Abstract
Recombinant clones of the carboxyl terminus of the major surface glycoprotein (MsgC) of Pneumocystis jirovecii are useful for analyzing serologic responses in humans. However, there is no standardized set of antigens in general use, which could lead to conflicting results. We have previously shown that human immunodeficiency virus type 1 (HIV-1)-infected patients with prior Pneumocystis pneumonia (PcP+) responded more frequently and more strongly to a clone of MsgC than did HIV-1-infected patients without PcP (PcP−). Here we test three new clones of MsgC to determine the effect of antigenic sequence variation on immune reactivity in blood donors and HIV-infected patients previously analyzed for reactivity to our original MsgC clone. In Western blot analyses, PcP+ patients exhibited the highest frequency of reactivity to each MsgC clone, and the frequency of reactivity with all four MsgC clones together was significantly higher in sera from PcP+ patients than in sera from the other patient groups. Furthermore, in an enzyme-linked immunosorbent assay we found that the PcP+ population had the highest level of reactivity to two of the four clones tested. One of the new clones could distinguish between PcP+ and PcP− populations, and two MsgC clones could distinguish blood donors from the other patient populations. The results show that inherent differences in MsgC amino acid sequence can affect recognition by antibodies independently of variations in protein length or patient population, and the utility of a clone depends on its sequence and on the populations tested.
Pneumocystis jirovecii is a fungal opportunistic pathogen of humans that causes Pneumocystis pneumonia (PcP) in immunocompromised people, including those infected with human immunodeficiency virus (HIV) (11, 31, 39, 42, 51, 52). The advent of highly active antiretroviral therapy for the treatment of HIV-infected (HIV+) patients, with the concomitant reconstitution of the immune response, has led to an interest in antigen-specific immune responses directed against P. jirovecii antigens in immunocompromised patients and healthy persons (9, 38).
The role of antibodies in infection with P. jirovecii is not well defined, and yet there is a high frequency of reactivity to Pneumocystis antigens in healthy adults (29, 34, 35, 40), probably due to exposure to the organism since childhood, since most children exhibit organism-specific antibodies in early childhood (34, 50). Furthermore, animal models of Pneumocystis infection suggest that antibodies are important in prevention of PcP (1, 13, 15, 17, 19, 25, 28, 33, 54). Little is known about the nature of the antigens recognized by human antibodies and the characteristics of the immune response to these antigens in health and disease. Many seroepidemiologic studies have been carried out by using crude antigen preparations derived from infected rodent lungs (4, 6, 27, 30, 34, 37, 40, 41). The potential antigenic variation in different ex vivo samples, the molecular complexity of Pneumocystis samples isolated ex vivo, the contamination of isolates with coinfecting pathogens, and the inability to clone and grow large numbers of Pneumocystis organisms in vitro make the isolation of standard antigen preparations difficult. Furthermore, Pneumocystis is species specific, suggesting that organisms or antigens derived from animal hosts are not optimal for experiments in the human system and that the use of host-specific reagents is more relevant (51). These problems, along with the difficulty in obtaining Pneumocystis samples of sufficient size from infected human lungs, suggest that recombinant proteins may be more appropriate for use in immunologic studies.
It is known that many Pneumocystis proteins can serve as antigens in both humans and rodents; however, the molecular identity of many of these antigens has not been determined (32, 40). One antigen that has been well studied is the major surface glycoprotein (Msg) or glycoprotein A, a family of related proteins encoded by multiple genes in the Pneumocystis genome (12, 14, 22, 26, 43). Msg elicits protective B-cell and T-cell responses and plays an important role in the interaction of Pneumocystis with its host (18, 46, 47, 48). It appears that expression of Msg is tightly regulated, and only one msg gene at a time can be transcribed in a Pneumocystis cell (23, 44, 45). Furthermore, the isoform of Msg that is expressed can change in response to interaction with an antibody, suggesting that Pneumocystis may switch Msg isoform as a mechanism of evasion of the immune system (16). Such antigenic variation is commonly seen in other organisms such as trypanosomes and the spirochete Borrelia (3, 49, 53).
We and others have generated recombinant fragments of Msg for use in immunological assays (2, 7, 8, 29). The carboxyl terminus of Msg, which we call MsgC, is the most antigenic fragment of Msg and has received much attention. We have demonstrated a higher frequency of reactivity by Western blotting and a higher magnitude of reactivity by enzyme-linked immunosorbent assay (ELISA) in HIV+ patients that had a prior episode of PcP (PcP+) compared to those who did not have PcP (PcP−) (7, 8). However, other workers have reported different results using recombinant Msg C-terminal fragments that are similar but not identical to the construct we used (2, 29). The observed discrepancies in the reported results could be due to differences in MsgC used or to differences in the patient populations sampled.
It is important to determine whether individual clones of MsgC behave differently in biological assays. Furthermore, Msg antigenic variation is important for basic science and clinical aspects of P. jirovecii infection. Here we have assessed the role of MsgC clone variation on serum antibody reactivity using patient samples with known reactivity patterns to our original MsgC fragment. We have generated a small panel of three new MsgC fragments that differ from one another in putative amino acid sequence and have analyzed by Western blotting and ELISA the serum antibody reactivity to these fragments in blood donors (BDs) and HIV+ patients that have previously been analyzed for reactivity to a single MsgC clone.
MATERIALS AND METHODS
Serum specimens.
Serum samples, as well as the demographic and clinical information about the human subjects used in the present study, were described in detail in our earlier reports (7, 8). Briefly, these specimens included 95 samples from healthy BDs at the Hoxworth Blood Center, Cincinnati, OH, and 94 specimens from HIV+ patients at the University of Cincinnati Infectious Disease Center. Of these, 33 were from PcP+ patients and 61 were from PcP− patients. PcP diagnosis was based on microscopic examination of a silver-stained preparation following bronchoscopy.
Isolation and expression of MsgC fragments.
