Abstract
We have used an Aspergillus nidulans macroarray carrying sequences of 2,787 genes from this fungus to monitor gene expression of both wild-type and uvsBATR (the homologue of the ATR gene) deletion mutant strains in a time course exposure to camptothecin (CPT). The results revealed a total of 1,512 and 1,700 genes in the wild-type and uvsBATR deletion mutant strains that displayed a statistically significant difference at at least one experimental time point. We characterized six genes that have increased mRNA expression in the presence of CPT in the wild-type strain relative to the uvsBATR mutant strain: fhdA (encoding a forkhead-associated domain protein), tprA (encoding a hypothetical protein that contains a tetratrico peptide repeat), mshA (encoding a MutS homologue involved in mismatch repair), phbA (encoding a prohibitin homologue), uvsCRAD51 (the homologue of the RAD51 gene), and cshA (encoding a homologue of the excision repair protein ERCC-6 [Cockayne's syndrome protein]). The induced transcript levels of these genes in the presence of CPT require uvsBATR. These genes were deleted, and surprisingly, only the ΔuvsC mutant strain was sensitive to CPT; however, the others displayed sensitivity to a range of DNA-damaging and oxidative stress agents. These results indicate that the selected genes when inactivated display very complex and heterogeneous sensitivity behavior during growth in the presence of agents that directly or indirectly cause DNA damage. Moreover, with the exception of UvsC, deletion of each of these genes partially suppressed the sensitivity of the ΔuvsB strain to menadione and paraquat. Our results provide the first insight into the overall complexity of the response to DNA damage in filamentous fungi and suggest that multiple pathways may act in parallel to mediate DNA repair.
The DNA damage response is a protective mechanism that ensures the maintenance of genome integrity during cellular reproduction. DNA damage takes several general forms, including single-strand breaks, double-strand breaks (DSBs), base damage, and DNA-protein cross-links, which cause replication fork progression blockage and can generate secondary lesions such as DSBs. Accumulation of mutations will eventually lead to genomic instability and, consequently, carcinogenesis in higher organisms. Cell cycle checkpoints and DNA repair are the primary defenses against genomic instability. If the DNA damage is extensive, apoptosis provides a mechanism to eliminate cells with high mutation potential (59). The two main signal transduction pathways that respond to DNA damage are conserved across evolution: the ATM (mutated in ataxia telangiectasia) and the ATR (ATM-Rad3-related) pathways (1, 62, 74, 76). The ATM pathway responds to the presence of DSBs. The ATR pathway also responds to DSBs but more slowly than ATM. In addition, the ATR pathway can respond to agents that interfere with the function of replication forks, such as hydroxyurea (HU), UV light, and DNA-alkylating agents such as methyl methanesulfonate (MMS) (55, 56). The ATM/ATR kinases phosphorylate and activate signal transduction pathways that ultimately interface with the Cdk/cyclin machinery (1).
Aspergillus nidulans has been used as a model genetic system for the study of cell cycle control and the DNA damage response (for a review, see references 5, 28, 58, and 66). The recent completion of the A. nidulans genome sequence (http://www-genome.wi.mit.edu/annotation/fungi/aspergillus/) (27) will facilitate a more systematic use of this fungus as a model system for DNA repair studies. A. nidulans can normally tolerate high concentrations of the antitopoisomerase drug camptothecin (CPT) (8). The basic mechanism of action for CPT is well characterized (26). Briefly, CPT generates replication-mediated DSBs, which in turn induce reversible or permanent cell cycle arrest in G2-M. For a comprehensive evaluation of genes that are transcriptionally modulated during growth in the presence of CPT, we performed a large-scale analysis of gene expression in A. nidulans by using a macroarray hybridization approach. The array technology can provide information about large-scale gene expression when cells are challenged by this DNA-damaging agent. Thus, we constructed a 3,619-element array containing expressed sequence tag (EST) sequences that have been functionally classified by similarity to known genes (2). Using this array, we identified genes whose expression displayed statistically significant modulation during growth of A. nidulans in the presence of CPT. The analysis of these data allowed us to identify specific genes and gene sets whose induction seem to be related to biochemical changes associated with the DNA damage response to this drug.
MATERIALS AND METHODS
Strains and media.
The A. nidulans strains used are described in Table 1. Media were of two basic types: (i) a complete medium with three variants, i.e., YAG (2% glucose, 0.5% yeast extract, 2% agar, trace elements), YUU (YAG supplemented with 1.2 g/liter each of uracil and uridine), and liquid YG or YG plus UU media of the same compositions (but without agar), and (ii) a modified minimal medium (1% glucose, original high-nitrate salts, trace elements, 2% agar, pH 6.5) or minimal medium without glucose. Trace elements, vitamins, and nitrate salts were as described previously (39) (appendix, available on request from the author). Standard genetic techniques for A. nidulans were used for all strain constructions (39). The sensitivity to DNA-damaging and oxidative stress agents and to UV light was evaluated by using the methodologies described previously (24). To evaluate differences, we used one-way analysis of variance and the Student-Newman-Keuls post hoc test. The data shown are the averages from three independent experiments, analyses were performed using the software package Sigma Stat (Jandel Scientifics), and the statistical significance was set at α = 0.05.
TABLE 1.
