Fe ferrous ascorbate solution (specific activity 0.152 × 106 cpm/nanoatom) and argon-equilibrated proteins according to the method of Mann et al. (21). The amounts of protein-bound 59Fe were determined by using the nitrocellulose filter binding assay of Söderman and Reichard (25) as modified by Nyholm et al. (26). The iron content in nonregenerated H6Rnr2p, H6Rnr2p/Rnr4p, and Rnr4p was determined by a colorimetric method (27). Five different amounts of H6Rnr2p (from 280 μg to 1120 μg), H6Rnr2p/Rnr4p (from 114 μg to 456 μg), or Rnr4p (from 145 μg to 1,450 μg) were analyzed.
CD Spectroscopy.
CD spectra were recorded at 20°C on a Jobin Yvon-spex CD 6 spectrometer (Longjumeau, France) by using a quartz cuvette with a 0.5-mm optical path. The spectra were measured in a 10 mM sodium phosphate buffer (pH 7.0) at several protein concentrations (1.7 μM to 17 μM). All CD spectra (averages of three scans) were corrected by a subtraction of a spectrum of a reference buffer, identical to the protein buffer. The ellipticity is reported as mean residue molar ellipticity ([θ], in degree cm2⋅dmol−1) according to [θ] = 100⋅[θ]obs/(C⋅L⋅N), where [θ]obs is the ellipticity (degrees), C is the concentration of polypeptide (M), L is the optical pathway (cm), and N is the number of amino acid residues in the polypeptide.
Electronic Paramagnetic Resonance (EPR) Spectroscopy.
EPR spectra were measured on a Bruker ESP 300 spectrometer as described earlier (26).
Sucrose Gradient Centrifugation.
Proteins were incubated in 0.1 ml of 20 mM Hepes-KOH, pH 7.4/100 mM KOAc/10 mM MgOAc/5 mM DTT/0.1 mM dTTP. After a 30-min incubation at 25°C, the samples were layered onto 4.0 ml of a 5–20% (vol/vol) linear gradient of sucrose in the same buffer as above and analyzed as described earlier (14).
Results
Protein Expression and Purification.
Expression and purification of Rnr4p yielded 18 mg of pure protein per g of bacterial cells (Fig. 1). The Rnr4p was present only in the soluble fraction of the crude extract.
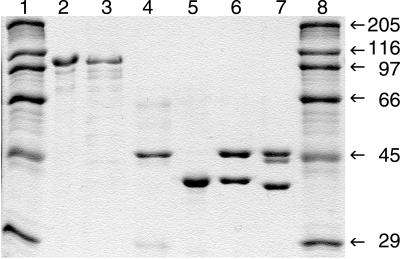
Figure 1.
SDS/PAGE analyses of purified recombinant yeast RNR subunits (1–2 μg per lane) isolated from E. coli. Lanes 1 and 8, molecular mass markers (kDa); lane 2, Rnr1p; lane 3, Rnr3p; lane 4, H6Rnr2p; lane 5, Rnr4p; lane 6, H6Rnr2p/Rnr4p complex; lane 7, H6Rnr2p/ΔRnr4p complex.
Expression and purification of Rnr1p or Rnr3p yielded 2 mg of highly purified protein per g of bacterial cells (Fig. 1). Binding of the yeast Rnr1p and Rnr3p to the dATP-Sepharose column was weaker than that of the mouse R1 protein, because the elution of the yeast proteins started already at 1 mM ATP, whereas the mouse protein elutes between 8 and 15 mM ATP (28).
Initial attempts to produce Rnr2p in bacteria or in yeast were unsuccessful. Under a variety of different growth and induction conditions, the protein expression was minute as detected by immunoblotting with the YL 1/2 antibodies (data not shown). Moreover, all of the Rnr2p was present in the insoluble fraction of the bacterial extract. Lowering the incubation temperature to 13°C resulted in limited amounts of soluble Rnr2p. Attempts to purify Rnr2p by ion-exchange chromatography on a Mono Q column eluted with a gradient of increasing NaCl concentrations did not produce any discrete peak of Rnr2p. Instead, Rnr2p was present in most fractions, producing several indistinct peaks. Similarly, no discrete peak of Rnr2p was obtained on a Mono Q ion-exchange column when we tried to purify Rnr2p overexpressed in yeast (data not shown). The endogenous Rnr4p, on the contrary, was present in only one peak. To circumvent these difficulties, we chose to use a modified Rnr2p, which had six additional histidine residues after the first methionine residue (H6Rnr2p); such a His-tailed Rnr2p could easily be purified by metal affinity chromatography in large amounts, yielding 0.8 mg of protein per g of bacterial cells (Fig. 1).