We had previously described the characterization of a fragment of Msg (MsgC) which we amplified by PCR from a cloned msg gene. Here we call that fragment MsgC1 to differentiate it from others that we have generated. New MsgC fragments called MsgC3, MsgC8, and MsgC9 were cloned, expressed, and purified as previously described (7, 8). Briefly, oligonucleotides were designed based on the known sequences of six msg genes of P. jirovecii (12, 29) and are specific for conserved regions of these genes. They were used in PCRs with DNA isolated from P. jirovecii-infected human lung as a template and Amplitaq enzyme (Applied Biosystems, Foster City, CA) to generate msgC gene segments. The sequences of the oligonucleotides were as follows: 5′-CGTTTCTCTTTTGTATGTGT-3′ (forward) and 5′-ATCATGAACGAAATAACC-3′ (reverse). The PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen, Madison WI) for restriction mapping and nucleotide sequencing and were then inserted into the pET30 vector (Novagen, Madison WI) in the correct orientation for the expression and purification of recombinant MsgC proteins in Escherichia coli. The recombinant proteins were expressed in inclusion bodies within E. coli and were purified by standard methods. Briefly, bacterial cultures expressing recombinant proteins were harvested by centrifugation, the cell pellet was sonicated and washed three times in binding buffer without urea (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]), and the final pellet was dissolved in binding buffer with 6 M urea. The recombinant preparations were purified by affinity chromatography using HIS-Binding resin (Novagen, Madison, WI), with the urea being removed during the wash stages. Eluted proteins were dialyzed overnight against phosphate-buffered saline (PBS; pH 7.4), divided into aliquots, and frozen at −70°C until use. Recombinant protein expressed from the pET30 vector without insert was used as a control antigen. Protein concentration was determined based on the A280 using a standard curve generated with bovine serum albumin.
Western blot analysis.
Western blot analysis was performed according to previously reported procedures (7). In brief, recombinant MsgC fragments were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, blocked in 1% nonfat milk in TTBS (20 mM Tris, 0.5 M NaCl, 0.05% Tween 20), cut into strips, and incubated with serum specimens (diluted 1/50 in TTBS). The strips were washed and incubated with horseradish peroxidase (HRP)-labeled goat anti-human immunoglobulin G (IgG; Kirkegaard & Perry Laboratories, Gaithersburg, MD), and color was developed with TMB (3,3′,5,5′-tetramethylbenzidine) substrate. HRP-labeled S protein, which reacts with the S-tag expressed by the pET30 vector, was used to identify the appropriate bands for analysis and served as a positive control. Each assay was read by two independent readers.
ELISA.
ELISA was performed according to previously reported procedures (8). Briefly, all serum specimens to be analyzed, as well as a standard reference serum, were tested against the following antigens: recombinant MsgC fragments, E. coli extract expressing the pET vector without insert as a vector control, and PBS without antigen (negative control). Duplicate wells of a 96-well plate were coated with antigen (5 μg/ml, 100 μl/well) in PBS (pH 7.4) overnight. The plates were washed in wash buffer (PBS with 0.05% Tween 20) and blocked with blocking buffer (wash buffer with 5% nonfat milk) (200 μl/well) for 2 h at room temperature. The plates were washed again, and human serum diluted 1/100 in blocking buffer was added to each well (100 μl/well). The plates were rocked overnight at 4°C and washed in wash buffer, and HRP-labeled goat anti-human IgG (heavy and light chain) was added to each well (100 μl/well at a 1/5,000 dilution in blocking buffer). The negative control (no antigen) and two positive controls (HRP-labeled S protein and a standard serum pool) were used on each plate. The plates were incubated at room temperature for 1 h, washed, and developed by adding TMB substrate (100 μl/well). Color was allowed to develop for 4 min, the reaction was stopped by the addition of 100 μl of 0.18 M H2SO4 to each well, and the plates were read at a wavelength of 450 nm. The reactivity of each serum specimen to Msg was corrected by subtraction of the reactivity of that serum to the pET protein (the mean optical density of Msg − the mean optical density of pET). Variations in assay results using a control serum were measured for MsgC1 on a per-plate, daily, and 4-day basis (the coefficients of variance [CV] were 3.6 to 7%, 4.8 to 7.4%, and 13.3%, respectively).
Statistics.
Statistical analysis for Western blot studies was performed by Graphpad Instat (Graphpad Software). The statistical significance (P < 0.05) of categorical variables was determined by the χ2 test or, when appropriate, by the Fisher exact test.
Mean differences in ELISA results by patient status were evaluated after subtracting the value of pET from each antigen level. The distributions of corrected antigen levels for MsgC1, MsgC3, MsgC8, and MsgC9 were visually assessed and tested for normality using the Shapiro-Wilk's statistic. The log transformation was applied in order to satisfy the assumption of normality, thus contributing to accurate interpretation of results of hypothesis testing. Preliminary analyses showed that antigen variability differed among patient groups. To satisfy the assumption of homogeneity of variance, weighted least-squares regressions were performed to evaluate differences between mean values for each antigen among patient groups: BDs, HIV PcP+ patients, and HIV PcP− patients. Differences between each pair of means was evaluated after the Tukey's adjustment was applied to the overall significance level of 0.05 to adjust for multiple comparisons. Analyses were carried out by using the SAS software PROC GLM (SAS for Windows, version 9.2; SAS Institute, Cary, NC).
GenBank accession numbers.
The GenBank accession numbers for MsgC3, MsgC8, and MsgC9 are DQ000981, DQ000982, and DQ000983, respectively.
RESULTS
Generation of a panel of different MsgC fragments.
