Abstract
The Dictyostelium rbrA gene encodes a putative Ariadne ubiquitin ligase. rbrA− cells form defective slugs that cannot phototax. Prestalk cell numbers are reduced in rbrA− slugs, and these prestalk cells do not localize to the tip of slugs. Chimeric slugs containing wild-type cells could phototax and form fruiting bodies.
The RBR family of ubiquitin ligases is a large and complex family with members present in animals, plants, fungi, and protists (17). RBR genes have been implicated in cancer and neurodegenerative diseases (e.g., parkin) (17-19, 21). Despite significant advances, much of the potential diversity in RBR protein function remains to be studied. Particularly interesting may be members of the Ariadne subfamily, since this subfamily appears to be the most ancient (1, 3, 17).
In this report, we identify a putative Ariadne ubiquitin ligase, RbrA, in Dictyostelium. Dictyostelium discoideum Ax4 and DH1 were used as wild-type and parental strains. All strains were grown in HL5 with glucose (Qbiogene, Carlsbad, CA) or with Klebsiella aerogenes on agar plates as described previously (12). Multicellular development, preparation of conditioned starvation medium, phototaxis studies, Northern blot analysis, and preparation of cDNA clones were done as previously described (12, 16, 20). Restriction enzyme-mediated integration (REMI) was carried out as described previously (14).
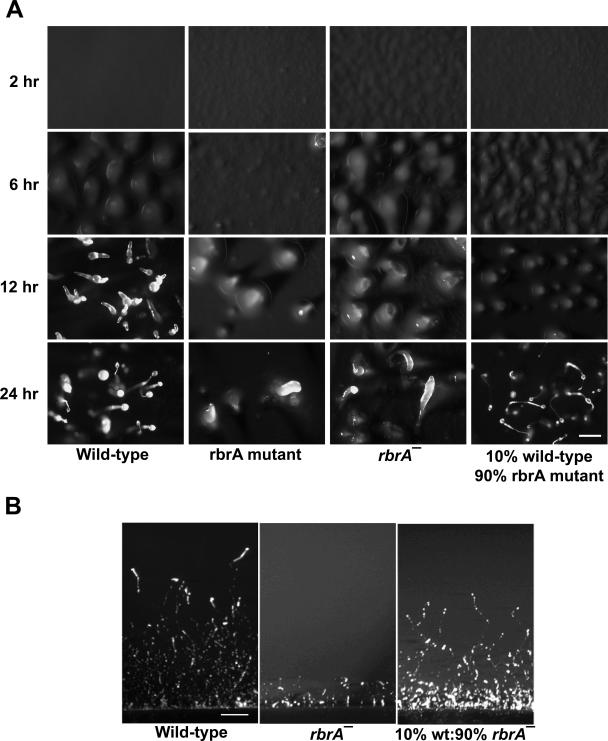
An rbrA mutant was found in a screen of REMI transformants as cells whose development is blocked at the finger/slug stage. rbrA mutant cells formed ripples by 2 h of development, which was earlier than wild type (Fig. 1); however, development then slowed compared to wild-type cells and fruiting bodies never formed. In mixtures of rbrA− (90%) and wild-type (10%) cells, early rippling was observed, but nearly all structures produced by the chimeras formed fruiting bodies (Fig. 1). The addition of wild-type conditioned medium to rbrA− cells did not cause a similar phenotype rescue. Thus, a small number of wild-type cells appeared to rescue the morphological developmental defect, and the rescue appeared to require cell-cell proximity.
FIG. 1.
Developmental phenotype of rbrA mutant cells. (A) Wild-type cells, rbrA mutant cells generated by REMI mutagenesis or rbrA mutant cells generated by replacement of a large fragment of the rbrA coding region with a blasticidin resistance cassette and rbrA mutant cells mixed with wild-type cells were plated for development on filters and photographed at the indicated hours of development. Bar is 0.5 mm. (B) Phototaxis phenotype. Wild-type cells, rbrA− cells, and rbrA− mutant cells mixed with wild-type cells were deposited onto 2% water agarose plates containing charcoal, and the plates were stored in a dark chamber with one end exposed to a light source. Slugs were photographed after 48 h of development. Bar, 1 cm.