Because the Rnr4p was shown to bind to the Rnr2p in a two-hybrid assay (9), we tried to coexpress these two proteins in the same bacterium hoping that binding of the highly soluble Rnr4p to the H6Rnr2p would increase the solubility of the latter. Indeed, we obtained 2 mg of coexpressed H6Rnr2p/Rnr4p per g of bacterial cells. Interestingly, the H6Rnr2p bound to a Ni-NTA agarose retained the Rnr4p in 2 M KCl, but not in 4 M urea, indicating that the complex of H6Rnr2p/Rnr4p formed in bacteria was stable and could not be disrupted by high salt concentrations.
Because the H6Rnr2p was present in approximately three times excess over Rnr4p in the eluates from the Ni-NTA columns, as judged from computer assisted scanning of Coomassie brilliant blue-stained SDS/PAGE gels, we tried to purify the complex further on a Mono Q ion exchange column. Elution with a gradient of increasing NaCl concentrations resulted in a discrete peak of a 1:1 molar ratio H6Rnr2p/Rnr4p complex, constituting about 50% of the loaded protein (Fig. 1, lane 6). The remaining 50% of protein contained only H6Rnr2p distributed among nearly all fractions in several indistinct peaks. The elution profile of the H6Rnr2p was similar to the elution profile of the Rnr2p overproduced in yeast, when it was purified on a Mono Q column (data not shown). The purified 1:1 H6Rnr2p/Rnr4p complex was used in all further studies. The same procedure was used, and the same results were obtained during the purification of the H6Rnr2p/ΔRnr4p complex (Fig. 1).
Activity of H6Rnr2p, Rnr4p, and H6Rnr2p/Rnr4p Complex.
No RNR activity was observed with H6Rnr2p alone or with Rnr4p alone in the presence of Rnr1p. In contrast, the specific activity of the coexpressed H6Rnr2p/Rnr4p complex reached 2,250 nmol per min per mg of H6Rnr2p at saturating amounts of the Rnr1p (Fig. 2A). We also assayed a mixture of separately produced H6Rnr2p (1.3 μg) and Rnr4p (4 μg) in the presence of 6 μg of Rnr1p. Now only 20 nmol of dCDP were formed per min per mg of H6Rnr2p. The high specific activity of the H6Rnr2p/Rnr4p complex indicated that it is the active form of the yeast small subunit.
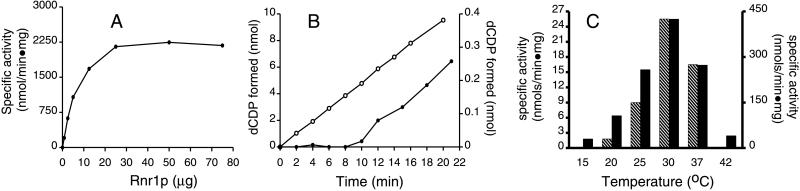
Figure 2.
Activity of the yeast small subunit. (A) Specific activity of the H6Rnr2p/Rnr4p complex (1 μg) in the presence of increasing amounts of Rnr1p. The specific activities were calculated per milligram of H6Rnr2p in the complex. (B) Activities of 2 μg of the H6Rnr2p/Rnr4p complex (○, left y axis) and of a mixture of 1 μg of H6Rnr2p and 1 μg of Rnr4p (●, right y axis), both times with 5 μg of Rnr1p. (C) Specific activities of the H6Rnr2p/Rnr4p complex (solid bars, right y axis) and of a H6Rnr2p and Rnr4p mixture (hatched bars, left y axis) in a yeast RNR assay at different temperatures.