We used PCR to generate a small panel of different MsgC clones as outlined in Materials and Methods. DNA sequence analysis showed that the three new MsgC clones differed from one another and from the existing MsgC clone in nucleotide sequence and in putative amino acid sequence (Fig. 1). The four MsgC variants, which each code for approximately 425 amino acids, exhibited 83 to 99% homology at the nucleotide level and 77 to 99% homology at the putative amino acid level. MsgC3 and MsgC9 were the most closely related, having just three amino acid substitutions, two of which resided within the amino-terminal 20 amino acid residues and resulted in charge differences.
FIG. 1.
Schematic and deduced amino acid sequences of MsgC fragments. (A) MsgC fragments are 1,338 nucleotides long and map to the carboxyl end of Msg. (B) The deduced amino acid sequences of MsgC1, MsgC3, MsgC8 and MsgC9. *, conserved amino acids across the four MsgC clones.
Western blot analysis of MsgC fragments.
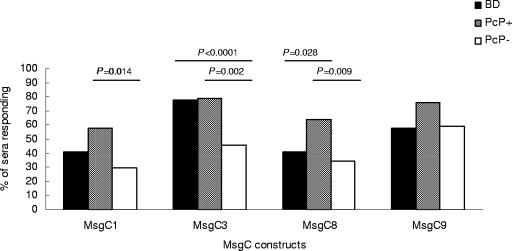
We initially tested 95 healthy BD sera and 94 samples from HIV+ patients, 33 of whom had had a previous bout of PcP, for the ability to recognize MsgC3, MsgC8, and MsgC9 in Western blot analysis. Figure 2 shows a Western blot of reactivity to MsgC3 in HIV+ patients and is typical of the results seen with all patient populations and all constructs. We then compared these results to those previously obtained for MsgC1 (Fig. 3 and Table 1). All patient populations were highly reactive to the MsgC clones, with 85 of the BDs (89.5%), 28 of the PcP+ patients (84.8%), and 45 of the PcP− patients (73.8%) reacting with at least one MsgC fragment. The frequency of reactivity to each individual MsgC clone varied with the clone tested and with the patient population; however, MsgC3 and MsgC9 were recognized more frequently than MsgC1 or MsgC8 in all populations. The PcP+ patient population exhibited the highest reactivity to each MsgC fragment, and PcP− patients exhibited the lowest frequency of reactivity to MsgC1, MsgC3, and MsgC8 of all of the patient groups.
FIG. 2.
Reactivity to MsgC3. A representative Western blot showing reactivity to MsgC3 is shown. Lanes: M, molecular mass marker with ladder sizes in kilodaltons; S, reactivity to S protein (+ control); −, negative control (no serum); +, positive control serum; 1 to 4, four different serum samples exhibiting “+” or “−” reactivity to MsgC3. The arrow indicates the expected molecular mass of MsgC3 in kilodaltons.
FIG. 3.
Reactivity to four MsgC fragments in a Western blot analysis. The frequencies of reactivity to MsgC fragments in healthy BDs and in HIV-infected PcP+ or PcP− patients were determined. Comparisons between the groups were done by using the Fisher exact test. Significant values (P < 0.05) are shown.
TABLE 1.
Western blot analysis of four different MsgC clones by using serum antibodies from BDs and PcP+ and PcP− patients
| MsgC clone(s) recognized | % of sera (no.) responding
|
||
|---|---|---|---|
| BD (n = 95) | PcP+ (n = 33) | PcP− (n = 61) | |
| MsgC1 alone | 6.3 (6) | 0.0 (0) | 3.3 (2) |
| MsgC3 alone | 15.8 (15) | 3.0 (1) | 6.6 (4) |
| MsgC8 alone | 0.0 (0) | 0.0 (0) | 3.3 (2) |
| MsgC9 alone | 3.2 (3) | 3.0 (1) | 13.1 (8) |
| MsgC1 + MsgC3 | 7.4 (7) | 3.0 (1) | 0.0 (0) |
| MsgC1 + MsgC8 | 0.0 (0) | 0.0 (0) | 1.6 (1) |
| MsgC1 + MsgC9 | 2.1 (2) | 3.0 (1) | 1.6 (1) |
| MsgC3 + MsgC8 | 0.0 (0) | 3.0 (1) | 0.0 (0) |
| MsgC3 + MsgC9 | 11.6 (11) | 6.1 (2) | 8.2 (5) |
| MsgC8 + MsgC9 | 0.0 (0) | 0.0 (0) | 4.9 (3) |
| MsgC1 + MsgC3 + MsgC8 | 2.1 (2) | 0.0 (0) | 0.0 (0) |
| MsgC1 + MsgC3 + MsgC9 | 2.1 (2) | 3.0 (1) | 6.6 (4) |
| MsgC3 + MsgC8 + MsgC9 | 17.9 (17) | 12.1 (4) | 8.2 (5) |
| MsgC1 + MsgC3 + MsgC8 + MsgC9 | 21.1 (20) | 48.5 (16) | 16.4 (10) |
| All MsgC1 | 41.1 (39) | 57.6 (19) | 29.5 (18) |
| All MsgC3 | 77.9 (74) | 78.8 (26) | 45.9 (28) |
| All MsgC8 | 41.1 (39) | 63.6 (21) | 34.4 (21) |
| All MsgC9 | 57.9 (55) | 75.8 (25) | 59.0 (36) |
| None | 10.5 (10) | 15.2 (5) | 26.2 (16) |
In BDs, MsgC3 was recognized by 74 of the samples (77.9%), whereas MsgC9 was recognized by 55 of the sera (57.9%) and MsgC1 and MsgC8 were recognized by 39 samples (41.1%). MsgC3 was also the most frequently recognized protein in PcP+ patients, with 26 sera (78.8%) exhibiting reactivity. MsgC9, MsgC8, and MsgC1 were recognized by 25 (75.8%), 21 (63.6%), and 19 (57.6%) PcP+ samples, respectively. In PcP− patients, MsgC9 was recognized at the highest frequency, with 36 of the sera reacting (59%). MsgC3, MsgC8 and MsgC1 were recognized by 28 (45.9%), 21 (34.4%) and 18 (29.5%) PcP− samples, respectively. Table 1 shows a summary of the reactivity of the sera from each patient population to each of the MsgC constructs used.