When developed on filters, we observed that slugs formed from rbrA− cells were capable of migrating at least short distances. However, slugs were unable to phototax when developed on agarose in a dark chamber with a directional light source (Fig. 1B). Slugs that formed from a mixture of rbrA− and wild-type (10%) cells migrated directionally toward the light source and traveled nearly as far as wild type (Fig. 1B). These are likely chimeric slugs because at least 50% of the slugs that formed showed positive phototaxis, and other experiments showed that chimeric slugs do form (see Fig. 4).
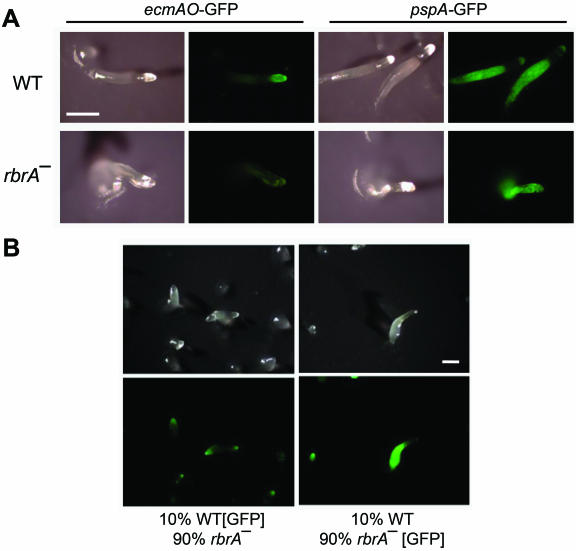
FIG. 4.
Cell sorting is altered in the rbrA mutant. (A) Wild-type and rbrA− cells were transformed with cell-type-specific GFP expression plasmids PsA-ubi-s65tGFP or 63-ubi-s65tGFP, which contained the GFP cDNA expressed by the prestalk-specific promoter from ecmAO (ecmAO-GFP) or the prespore-specific promoter from pspA (pspA-GFP), respectively. Cells were developed to the slug stage on filters and photographed by using light and fluorescence imaging. The slugs shown are representative of those seen in two independent experiments. (B) In chimeric slugs, rbrA− cells preferentially sort to prespore region. Wild-type (WT) and rbrA− cells were transformed with a plasmid carrying the GFP cDNA expressed under the control of the constitutively expressed actin-15 promoter (pTX-GFP). GFP-expressing cells of one strain were mixed with non-GFP-expressing cells of the other strain and developed on filters. Each mixture contained 10% wild-type cells. The chimeric slugs were photographed by using light and fluorescence imaging. Bar, 0.5 mm.
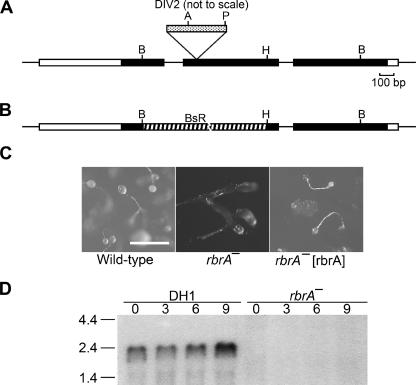
Genomic rbrA DNA was cloned by digestion with BglII to generate plasmid pM10B (Fig. 2A). The pM10B plasmid was linearized with BglII and used to transform a wild-type host strain (DH1). The resulting gene disruption by homologous recombination successfully recapitulated the original rbrA mutant phenotype. A second rbrA mutant was constructed by using a gene targeting plasmid, pRbrBsr, to replace a portion of the rbrA gene with a blasticidin-resistance cassette (Fig. 2B). This rbrA− strain demonstrated a mutant phenotype nearly identical to the original rbrA mutant (Fig. 1). Expression of the rbrA cDNA complemented the defective developmental phenotype of rbrA− cells (Fig. 2C). A 2.2-kb rbrA mRNA was present in vegetatively growing cells (0 h), increased in level at 9 h (Fig. 2D), and remained at the 9-h level throughout the remainder of development (data not shown). rbrA mRNA was not present in rbrA− cells (Fig. 2D).
FIG.2.