The CDP reduction with the H6Rnr2p/Rnr4p complex was linear with time in the interval from 0 to 20 min (Fig. 2B). However, under the same conditions, a mixture of separately produced H6Rnr2p and Rnr4p showed no activity during the first 10 min; then the activity increased and remained linear, but at a very low level compared with the activity of the coexpressed H6Rnr2p/Rnr4p complex (Fig. 2B). This result indicated that the formation of the active H6Rnr2p/Rnr4p complex is time dependent.
Furthermore, when we tested the temperature dependence of the yeast RNR activity in an interval from 0 to 50°C, we found that the formation of the active H6Rnr2p/Rnr4p complex is also temperature dependent (Fig. 2C). The RNR activity in the assays with both the H6Rnr2p/Rnr4p complex and a mixture of separately produced H6Rnr2p and Rnr4p had a temperature optimum at around 30°C. At this temperature, the specific activity of the H6Rnr2p and Rnr4p mixture was 16 times lower than that of the H6Rnr2p/Rnr4p complex. However, at 25°C the specific activity of the H6Rnr2p and Rnr4p mixture was 29 times lower that that of the complex, and at 20°C, it was 59 times lower. The activity of the H6Rnr2p and Rnr4p mixture at 15°C was close to background values with the protein amounts tested (Fig. 2C).
The specific activity of a H6Rnr2p/ΔRnr4p complex lacking the last 8 C-terminal amino acid residues of Rnr4p was drastically lower compared with that of the H6Rnr2p/Rnr4p complex (less than 1%), indicating that the C-terminal part of the Rnr4p is essential for the interaction between the Rnr1p and H6Rnr2p/Rnr4p.
Iron Content and Regeneration of the Iron Center.
The H6Rnr2p/Rnr4p complex, H6Rnr2p, and Rnr4p were analyzed for iron content. No iron was detected in Rnr4p. The H6Rnr2p/Rnr4p complex contained 60.5 ± 0.9%, and H6Rnr2p contained 7.0 ± 0.4% of iron, assuming that maximal occupancy (100%) is two irons per H6Rnr2p polypeptide chain.
We then used 59Fe in an anaerobic regeneration procedure to determine whether additional iron could bind to the different yeast small RNR subunits. In contrast to the 25% increase of the iron content observed in a control experiment with mouse apo-R2 protein, we could detect only a 10% increase both in H6Rnr2p and H6Rnr2p/Rnr4p; no iron could be bound to the Rnr4p.
EPR Spectroscopy.
No organic radical could be detected in preparations of H6Rnr2p or Rnr4p by EPR before or after the iron center regeneration procedure. In contrast, the complex of coexpressed H6Rnr2p/Rnr4p (36 μM) contained 14 μM tyrosyl radical or 39% assuming that the maximal value (100%) is one radical per H6Rnr2p polypeptide. The shape of the radical spectrum (data not shown) was the same as the one obtained earlier in permeabilized yeast cells (29) and was characteristic of the tyrosyl radical conformation in RNRs of higher organisms like mouse (30). High-field EPR results on the yeast radical (to be published) also indicate the presence of a hydrogen bond to the tyrosyl radical phenoxy group, as in the mouse RNR. The tyrosyl radical signal did not increase after attempts to regenerate the iron center. Interestingly, we were able to generate a tyrosyl radical also in a mixture of separately produced H6Rnr2p and Rnr4p. In this experiment, we incubated a 52 μM solution of H6Rnr2p in buffer A containing 16 mM DTT and 150 μM FeCl3 during 25 min at 25°C either in the presence or in the absence of Rnr4p (72 μM final concentration). After the incubation, the tube with Rnr4p contained 0.2 μM tyrosyl radical or 0.4%, whereas no radical was detected in the tube incubated in the absence of Rnr4p.
Free H6Rnr2p and H6Rnr2p Bound to Rnr4p Have Different Conformations as Detected by CD.