As previously described, the frequency of reactivity to MsgC1 was significantly higher in PcP+ patients than in PcP− patients (P = 0.014). This was also true for MsgC3 and MsgC8 but not for MsgC9 (Fig. 3). MsgC3 was recognized by 26 PcP+ sera (78.8%) compared to 28 of PcP− sera (45.9%) (P = 0.002). Twenty-one PcP+ sera (63.6%) recognized MsgC8 compared to 21 PcP− sera (34.4%) (P = 0.009). MsgC9 was recognized by 25 PcP+ sera (75.8%) and 36 PcP− sera (59%) (P > 0.05).
Recognition levels of MsgC3 were also significantly different between BD and PcP− sera, with 74 BD sera (77.9%) and 28 PcP− sera (45.9%) reacting positively (P < 0.0001). Reactivities to MsgC1, MsgC8, and MsgC9 were not significantly different between BDs and PcP− populations. The frequency of recognition of MsgC8 could differentiate between BDs and PcP+ patients, with 39 BD sera (41.1%) and 21 PcP+ sera (63.6%) reacting (P = 0.028). Reactivities to MsgC1, MsgC3, or MsgC9 were not significantly different between BDs and PcP+ patients.
Recognition of multiple MsgC fragments in Western blot analyses.
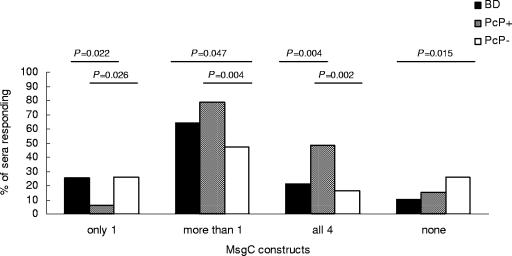
The frequency of reactivity to each MsgC clone dictates that some serum samples must recognize more than one MsgC fragment. Figure 4 shows that there was a bias toward recognition of multiple MsgC clones in all populations; this bias was strongest in PcP+ patients and weakest in PcP− patients. Of 95 BD sera tested, 61 (64.2%) reacted with more than one clone, and 20 (21%) reacted with all clones. Similarly, 26 of 33 of PcP+ sera (78.8%) recognized more than one MsgC clone, and 16 (48.5%) recognized all clones. Sera from PcP− patients exhibited the highest frequency of single-MsgC reactivity, with 16 of 61 sera (26.2%) recognizing a single MsgC clone and only 10 (16.4%) recognizing all clones.
FIG. 4.
Western blot analysis of coreactivity to multiple MsgC fragments in healthy BDs and in HIV-infected PcP+ or PcP− patients. Comparisons between the groups were done by using the Fisher exact test. Significant values (P < 0.05) are shown.
Recognition of MsgC clones in ELISA.
We next compared the abilities of BD and PcP+ and PcP− patient populations to recognize each MsgC clone in ELISA. The response to MsgC1 and MsgC3 was higher than the response to MsgC8 and MsgC9 in all populations tested, and the response to MsgC8 was the lowest in all groups (Table 2). The patterns of recognition of the clones were similar in PcP+ and PcP− sera, with the highest reactivity recorded against MsgC1, followed by MsgC3, MsgC9, and MsgC8. However, the hierarchy of recognition of the clones was different for BDs (MsgC3 > MsgC1 > MsgC9 > MsgC8). Furthermore, the magnitudes of the responses were similar for all clones in the BD population, whereas the responses seen in PcP+ and PcP− sera were more widespread.
TABLE 2.
Geometric mean results of ELISA by blood donor and PcP status for each antigen
| Patient group | Mean titer (95% confidence interval)a
|
|||
|---|---|---|---|---|
| MsgC1* | MsgC3† | MsgC8‡ | MsgC9‡ | |
| BD | 0.24 (0.18-0.32) | 0.29 (0.22-0.39) | 0.14 (0.10-0.19) | 0.15 (0.11-0.20) |
| PcP+ | 0.83 (0.59-1.17) | 0.50 (0.34-0.74) | 0.05 (0.03-0.08) | 0.11 (0.07-0.20) |
| PcP− | 0.46 (0.37-0.58) | 0.25 (0.18-0.34) | 0.03 (0.02-0.04) | 0.05 (0.03-0.06) |
P values were obtained by using Tukey's adjustment for multiple comparisons. Mean values were obtained by using weighted least-squares regression. *, significant (P ≤ 0.01) differences between all means; †, significant (P ≤ 0.01) difference between means (PcP− versus PcP+); ‡, significant (P ≤ 0.01) differences between means (BD versus PcP−, BD versus PcP).
The response to MsgC1 in ELISA was significantly higher in the PcP+ patient population than in the BD or PcP− groups and was higher in the PcP− group than in BDs (Table 2). The response to MsgC3 could differentiate between PcP+ and PcP− populations, and the responses to MsgC8 and MsgC9 could differentiate the BD from both the PcP+ and PcP− cohorts.
DISCUSSION
We had previously shown that we could differentiate between HIV+ patient cohorts that had PcP and those that did not have PcP on the basis of reactivity to a cloned fragment of Msg called MsgC1 in Western blot analyses and ELISAs. Here we expand that study to include three new clones of MsgC. Our goal was to determine whether the ability to distinguish patient populations was a characteristic of MsgC1 alone or if other MsgC clones had similar properties. The results show that the ability to differentiate between PcP+ and PcP− patient groups was not restricted to MsgC1, but neither was it a property common to all MsgC clones. Using the four clones of MsgC tested here, we were able to find significant differences in serum reactivity in BDs and in PcP+ and PcP− patient populations.