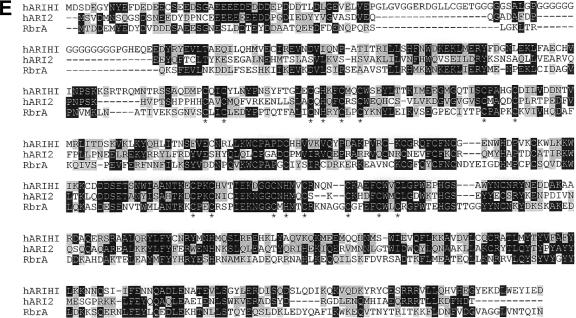
rbrA gene. (A) Genomic map and REMI-generated mutation. REMI was carried out by electroporating EcoRI-linearized DIV2 (the mutagenic plasmid carrying the Dictyostelium pyr5-6 gene) into DH1 (pyr5-6−) cells along with the restriction enzyme MunI and selecting for uracil prototrophs. The thick bar represents exons of rbrA, and the black portion of the thick bar designates the open reading frame. Restriction sites: A, Asp718; B, BglII; H, HindIII; P, PvuII. The location of the DIV2 insertion is identified (spotted bar; not drawn to scale). (B) Diagram of rbrA disrupted by blasticidin-resistance gene (striped bar) replacement. (C) Rescue of rbrA mutant development phenotype. Wild-type cells, rbrA− cells, and rbrA− cells carrying an rbrA expression plasmid (rbrA− [rbrA]) were grown on nutrient agar plates in association with Klebsiella aerogenes and then allowed to starve on the agar plates. Bar is 0.5 mm. (D) Expression of rbrA. RNA from the indicated cell lines developed for the indicated time in hours was resolved by electrophoresis through 1.2% agarose-formaldehyde gels, blotted onto nylon membranes, and hybridized with a probe specific for rbrA. Size standards are indicated on the left in kilobases. (E) Comparison of the predicted amino acid sequences of RbrA with human ARIHI and ARI2. The amino acid identities between RbrA (AAR10851; dictyBase DDB0191418), human ARIH1 (AAH51877), and human ARI2 (CAA10276) are in black, and amino acid similarities are in highlighted in gray. Conserved cysteine and histidine residues of the RING fingers are marked by an asterisk.
rbrA genomic and cDNA clones revealed a single long open reading frame interrupted by two introns (Fig. 2A). The deduced amino acid sequence predicts a protein of 520 residues with a molecular mass of 61 kDa (Fig. 2E). RbrA has strong sequence similarity to the Ariadne subfamily of the RBR family of E3 ubiquitin ligases (17). The characteristic series of acid-rich, RBR (for “RING, between rings, RING”), and coiled-coil/leucine rich motifs found in Ariadne subfamily members are present in RbrA (Fig. 2E). RbrA has 34% identity (54% similarity) to human HHARI. These data suggest that rbrA encodes an E3 ligase.
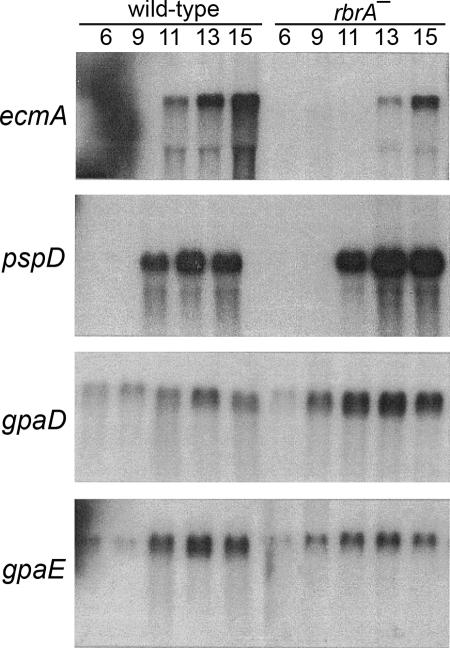
To investigate the function of RbrA, we examined the expression of developmentally regulated genes. Transcript levels for the prestalk-cell-specific ecmA gene (23) were lower in rbrA− cells compared to wild-type cells (Fig. 3). The prespore-specific pspD (25) was regulated temporally in developing rbrA− cells as in wild-type cells, but at much higher than wild-type levels (Fig. 3). In addition, transcript levels for gpaD (required for spore formation) (10) were elevated and gpaE (tip formation) (11) levels were lower during rbrA− cell development (Fig. 3).
FIG. 3.