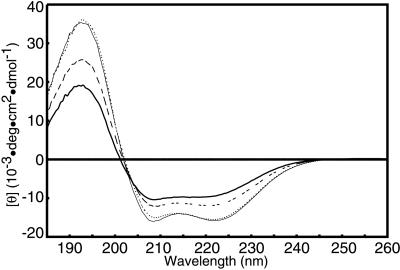
The CD spectra of the H6Rnr2p, Rnr4p, and coexpressed H6Rnr2p/Rnr4p showed characteristics of an α-helical structure with minima at 208 and 222 nm (Fig. 3). The mean residue ellipticity at 222 nm, [θ]222, which is significantly different in the spectra, may be used as an indicator of the amount of α-helical secondary structure (31), because the contribution from other secondary structures is minimal at this wavelength. Compared with the H6Rnr2p, Rnr4p and the coexpressed H6Rnr2p/Rnr4p complex had significantly more negative [θ]222 and therefore should have a higher content of α-helical secondary structure than H6Rnr2p. The isodichroic point near 203 nm indicates a two-state equilibrium and confirms that the change in the H6Rnr2p on complexing with Rnr4p is indeed mainly a gain in α-helical content (31). We also measured the CD spectrum of an equimolar mixture of separately produced H6Rnr2p and Rnr4p (Fig. 3). It nearly matched the average of the separately measured H6Rnr2p spectrum and Rnr4p spectrum and did not change significantly over 2 h.
Figure 3.
CD spectra of H6Rnr2p (thick line), Rnr4p (thin line), a mixture of H6Rnr2p and Rnr4p (dashed line) and the H6Rnr2p/Rnr4p complex (dotted line).
Complex Formation Between Rnr1p and Coexpressed H6Rnr2p/Rnr4p Assayed by Sucrose Gradient Centrifugation.
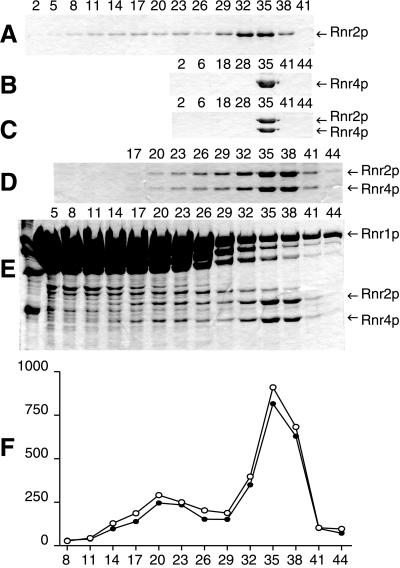
Analyses of the oligomeric structure of the small RNR subunits composed of H6Rnr2p (Fig. 4A), Rnr4p, or coexpressed H6Rnr2p/Rnr4p complex by sucrose gradient centrifugation showed that all three proteins clearly sedimented as dimers with sedimentation coefficients of 5.6 S. However, a fraction of the H6Rnr2p also trailed to higher molecular mass positions without producing discrete peaks (Fig. 4 A–C).
Figure 4.
Analyses of the oligomeric structure of yeast RNR by sucrose gradient centrifugation. The numbers correspond to the different fractions, which were around 50 in all experiments. The fractions were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. (A) 240 μg of H6Rnr2p. (B) 220 μg of Rnr4p. (C) 140 μg of the H6Rnr2p/Rnr4p complex. (D) 140 μg of the H6Rnr2p/Rnr4p complex and 480 μg of Rnr1p. (E) 140 μg of the H6Rnr2p/Rnr4p complex and 480 μg of Rnr1p in the presence of 4 mg/ml of Rnr1p throughout the gradient. (F) Computer assisted quantification of the amounts of H6Rnr2p (o) and Rnr4p (●) in (E). The ordinate is pixels per mm2. In D, E and F, fraction 20 corresponds to 9.7S and fraction 35 corresponds to 5.6S.
Next, we determined the oligomeric structure of the active yeast RNR complex with Rnr1p and H6Rnr2p/Rnr4p. Both polypeptides of the H6Rnr2p/Rnr4p heterodimer sedimented up to the position of an α2ββ′ heterotetramer (9.7 S), where α is Rnr1p, β is H6Rnr2p, and β′ is Rnr4p (Fig. 4D). The interaction between the large and small RNR subunits seemed to be very weak, because only a small portion of the heterodimer shifted to the α2ββ′ position. In addition, the H6Rnr2p and Rnr4p were present in all the fractions sedimenting between the positions of the α2ββ′ heterotetramer and the ββ′ heterodimer, indicating that the RNR complex easily disassociates. The addition of 4 mg/ml Rnr1p throughout the gradient resulted in the appearance of a peak of H6Rnr2p/Rnr4p sedimenting at the position of α2ββ′ (Fig. 4 E and F).