The spectrum of results is interesting for two main reasons. First, the utility of any given MsgC clone is an intrinsic property of the amino acid sequence of the molecule, since the same patient sera were tested on each MsgC clone. The fact that individual MsgC clones have different characteristics within the same test populations is one explanation for the difference in results obtained by different labs using recombinant Msg fragments in immunological assays. Second, the choice of MsgC to be studied can be made to suit the patient cohorts studied. For example, in Western blot analyses, MsgC3 would be useful for comparing BD and PcP− sera or PcP+ and PcP− specimens, but MsgC8 would be useful for studying BD and PcP+ sera.
Each MsgC was recognized independently of the others in Western blot analyses. This suggests that there are clone-specific antibody epitopes on each MsgC molecule and that these epitopes are responsible for the discriminatory power of the MsgC clone. There is, however, a bias toward the recognition of multiple clones in a given serum sample, which is evident in both methods of assay and in all patient populations examined. The ability to recognize more than one MsgC clone differentiates PcP+ patients as a group from either PcP− patients or BDs in Western blot analysis in that there is a significantly higher frequency of recognition of multiple clones in the PcP+ cohort. The recognition of more than one MsgC molecule could be explained by the presence of separate pools of MsgC clone-specific antibodies, a single pool of cross-reactive antibodies, or both. Although cross-reactive antibodies may exist, they do not mask the presence of unique epitopes on MsgC molecules, allowing differences in reactivity in patient populations to be studied.
In an ELISA each MsgC could be recognized by each patient population, but the magnitudes of the responses varied with the clone used and the population assayed. The response to MsgC1 and MsgC3 was highest in the PcP+ patients and was significantly higher than the corresponding response in PcP− patients. The response to MsgC1 was also significantly higher in the PcP+ patient population than in the BDs.
In the BD group, the levels of response to each of the four clones tested were very similar. This was not the case for either the PcP+ or PcP− cohort, which had a much broader spectrum of responses. Interestingly, the hierarchy in responses seen in PcP+ and PcP− patient populations (MsgC1 > MsgC3 > MsgC9 > MsgC8) was different for healthy BDs, where MsgC3 showed the highest level of reactivity.
The results of ELISA and Western blot analyses are quite distinct and are not fully concordant, as we had previously reported for the recombinant Msg fragments MsgA, MsgB, and MsgC1. We interpret this as meaning that the epitopes that are recognized in both assays are different. It is likely that Western blotting selects for linear epitopes, since the process involves reduction and denaturation of antigens. However, in an ELISA the antigens are not reduced or denatured and may represent a mixture of linear and conformational epitopes.
The PcP+ patient population is interesting in that it exhibits an increased frequency of response to multiple MsgC clones in Western blot analyses compared to BDs or PcP− patients, has the highest magnitude of response to two of the four clones tested by ELISA, and demonstrates a different hierarchy of responses in ELISA than do healthy BDs. One possible explanation for this is that healthy BDs who are exposed to Pneumocystis probably clear the organism from the body faster than PcP+ patients. The longer exposure time in PcP+ patients may allow a wider spectrum of Msg variants to be expressed by the organism and to serve as antigens for the immune system. Healthy BD and PcP− patients would not have such an exposure and would be less likely to recognize multiple MsgC clones.
One caveat in studying reactivity to multiple clones of MsgC is the diversity of the clones used in the study. It is likely that if clones of an antigen are closely related in linear amino acid sequence, they will behave similarly in immunological assays, thereby increasing the frequency of reactivity to multiple clones. Our MsgC clones exhibit 77 to 99% homology at the putative amino acid level, and it is probable that results would vary if different clones were used. The choice of oligonucleotides used for PCR may be a factor in generating diversity in a panel of MsgC clones. The oligonucleotides that we designed were chosen because they are specific for a highly conserved region of six different msg genes whose sequences are available in the scientific literature. Five of the six genes analyzed were identical in nucleotide sequence, and all six were identical in putative amino acid sequence, at the sites where our primers anneal. Our primers were designed based on the frequency of nucleotide sequences in these conserved regions and, to increase the chances of obtaining products, favor the most frequent sequence. It is possible that choosing a different set of primers would result in a different set of MsgC fragments; however, this potential for bias in the generation of MsgC clones does not affect the results of this work, since the four clones we have generated at random all have different nucleotide sequences and behave differently in antibody recognition assays.
The major surface glycoprotein of Pneumocystis is a glycosylated protein, and it is likely that sugar moieties play a role in the formation of antigenic epitopes in the Msg molecule. The Msg fragments that we have used here were generated as recombinant proteins expressed in E. coli and are therefore not posttranslationally modified by glycosylation. The epitopes that we address here and in previous studies are contained within the protein backbone of Msg and do not include any effects of posttranslational modification. Therefore, it is likely that we will not be able to study certain antibody epitopes of Msg; however, the fact that these recombinants are recognized specifically by antibodies and that reactivity to recombinant MsgC fragments can differentiate between different patient populations suggests that this system of study is valid and will yield interesting results. Other expression systems that allow for glycosylation to occur, for example, Pichia pastoris, would allow us to test whether proteins expressed in different glycosylation states behave differently in immunological assays. To date, we have not expressed our Msg recombinant antigens in such systems, and the validity of such systems may be doubtful since glycosylation patterns may vary on an organism-specific basis.
It is likely that MsgC molecules can consist of more than one epitope, which complicates serological analysis of immunoreactivity. The nature of individual epitopes of Msg has not been studied, and a comparison of the linear sequences of different Msg molecules shows too many differences to be useful for mapping individual epitopes. However, the present study has identified two MsgC clones—MsgC3 and MsgC9—that differ in only three amino acids close to the amino-terminal end of the molecule. These clones behave differently from one another in both Western blot analyses and ELISAs, and each clone can be recognized independently of the other, suggesting that a small number of amino acids can affect the recognition of MsgC by antibodies. We do not know whether these truly represent MsgC clones encoded by the Pneumocystis genome or if they are variants due to PCR errors; however, they do serve as valid antigens with which to probe the breadth of the immune response to an Msg molecule and can test the extent of variability that is tolerated by antibodies during immune recognition. The small number of polymorphic amino acids in these two clones makes it feasible to map the contribution of individual amino acids to the epitope responsible for each phenotype. We are currently using site-directed mutagenesis to make and express MsgC3/MsgC9 chimeric molecules to further dissect the epitopes recognized in these molecules. The identification of individual epitopes on these molecules will be very useful for studying regions of Msg that may be involved in protection from infection or recovery from disease.