Gene expression in rbrA− cells. Total RNA was prepared from wild-type cells (Ax4) at the indicated stages of development (in hours). RNA (5 μg) was resolved by electrophoresis through 1.2% agarose-formaldehyde gels, blotted onto nylon membranes, and hybridized with probes specific for ecmA, pspD (PL3), gpaD (Gα4), and gpaE (Gα5).
To investigate cell-type proportioning and cell sorting, rbrA− and parental cells were transformed with cell-type-specific green fluorescent protein (GFP) expression plasmids (5). When the ecmAO promoter was used to express GFP in wild-type cells, fluorescence appeared in the anterior prestalk region, as expected (6, 13) (Fig. 4A); however, rbrA− slugs showed a very low level of fluorescence throughout the slug. GFP expressed from the pspA promoter marked the posterior prespore region in wild-type slugs, whereas the entire rbrA− slug was marked (Fig. 4A). Thus, prespore cells were prevalent and appeared to dominate the entire rbrA− slug.
To determine whether the initial cell-autonomous differentiation was altered in rbrA− cells (2, 8, 22), cells were starved at low cell density and the numbers of CP2-positive prestalk cells and SP70-positive prespore cells were counted (9). The percentage of CP2- and SP70-positive wild-type cells was similar to what we previously observed (4, 24). There was essentially no difference in the initial differentiation of rbrA− cells compared to wild-type cells (Table 1).
TABLE 1.
Cell-type proportioning is altered in rbrA− slugsa
| Strain | Mean % ± SEM
|
|||
|---|---|---|---|---|
| Low cell density
|
Dissociated slugs
|
|||
| SP70 (prespore) positive | CP2 (prestalk) positive | SP70 (prespore) positive | CP2 (prestalk) positive | |
| Wild type | 33 ± 1 | 10 ± 1 | 71 ± 3 | 13 ± 1 |
| rbrA− | 31 ± 1 | 10 ± 1 | 70 ± 8 | 4 ± 1 |
To assay differentiation at low cell density, wild-type and rbrA− cells were starved in wild-type conditioned medium at low cell density and then fixed and stained for SP70 or CP2. To assay differentiation in slugs, cells were developed on filters to the slug stage. Slugs were dissociated to single cells, and the cells were fixed and stained for SP70 or CP2. Total cells and stained cells were counted. The results are means from eight experiments (low-cell-density assay) or four experiments (dissociated slugs). The difference between the percentage of CP2-positive cells in dissociated slugs between wild-type and rbrA− is significant (P < 0.005 [Student t test]).
To determine the percentage of prestalk and prespore cells in slugs, wild-type and rbrA− slugs were dissociated into single cells, fixed, and stained for prespore (SP70) or prestalk (CP2) markers (7). The percentage of CP2- and SP70-positive wild-type cells was similar to what we previously observed (4). rbrA− slugs had a percentage of SP70-positive prespore cells that was similar to that of the wild type but had threefold-fewer CP2-positive prestalk cells than did the wild type (Table 1). Together, the data suggest that the initial cell-autonomous differentiation of rbrA− cells into CP2-positive prestalk cells and SP70-positive prespore cells is normal and that rbrA− slugs contain an abnormally low number of CP2-positive prestalk cells.
To determine the effect of wild-type cells on rbrA− cell-fate choice, mixtures of wild-type and rbrA− cells were prepared in which one of the strains contained the GFP gene driven by the constitutively expressed actin-15 promoter (15). Wild-type and rbrA− cells were mixed in various proportions and developed on filters. Fluorescence imaging of chimeric slugs indicated that the wild-type cells preferentially located to the tip region, which is occupied by prestalk cells in pure wild-type slugs (Fig. 4B). The rbrA− cells preferentially located to the posterior of the slug, which normally is populated with prespore cells.
To investigate the ability of rbrA− cells to form spores during chimeric development, spore assays were carried out on fruiting bodies produced by chimeras (6). Experiments with chimeras containing 3 to 40% rbrA− cells produced less than 1% detergent-resistant spores with the rbrA− phenotype, suggesting that rbrA− spores were sensitive to detergent treatment. We therefore carried out additional experiments in which spores were plated before and after detergent treatment (Table 2). The results showed that rbrA− cells did not efficiently survive the terminal differentiation stages.