Discussion
The unique feature of the yeast RNR is that the active form of the small subunit consists of two polypeptide chains encoded by two different genes, the RNR2 and RNR4. We provide strong experimental evidence for this heterodimeric structure. First, the Rnr2p and Rnr4p form a stable 1:1 complex, which sediments as a heterodimer in a sucrose gradient. Second, the specific activity of the Rnr2p/Rnr4p heterodimer assayed in the presence of the Rnr1p is around 2,250 nmol of dCDP formed per min per mg of Rnr2p; this value is much higher than the specific activity obtained for the recombinant mouse R2 protein (350 nmol/mg⋅min; ref. 32) and close to the specific activity of the E. coli R2 protein (2,500 nmol/mg⋅min; ref. 33). In contrast, the homodimers of the Rnr2p or Rnr4p have no measurable activity under the same conditions. Third, as part of the heterodimer, the Rnr4p apparently interacts with the Rnr1p via its C-terminal tail, which is a characteristic feature of all other proteins of the class Ia RNR R2 subunits (4). This conclusion is supported by a very low activity of an Rnr2p/ΔRnr4p heterodimer, where the C-terminal tail of the Rnr4p is truncated. Furthermore, both polypeptides of the Rnr2p/Rnr4p heterodimer cosediment in a sucrose gradient containing Rnr1p at the position expected for a α2ββ′complex. Because the Rnr4p lacks iron center and tyrosyl radical, in the α2ββ′ complex, only one of the two possible active sites of the α2 dimer is active at any given time, a type of half-site reactivity similar to what was once proposed for the E. coli RNR (34).
Finally, the yeast Rnr2p distributes among nearly all different fractions on ion-exchange chromatography, producing several indistinct peaks, thus indicating conformational inhomogeneity. The same result was obtained with Rnr2p purified from yeast and bacterial cells, excluding that the properties of the recombinant protein are different in different expression systems. The CD spectroscopy indicated that the pure Rnr2p has an α-helical content significantly lower than other class Ia RNR small subunits. However, after binding to Rnr4p, the protein acquires an α-helical content typical of other class Ia small subunits and chromatographs in a distinct peak. These results strongly indicate that the function of the Rnr4p is to fold correctly and stabilize the Rnr2p. For this purpose, the Rnr4p needs neither an iron center nor a tyrosyl radical as evident from in vivo studies, in which an Rnr4p with an Y131F substitution allowed normal growth of rnr4 yeast cells (8). Our results suggest that Rnr2p and Rnr4p should be translated simultaneously. Otherwise, both proteins form stable homodimers, which are inactive and, at higher concentrations, inhibitory in the RNR assay, probably because they bind to Rnr1p (data not shown). Alternatively, there may be cellular factors facilitating the formation of the Rnr2p/Rnr4p heterodimers from the Rnr2p and Rnr4p homodimers. The small amounts of active small subunit formed on mixing separately produced Rnr2p and Rnr4p in vitro might be a consequence of the low concentrations of monomers present in a mixture mainly consisting of stable homodimers. Attempts to promote the formation of active H6Rnr2p/Rnr4p heterodimer in vitro by increasing the monomer concentration in a mixture of separately produced H6Rnr2p and Rnr4p by the addition of urea or imidazole have been unsuccessful (data not shown).
Deletion of the RNR4 gene either is lethal or leads to different mutant phenotypes, such as slow growth and cold-sensitivity, depending on the genetic background of the yeast strain. Therefore, Rnr2p can function without Rnr4p in yeast cells but at a critically low level. This can be due to the presence of chaperons, which may to some extent assist Rnr2p folding. We have shown that the specific activity of the yeast RNR small subunit depends highly on the concentration of the large subunit (Fig. 2A), because the subunit interaction is weak (Fig. 4 D–F). This weak interaction may explain why the impaired activity of the yeast RNR small subunit lacking Rnr4p can be increased by overexpression of the large subunit genes RNR1 or RNR3 (9).