The results of the present study show significant differences in frequency and magnitude of serum antibody responses in BDs and in PcP+ and PcP− patient populations to different MsgC clones. We have examined the total IgG response to Msg antigens and, while the results are interesting, the present study is a preliminary one, and the analysis of IgG subtypes may yield data relevant to the disease state. We have not yet examined the reactivity of individual IgG subtypes to Msg fragments in any patient population. The results of such an analysis may help define a clinically relevant role for immune reactivity to Msg constructs in healthy donors and patients at risk for developing PcP.
Although the role of P. jirovecii-specific antibodies in recovery from or susceptibility to PcP has not been clearly defined, antibody titers do vary with onset and recovery from PcP (5, 10, 20, 21, 24, 36). The data reported here suggest that study of the immune responses to MsgC fragments may be a way of identifying epitopes important in PcP outcome and suggest that these reagents will be useful for describing serological parameters that may change with disease state or HIV infection. An understanding of these responses and the role they play in Pneumocystis pneumonia may help identify at-risk patients that have strong immune reactivity to Pneumocystis antigens and may be predictive of the outcome of disease. Our goal is to assay the immune responses of individual HIV-infected patients, both PcP+ and PcP−, over time to determine whether there is a change in the serological reactivity to Msg fragments over time and how that affects the outcome of infection.
Acknowledgments
We gratefully acknowledge the excellent assistance of Rongrong Wang.
This study was funded by the Medical Research Service, Department of Veterans Affairs, and NIH grants AI06527, AI25467, AI062492, and HL64570.
REFERENCES
- 1.Bartlett, M. S., W. C. Angus, M. M. Shaw, P. J. Durant, C. H. Lee, J. M. Pascale, and J. W. Smith. 1998. Antibody to Pneumocystis carinii protects rats and mice from developing pneumonia. Clin. Diagn. Lab. Immunol. 5:74-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop, L. R., and J. A. Kovacs. 2003. Quantitation of anti-Pneumocystis jiroveci antibodies in healthy persons and immunocompromised patients. J. Infect. Dis. 187:1844-1848. [DOI] [PubMed] [Google Scholar]
- 3.Borst, P. 1991. Molecular genetics of antigenic variation. Immunol. Today 12:A29-A33. [DOI] [PubMed] [Google Scholar]
- 4.Buhl, L., O. P. Settnes, and P. L. Andersen. 1993. Antibodies to Pneumocystis carinii in Danish blood donors and AIDS patients with and without Pneumocystis carinii pneumonia. APMIS 101:707-710. [PubMed] [Google Scholar]
- 5.Burns, S. M., J. A. Read, P. L. Yap, and R. P. Brettle. 1990. Reduced concentrations of IgG antibodies to Pneumocystis carinii in HIV-infected patients during active Pneumocystis carinii infection and the possibility of passive immunization. J. Infect. 20:33-39. [DOI] [PubMed] [Google Scholar]
- 6.Chatterton, J. M., A. W. Joss, T. H. Pennington, and D. O. Ho-Yen. 1999. Usefulness of rat-derived antigen in the serodiagnosis of Pneumocystis carinii infection. J. Med. Microbiol. 48:681-687. [DOI] [PubMed] [Google Scholar]
- 7.Daly, K. R., C. J. Fichtenbaum, R. Tanaka, M. J. Linke, R. O'Bert, T. D. Thullen, M. S. Hui, A. G. Smulian, and P. D. Walzer. 2002. Serologic responses to epitopes of the major surface glycoprotein of Pneumocystis jiroveci differ in human immunodeficiency virus-infected and uninfected persons. J. Infect. Dis. 186:644-651. [DOI] [PubMed] [Google Scholar]
- 8.Daly, K. R., J. Koch, L. Levin, and Peter D. Walzer. 2004. Enzyme-linked immunosorbent assay and serologic responses to Pneumocystis jiroveci. Emerg. Infect. Dis. 10:848-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dworkin, M. S., D. L. Hanson, J. E. Kaplan, J. L. Jones, and J. W. Ward. 2000. Risk for preventable opportunistic infections in persons with AIDS after antiretroviral therapy increases CD4+ T lymphocyte counts above prophylaxis thresholds. J. Infect. Dis. 182:611-615. [DOI] [PubMed] [Google Scholar]
- 10.Elvin, K., A. Bjorkman, N. Heurlin, B. M. Eriksson, L. Barkholt, and E. Linder. 1994. Seroreactivity to Pneumocystis carinii in patients with AIDS versus other immunosuppressed patients. Scand. J. Infect. Dis. 26:33-40. [DOI] [PubMed] [Google Scholar]
- 11.Frenkel, J. K. 1999. Pneumocystis pneumonia, an immunodeficiency-dependent disease (IDD): a critical historical overview. J. Eukaryot. Microbiol. 46:89S-92S. [PubMed] [Google Scholar]
- 12.Garbe, T. R., and J. R. Stringer. 1994. Molecular characterization of clustered variants of genes encoding major surface antigens of human Pneumocystis carinii. Infect. Immun. 62:3092-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garvey, B. A., J. A. Wiley, F. Gigliotti, and A. G. Harmsen. 1997. Protection against Pneumocystis carinii pneumonia by antibodies generated from either T helper 1 or T helper 2 responses. Infect. Immun. 65:5052-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gigliotti, F. 1992. Host species-specific antigenic variation of a mannosylated surface glycoprotein of Pneumocystis carinii. J. Infect. Dis. 165:329-336. [DOI] [PubMed] [Google Scholar]