TABLE 2.
rbrA− cells do not efficiently produce viable sporesa
| Expt | % rbrA
|
||
|---|---|---|---|
| Input cells | Viable cells in sori | Spores | |
| 1 | 10.0 | 0.6 | 0.4 |
| 47.6 | 0.9 | 0.0 | |
| 2 | 3.6 | 0.7 | 0.1 |
| 42.8 | 1.4 | 0.7 | |
| 3 | 4.6 | 0 | 0.3 |
| 32.4 | 5.8 | 0.3 | |
Wild-type and rbrA cell mixtures containing about 3 and 40% rbrA− cells (the actual ratio was determined by plating input cells) were allowed to develop, and the percentage of rbrA− cells in sori was determined before (viable cells) and after (spores) detergent treatment.
The predicted amino acid sequence of Dictyostelium RbrA suggests that it is a member of the Ariadne subfamily of ubiquitin ligases. RbrA appears to be required for normal cell-type proportioning and cell sorting during multicellular development. Prestalk cell numbers are reduced in rbrA− slugs and these prestalk cells do not localize to the tip of slugs. Development terminates at the finger or slug stage, and these slugs do not phototax, possibly because the tip region does not properly form. In addition to being necessary for a normal percentage of prestalk cells and the organization of the slug, RbrA is also necessary for spore cell viability. RbrA thus affects multiple processes such as prestalk cell differentiation, pattern formation, and spore cell maturation.
Acknowledgments
We thank Ed Frank for library screening; Vikram Penumali, Lindsey Simon, and Crystal Leanza for assistance with experiments; and Diane Hatton for assistance with sequence analysis. We thank the following coworkers for kindly providing plasmids: Thomas Egelhoff for pTX-GFP, Harry MacWilliams for cell-type-specific GFP expression plasmids, Rick Firtel for pSL17 and pJH198, Jeff Williams for pDd63, and Daphne Blumberg for plasmid PL3. We also thank Dale Hereld and Peter Devreotes for D. discoideum strain DH1.
This study was supported by an M. J. Murdock Charitable Trust grant to D.F.L.
REFERENCES
- 1.Aguilera, M., M. Oliveros, M. Martinez-Padron, J. A. Barbas, and A. Ferrus. 2000. Ariadne-1: a vital Drosophila gene is required in development and defines a new conserved family of RING-finger proteins. Genetics 155:1231-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Araki, T., H. Nakao, I. Takeuchi, and Y. Maeda. 1994. Cell cycle-dependent sorting in the development of Dictyostelium cells. Dev. Biol. 162:221-228. [DOI] [PubMed] [Google Scholar]
- 3.Ardley, H. C., N. G. Tan, S. A. Rose, A. F. Markham, and P. A. Robinson. 2001. Features of the Parkin/Ariadne ubiquitin ligase, HHARI, which regulate its interaction with the ubiquitin-conjugating enzyme, UbcH7. J. Biol. Chem. 276:19640-19647. [DOI] [PubMed] [Google Scholar]
- 4.Clay, J. L., R. A. Ammann, and R. H. Gomer. 1995. Initial cell type choice in a simple eukaryote: a cell-autonomous or morphogen-gradient dependent? Dev. Biol. 172:665-674. [DOI] [PubMed] [Google Scholar]
- 5.Deischsel, H., S. Friedel, A. Detterbeck, C. Coyne, U. Hamker, and H. K. MacWilliams. 1999. Green fluorescent proteins with short half-lives as reporters in Dictyostelium discoideum. Dev. Genes Evol. 209:63-68. [DOI] [PubMed] [Google Scholar]
- 6.Ennis, H. L., D. N. Dao, S. U. Pukatzki, and R. H. Kessin. 2000. Dictyostelium amoebae lacking an F-box protein form spores rather than stalk in chimeras with wild type. Proc. Natl. Acad. Sci. USA 97:3292-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomer, R. H. 1987. A strategy to study development and pattern formation: use of antibodies against products of cloned genes. Methods Cell Biol. 28:471-487. [DOI] [PubMed] [Google Scholar]