Interestingly, there may be a parallel between the Rnr1p/Rnr3p and the Rnr2p/Rnr4p. Highly purified recombinant Rnr3p, with an amino acid sequence identity of 80% compared with the Rnr1p, is almost inactive when assayed together with the H6Rnr2p/Rnr4p heterodimer. However, on the addition of Rnr1p to an assay mixture containing Rnr3p, more RNR activity is observed compared with when Rnr1p is added to an identical assay mixture lacking the Rnr3p (data not shown).
From the SDS/PAGE analysis and Western blots published by Stubbe and coworkers (18), it is clear that their highly purified Rnr2p, which they expressed in yeast, contains Rnr4p. This fact can explain the observed RNR activity. They also described an Rnr2p mutant, K387N, which had a higher specific activity (300 nmol of dCDP per min per mg) than their wild-type Rnr2p. We believe that the higher activity of this mutant can be attributed to the higher Rnr4p content in this preparation, as can be seen in the SDS/PAGE analysis, and not to the mutation itself. Their Rnr2p specific activity should be compared with the specific activity of 2,250 nmol of dCDP per min per mg for our H6Rnr2p complexed 1:1 with Rnr4p. The Rnr2p produced in bacteria and therefore completely lacking Rnr4p is inactive when assayed together with Rnr1p, both in our and Stubbe's studies.
Stubbe and coworkers (18) proposed that Rnr4p binds iron and delivers it to Rnr2p in a process similar to the activation of the copper zinc superoxide dismutase by Lys-7. In contrast to this hypothesis, our data suggest that the Rnr4p does not function transiently in the activation of the Rnr2p. Instead, it directs the folding of the Rnr2p and is then required in the heterodimeric complex to stabilize an Rnr2p structure, which allows binding of iron and generation of tyrosyl radical.
Our results show that there are significant differences between the yeast RNR and other class Ia RNRs concerning the structure of the active enzyme complex. Hopefully, further studies will reveal whether there are other differences and why this highly conserved enzyme has developed in different directions in yeast and other eukaryotes.
Acknowledgments
We thank S. Elledge for providing plasmids containing RNR1, RNR2, and RNR3 and Y. Andreichuk for helping with the expression of Rnr2p in yeast. This work was supported by the Swedish Natural Sciences Research Council, by the Kempe foundation, and by fellowships to V.D. from the Wenner-Grenska Samfundet and The Royal Swedish Academy of Sciences. The European Training and Mobility of Researchers program under contract nos. ERBFMGECT95077 and ERBFMRXCT98027 is also acknowledged.
Abbreviations
- RNR
ribonucleotide reductase
- AS
ammonium sulfate
- CDP
cytidine 5′-diphosphate
- EPR
electronic paramagnetic resonance
References
- 1.Reichard P. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 2.Reichard P. Science. 1993;260:1773–1777. doi: 10.1126/science.8511586. [DOI] [PubMed] [Google Scholar]
- 3.Thelander L, Reichard P. Annu Rev Biochem. 1979;48:133–158. doi: 10.1146/annurev.bi.48.070179.001025. [DOI] [PubMed] [Google Scholar]
- 4.Lycksell P-O, Ingemarson R, Davis R, Gräslund A, Thelander L. Biochemistry. 1994;33:2838–2842. doi: 10.1021/bi00176a013. [DOI] [PubMed] [Google Scholar]
- 5.Elledge S J, Davis R W. Genes Dev. 1990;4:740–751. doi: 10.1101/gad.4.5.740. [DOI] [PubMed] [Google Scholar]