- 15.Gigliotti, F., B. A. Garvey, C. G. Haidaris, and A. G. Harmsen. 1998. Recognition of Pneumocystis carinii antigens by local antibody-secreting cells following resolution of P. carinii pneumonia in mice. J. Infect. Dis. 178:235-242. [DOI] [PubMed] [Google Scholar]
- 16.Gigliotti, F., B. A. Garvy, and A. G. Harmsen. 1996. Antibody-mediated shift in the profile of glycoprotein phenotypes observed in a mouse model of Pneumocystis carinii pneumonia. Infect. Immun. 64:1892-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gigliotti, F., and A. G. Harmsen. 1997. Pneumocystis carinii host origin defines the antibody specificity and protective response induced by immunization. J. Infect. Dis. 176:1322-1326. [DOI] [PubMed] [Google Scholar]
- 18.Gigliotti, F., and W. T. Hughes. 1998. Passive immunoprophylaxis with specific monoclonal antibody confers partial protection against Pneumocystis carinii pneumonitis in animal models. J. Clin. Investig. 81:1666-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen, A. G., W. Chen, and F. Gigliotti. 1995. Active immunity to Pneumocystis carinii reinfection in T-cell-depleted mice. Infect. Immun. 63:2391-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann, B., P. B. Nielsen, N. Odum, J. Gerstoft, P. Platz, L. P. Ryder, A. G. Poulsen, L. Mathiesen, E. Dickmeiss, B. Norrild, H. K. Andersen, B. F. Westergaard, C. M. Nielsen, W. Holten-Andersen, M. Mojon, J. O. Nielsen, and A. Svejgaard. 1988. Humoral and cellular responses to Pneumocystis carinii, CMV, and herpes simplex in patients with AIDS and in controls. Scand. J. Infect. Dis. 20:389-394. [DOI] [PubMed] [Google Scholar]
- 21.Hofmann, B., N. Odum, P. Platz, L. P. Ryder, A. Svejgaard, P. B. Nielsen, W. Holten-Andersen, J. O. Gerstoft, J. Nielsen, and M. Mojon. 1985. Humoral responses to Pneumocystis carinii in patients with acquired immunodeficiency syndrome and in immunocompromised homosexual men. J. Infect. Dis. 152:838-840. [DOI] [PubMed] [Google Scholar]
- 22.Kovacs, J. A., F. Powell, J. C. Edman, B. Lundgren, A. Martinez, B. Drew, and C. W. Angus. 1993. Multiple genes encode the major surface glycoprotein of Pneumocystis carinii. J. Biol. Chem. 268:6034-6040. [PubMed] [Google Scholar]
- 23.Kutty, G., L. Ma, and J. A. Kovacs. 2001. Characterization of the expression site of the major surface glycoprotein of human-derived Pneumocystis carinii. Mol. Microbiol. 42:183-193. [DOI] [PubMed] [Google Scholar]
- 24.Laursen, A. L., and P. L. Andersen. 1998. Low levels of IgG antibodies against Pneumocystis carinii among HIV-infected patients. Scand. J. Infect. Dis. 30:495-499. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M., S. R. Cho, Y. K. Park, M. H. Choi, and S. T. Hong. 1998. The effect of heterogeneous hyperimmune IgG antibody on prophylaxis and treatment of Pneumocystis carinii infection in rats. Korean J. Parasitol. 36:127-132. [DOI] [PubMed] [Google Scholar]
- 26.Linke, M. J., S. M. Sunkin, R. P. Andrews, J. R. Stringer, and P. D. Walzer. 1998. Expression, structure, and location of epitopes of the major surface glycoprotein of Pneumocystis carinii f. sp. carinii. Clin. Diagn. Lab. Immunol. 5:50-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddison, S. E., G. V. Hayes, M. H. Ivey, V. C. Tsang, S. B. Slemenda, and L. G. Norman. 1982. Fractionation of Pneumocystis carinii antigens used in an enzyme-linked immunosorbent assay for antibodies and in the production of antiserum for detecting Pneumocystis carinii antigenemia. J. Clin. Microbiol. 15:1029-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcotte, H., D. Levesque, K. Delanay, A. Bourgeault, R. de la Durantaye, S. Brochu, and M. C. Lavoie. 1996. Pneumocystis carinii infection in transgenic B cell-deficient mice. J. Infect. Dis. 173:1034-1037. [DOI] [PubMed] [Google Scholar]
- 29.Mei, Q., R. E. Turner, V. Sorial, D. Klivington, C. W. Angus, and J. A. Kovacs. 1998. Characterization of major surface glycoprotein genes of human Pneumocystis carinii and high-level expression of a conserved region. Infect. Immun. 66:4268-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen, P. B., and M. Mojon. 1988. Enzyme-linked immunosorbent assay compared with indirect immunofluorescence test for detection of Pneumocystis carinii specific immunoglobulins G, M, and A. APMIS 96:649-654. [DOI] [PubMed] [Google Scholar]
- 31.Nuesch, R., C. Bellini, and W. Zimmerli. 1999. Pneumocystis carinii pneumonia in human immunodeficiency virus (HIV)-positive and HIV-negative immunocompromised patients. Clin. Infect. Dis. 29:1519-1523. [DOI] [PubMed] [Google Scholar]