- 8.Gomer, R. H., and R. A. Firtel. 1987. Cell-autonomous determination of cell-type choice in Dictyostelium development by cell-cycle phase. Science 237:758-762. [DOI] [PubMed] [Google Scholar]
- 9.Gomer, R. H., I. S. Yuen, and R. A. Firtel. 1991. A secreted 80 × 103 Mr protein mediates sensing of cell density and the onset of development in Dictyostelium. Development 112:269-278. [DOI] [PubMed] [Google Scholar]
- 10.Hadwiger, J. A., and R. A. Firtel. 1992. Analysis of G alpha 4, a G-protein required for multicellular development in Dictyostelium. Genes Dev. 6:38-49. [DOI] [PubMed] [Google Scholar]
- 11.Hadwiger, J. A., K. Natarajan, and R. A. Firtel. 1996. Mutations in the Dictyostelium heterotrimeric G protein α subunit Gα5 alter the kinetics of tip morphogenesis. Development 122:1215-1224. [DOI] [PubMed] [Google Scholar]
- 12.Jain, R., I. S. Yuen, C. R. Taphouse, and R. H. Gomer. 1992. A density-sensing factor controls development in Dictyostelium. Genes Dev. 6:390-400. [DOI] [PubMed] [Google Scholar]
- 13.Jermyn, K. A., K. T. I. Duffy, and J. G. 1989. Williams. A new anatomy of prestalk zone in Dictyostelium. Nature 340:144-146. [DOI] [PubMed] [Google Scholar]
- 14.Kuspa, A., and W. F. Loomis. 1992. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl. Acad. Sci. USA 89:8803-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levi, S., M. Polyakov, and T. T. Egelhoff. 2000. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid 44:231-238. [DOI] [PubMed] [Google Scholar]
- 16.Lindsey, D. F., A. Amerik, W. J. Deery, J. D. Bishop, M. Hochstrasser, and R. H. Gomer. 1998. A deubiquitinating enzyme that disassembles free polyubiquitin chains is required for development but not growth in Dictyostelium. J. Biol. Chem. 273:29178-29187. [DOI] [PubMed] [Google Scholar]
- 17.Marin, I., J. I. Lucas, A. Gradilla, and A. Ferrus. 2004. Parkin and relatives: the RBR family of ubiquitin ligases. Physiol. Genomics 17:253-263. [DOI] [PubMed] [Google Scholar]
- 18.Nikolaev, A. Y., M. Li, N. Puskas, J. Qin, and W. Gu. 2003. Parc: a cytoplasmic anchor for p53. Cell 112:29-40. [DOI] [PubMed] [Google Scholar]
- 19.Staropoli, J. F., C. McDermott, C. Martinat, B. Schulman, E. Demireva, and A. Abeliovich. 2003. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainite excitotoxicity. Neuron 37:735-749. [DOI] [PubMed] [Google Scholar]
- 20.Sussman, M. 1987. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions, p. 9-29. In J. A. Spudich (ed.), Methods in cell biology. Academic Press, Inc., Orlando, Fla. [DOI] [PubMed]
- 21.Tan, N. G., H. A. Ardley, G. B. Scott, S. A. Rose, A. F. Markham, and P. A. Robinson. 2003. Human homologue of ariadne promotes the ubiquitylation of translation initiation factor 4E homologous protein, 4EHP. FEBS Lett. 554:501-504. [DOI] [PubMed] [Google Scholar]
- 22.Weijer, C. J., G. Duschl, and C. N. David. 1984. Dependence of cell-type proportioning and sorting on cell cycle phase in Dictyostelium discoideum. J. Cell Sci. 70:133-145. [DOI] [PubMed] [Google Scholar]
- 23.Williams, J. G., A. Ceccarelli, S. McRobbie, H. Mahbubani, R. R. Kay, A. Early, M. Berks, and K. A. Jermyn. 1987. Direct induction of Dictyostelium prestalk gene expression by DIF provides evidence that DIF is a morphogen. Cell 49:185-192. [DOI] [PubMed] [Google Scholar]
- 24.Wood, S. A., R. R. Ammann, D. A. Brock, L. Li, T. P. Spann, and R. H. Gomer. 1996. RtoA links initial cell type choice to the cell cycle in Dictyostelium. Development 122:3677-3685. [DOI] [PubMed] [Google Scholar]
- 25.Yoder, B. K., J. Mao, G. W. Erdos, C. M. West, and D. D. Blumberg. 1994. Identification of a new spore coat protein gene in the cellular slime mold Dictyostelium discoideum. Dev. Biol. 163:49-65. [DOI] [PubMed] [Google Scholar]