- 6.Elledge S J, Davis R W. Mol Cell Biol. 1987;7:2783–2793. doi: 10.1128/mcb.7.8.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurd H K, Roberts C W, Roberts J W. Mol Cell Biol. 1987;7:3673–3677. doi: 10.1128/mcb.7.10.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P J, Chabes A, Casagrande R, Tian X C, Thelander L, Huffaker T C. Mol Cell Biol. 1997;17:6114–6121. doi: 10.1128/mcb.17.10.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang M, Elledge S J. Mol Cell Biol. 1997;17:6105–6113. doi: 10.1128/mcb.17.10.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang M, Zhou Z, Elledge S J. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 11.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 12.Weinert T. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 13.Zhao X, Muller E G, Rothstein R. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 14.Chabes A, Domkin V, Thelander L. J Biol Chem. 1999;274:36679–36683. doi: 10.1074/jbc.274.51.36679. [DOI] [PubMed] [Google Scholar]
- 15.Desany B A, Alcasabas A A, Bachant J B, Elledge J E. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitols E, Bauer V A, Stanbrough E C. Biochem Biophys Res Commun. 1970;41:71–77. doi: 10.1016/0006-291x(70)90470-5. [DOI] [PubMed] [Google Scholar]
- 17.Lammers M, Follmann H. Eur J Biochem. 1984;140:281–287. doi: 10.1111/j.1432-1033.1984.tb08099.x. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen H-H T, Ge J, Perlstein D L, Stubbe J. Proc Natl Acad Sci USA. 1999;96:12339–12344. doi: 10.1073/pnas.96.22.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis R, Thelander M, Mann G J, Behravan G, Soucy F, Beaulieu P, Lavallee P, Gräslund A, Thelander L. J Biol Chem. 1994;269:23171–23176. [PubMed] [Google Scholar]
- 20.Andreichuk I V, Shabes A V, Ryzhova T A, Kotova I A, Domkin V D. Mol Gen Mikrobiol Virusol. 1995;1:21–28. [PubMed] [Google Scholar]
- 21.Mann G J, Gräslund A, Ochiai E, Ingemarson R, Thelander L. Biochemistry. 1991;30:1939–1947. doi: 10.1021/bi00221a030. [DOI] [PubMed] [Google Scholar]
- 22.Thelander L, Eriksson S, Åkerman M. J Biol Chem. 1980;255:7426–7432. [PubMed] [Google Scholar]
- 23.Thelander M, Gräslund A, Thelander L. J Biol Chem. 1985;260:2737–2741. [PubMed] [Google Scholar]
- 24.Engström Y, Eriksson S, Thelander L, Åkerman M. Biochemistry. 1979;18:2941–2948. doi: 10.1021/bi00581a004. [DOI] [PubMed] [Google Scholar]
- 25.Söderman K, Reichard P. Anal Biochem. 1986;152:89–93. doi: 10.1016/0003-2697(86)90124-7. [DOI] [PubMed] [Google Scholar]
- 26.Nyholm S, Mann G J, Johansson A G, Bergeron R J, Gräslund A, Thelander L. J Biol Chem. 1993;268:26200–26205. [PubMed] [Google Scholar]
- 27.Atkin C L, Thelander L, Reichard P, Lang G. J Biol Chem. 1973;248:7464–7472. [PubMed] [Google Scholar]
- 28.Ullman B, Gudas L J, Caras I W, Eriksson S, Weinberg G L, Wormsted M A, Martin D W., Jr J Biol Chem. 1981;256:10189–10192. [PubMed] [Google Scholar]
- 29.Harder J, Follmann H. Free Radical Res Commun. 1990;10:281–286. doi: 10.3109/10715769009149896. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt P, Andersson K, Barra A-L, Thelander L, Gräslund A. J Biol Chem. 1996;271:23615–23618. doi: 10.1074/jbc.271.39.23615. [DOI] [PubMed] [Google Scholar]
- 31.Greenfield N, Fasman G D. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 32.Rova U, Goodtzova K, Ingemarson R, Behravan G, Gräslund A, Thelander L. Biochemistry. 1995;34:4267–4275. doi: 10.1021/bi00013a016. [DOI] [PubMed] [Google Scholar]
- 33.Ormö M, Regnström K, Wang Z, Que L, Jr, Sahlin M, Sjöberg B-M. J Biol Chem. 1995;270:6570–6576. doi: 10.1074/jbc.270.12.6570. [DOI] [PubMed] [Google Scholar]
- 34.Sjöberg B M, Karlsson M, Jörnvall H. J Biol Chem. 1987;262:9736–9743. [PubMed] [Google Scholar]