- 32.Park, H. Y., S. U. Lee, S. W. Chae, S. Huh, J. R. Yu, J. Kim, and S. T. Hong. 1999. Variation of antigenicity and serological reaction to Pneumocystis carinii in Korea. Korean J. Parasitol. 37:109-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pascale, J. M., M. M. Shaw, P. J. Durant, A. A. Amador, M. S. Bartlett, J. W. Smith, R. L. Gregory, and G. L. McLaughlin. 1999. Intranasal immunization confers protection against murine Pneumocystis carinii lung infection. Infect. Immun. 67:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peglow, S. L., A. G. Smulian, M. J. Linke, C. L. Pogue, S. Nurre, J. Crisler, J. Phair, J. W. Gold, D. Armstrong, and P. D. Walzer. 1990. Serologic responses to specific Pneumocystis carinii antigens in health and disease. J. Infect. Dis. 161:296-306. [DOI] [PubMed] [Google Scholar]
- 35.Pifer, L. L., W. T. Hughes, S. Stagno, and D. Woods. 1978. Pneumocystis carinii infection: evidence for high prevalence in normal and immunosuppressed children. Pediatrics 61:35-41. [PubMed] [Google Scholar]
- 36.Pifer, L. L., H. B. Niell, S. B. Langdon, S. Baltz, S. T. Clark, C. C. Edwards, and D. R. Woods. 1987. Evidence for depressed humoral immunity to Pneumocystis carinii in homosexual males, commercial plasma donors, and patients with acquired immunodeficiency syndrome. J. Clin. Microbiol. 25:991-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pifer, L. L., D. R. Woods, C. C. Edwards, R. E. Joyner, F. J. Anderson, and K. Arheart. 1988. Pneumocystis carinii serologic study in pediatric acquired immunodeficiency syndrome. Am. J. Dis. Child. 142:36-39. [PubMed] [Google Scholar]
- 38.Pontesilli, O., S. Kerkhof-Garde, D. W. Notermans, N. A. Foudraine, M. T. Roos, M. R. Klein, S. A. Danner, J. M. Lange, and F. Miedema. 1999. Functional T-cell reconstitution and human immunodeficiency virus-1-specific cell-mediated immunity during highly active antiretroviral therapy. J. Infect. Dis. 180:76-86. [DOI] [PubMed] [Google Scholar]
- 39.Redhead, S. A., M. T. Cushion, J. K. Frenkel, and J. R. Stringer. 2006. Pneumocystis and Trypanosoma cruzi: nomenclature and typifications. J. Eukaryot. Microbiol. 53:2-11. [DOI] [PubMed] [Google Scholar]
- 40.Smulian, A. G., D. W. Sullivan, M. J. Linke, N. A. Halsey, T. C. Quinn, A. P. MacPhail, M. A. Hernandez-Avila, S. T. Hong, and P. D. Walzer. 1993. Geographic variation in the humoral response to Pneumocystis carinii. J. Infect. Dis. 67:1243-1247. [DOI] [PubMed] [Google Scholar]
- 41.Smulian, A. G., and P. D. Walzer. 1994. Serological studies of Pneumocystis carinii infection, p. 141-151. In P. D. Walzer (ed.), Pneumocystis carinii pneumonia. Marcel Dekker, New York, N.Y.
- 42.Stringer, J. R., C. B. Beard, R. F. Miller, and A. E. Wakefield. 2002. A new name (Pneumocystis jiroveci) for Pneumocystis from humans. Emerg. Infect. Dis. 8:891-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stringer, J. R., and S. P. Keely. 2001. Genetics of surface antigen expression in Pneumocystis carinii. Infect. Immun. 69:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunkin, S. M., and J. R. Stringer. 1996. Translocation of surface antigen genes to a unique telomeric expression site in Pneumocystis carinii. Mol. Microbiol. 19:283-295. [DOI] [PubMed] [Google Scholar]
- 45.Sunkin, S. M., and J. R. Stringer. 1997. Residence at the expression site is necessary and sufficient for the transcription of surface antigen genes of Pneumocystis carinii. Mol. Microbiol. 25:147-160. [DOI] [PubMed] [Google Scholar]
- 46.Theus, S. A., R. P. Andrews, P. Steele, and P. D. Walzer. 1995. Adoptive transfer of lymphocytes sensitized to the major surface glycoprotein of Pneumocystis carinii confers protection in the rat. J. Clin. Investig. 95:2587-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theus, S. A., N. Sawhney, A. G. Smulian, and P. D. Walzer. 1998. Proliferative and cytokine responses of human T lymphocytes isolated from human immunodeficiency virus-infected patients to the major surface glycoprotein of Pneumocystis carinii. J. Infect. Dis. 177:238-241. [DOI] [PubMed] [Google Scholar]
- 48.Theus, S. A., A. G. Smulian, P. Steele, M. J. Linke, and P. D. Walzer. 1998. Immunization with the major surface glycoprotein of Pneumocystis carinii elicits a protective response. Vaccine 16:1149-1157. [DOI] [PubMed] [Google Scholar]
- 49.Van der Ploeg, L. H. T., K. Gottesdiener, and M. G. S. Lee. 1992. Antigenic variation in African trypanosomes. Trends Genet. 8:452-457. [DOI] [PubMed] [Google Scholar]
- 50.Vargas, S. L., W. T. Hughes, M. E. Santolaya, A. V. Ulloa, C. A. Ponce, C. E. Cabrera, F. Cumsille, and F. Gigliotti. 2001. Search for primary infection by Pneumocystis carinii in a cohort of normal, healthy infants. Clin. Infect. Dis. 32:855-861. [DOI] [PubMed] [Google Scholar]
- 51.Wakefield, A. E. 2002. Pneumocystis carinii. Br. Med. Bull. 61:175-188. [DOI] [PubMed] [Google Scholar]
- 52.Walzer, P. D. 1999. Immunological features of Pneumocystis carinii infection in humans. Clin. Diagn. Lab. Immunol. 6:149-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 54.Zheng, M., J. E. Shellito, L. Marrero, Q. Zhong, S. Julian, P. Ye, V. Wallace, P. Schwarzenberger, and J. K. Kolls. 2001. CD4+ T cell-independent vaccination against Pneumocystis carinii in mice. J. Clin. Investig. 108:1469-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]