A. nidulans strains used in this work
| Strain | Genotype | Reference |
|---|---|---|
| A4 | Glasgow wild type (veA+) | FGSC |
| AAH14 | pyrG89 pabaA1 yA2 ΔuvsB::argB | 24 |
| ATr60cd1 | pyrG89 argB2 chaA1 uvsC::pyr4 | 35 |
| ATr60cd1-14 | pyrG89 argB2 chaA1 uvsC::pyr4 ΔuvsB::argB | This work |
| DC1 | pyrG89 argB2 chaA1 | 35 |
| GR5 | pyrG89 wA3 pyroA4 | FGSC A773 |
| IM70 | pyrG89 yA2 argB2 ΔphbA::pyrG | This work |
| IM71 | pyrG89 yA2 argB2 ΔmshA::pyrG | This work |
| IM72-3 | pyrG89 argB2 chaA1 ΔcshA::pyrG | This work |
| IM73-104 | pyrG89 argB2 chA1 ΔfhdA::pyrG | This work |
| IM74-10 | pyrG89 argB2 chA1 ΔtprA::pyrG | This work |
| IM70-14 | pyrG89 yA2 argB2 ΔphbA::pyrG ΔuvsB::argB | This work |
| IM71-14 | pyrG89 yA2 argB2 ΔmshA::pyrG ΔuvsB::argB | This work |
| IM72-2.14 | pyrG89 yA2 argB2 ΔcshA::pyrG ΔuvsB::argB | This work |
| IM73-104.14A | pyrG89 yA2 argB2 ΔfhdA::pyrG ΔuvsB::argB | This work |
| IM74-10.14A | pyrG89 yA2 argB2 ΔtprA::pyrG ΔuvsB::argB | This work |
| TNO2A3 | pyrG89 pyroA4 chaA1 ΔnKuA::argB | 53 |
| UI224 | pyrG89 yA2 argB2 | FGSC |
DNA manipulations and construction of the inactivation strains.
DNA manipulations were as described previously (63). DNA fragment probes for Southern blots were labeled with [α-32P]dCTP by using the random primers DNA labeling system kit (Invitrogen). PCR primers were designed for amplifying each DNA fragment necessary for PCR-mediated technique (42) by using Primer Express version 1.0 (Applied Biosystems) design software. In the deletion constructions, the Aspergillus fumigatus pyrG gene was amplified from the pCDA21 plasmid (10), and this is referred to as the zeo-pyrG cassette because the amplified fragment also contains the zeocin resistance gene. The PCR-mediated constructions for the phbA (accession no. AN6073.2; encoding prohibitin), mshA (AN1708.2; the homologue of the mutS family DNA mismatch repair protein gene msh6), cshA (AN7103.2; the homologue of the CSB Cockayne's syndrome protein gene), fhdA (AN2893.2; encoding a protein containing a forkhead-associated domain), and tprA (AN3617.2; encoding a protein with tetrarico peptide repeats) genes consisted of three initial amplifications that generated 5′- and 3′-flanking regions of the target genes and final fusion PCRs. For the DNA fragments containing the flanking regions, genomic DNA of the FGSC A4 strain was used as a template. Table S1 in the supplemental material shows the primer sequences and the fragment sizes of the 5′- and 3′-flanking regions for each gene. Primers numbered −1 and −2 preceded by the name of the gene were used for the 5′-flanking region, and primers numbered −3 and −4 preceded by the gene name were used to amplify the 3′-flanking region; primers −2 and −3 have homology to the zeo-pyrG gene. Primers for the zeo-pyrG gene amplification also contained regions of homology to the 5′ and 3′ ends for each corresponding gene flanking regions, yielding a 2,417-bp fragment. The final fusion PCR fragments were independently generated using the three previous DNA fragments as templates and the outermost primers −1 and −4. After the reaction, the fusion PCR products were gel purified with Perfectprep Gel Cleanup (Eppendorf) according to the manufacturer's instructions. Transformation of A. nidulans strain TNO2A3 (ΔnkuA) (53) was according to the procedure described previously (57), using approximately 5 μg of DNA fragments. Transformants were scored for their ability to grow on YAG medium. Southern analysis demonstrated that the deletion cassette had integrated at each gene locus and that a single integration event had occurred. For all the Southern analyses, each gene open reading frame (ORF) was used as probe, and the absence of a radioactive signal indicated that the entire gene was replaced. In order to test whether additional integrative events had occurred, the membranes were stripped and subjected to another hybridization using the pyrG gene as a probe. The deleted transformant was crossed with strain UI224 in order to eliminate the ΔnkuA mutation; accordingly, all the deletion strains are wild type for this locus.
RNA isolation and cDNA library construction.
A. nidulans strain GR5 or AAH14 (1.0 × 107 conidia/ml) was used to inoculate 50 ml of liquid cultures, which were incubated in a reciprocal shaker at 37°C for 16 h. Mycelia were aseptically transferred to fresh YG medium in the presence or absence of 25 μM of CPT. Mycelia were harvested by filtration through a Whatman no. 1 filter, washed thoroughly with sterile water, quickly frozen in liquid nitrogen, and disrupted by grinding, and total RNA was extracted with Trizol reagent (Life Technologies). Ten micrograms of RNA from each treatment was then fractionated in a 2.2 M formaldehyde-1.2% agarose gel, stained with ethidium bromide, and then visualized with UV light. The presence of intact 25S and 17S rRNA bands was used as a criterion to assess the integrity of the RNA. RNase-free DNase treatment was done as previously described (65). Polyadenylated RNA was purified using oligo(dT)-cellulose (Oligotex mRNA kit; QIAGEN, Germany), and a unidirectional cDNA library was constructed in plasmid pSPORT-1 (Ampr) by using the Superscript plasmid system for cDNA synthesis and plasmid cloning (Life Technologies) according to the manufacturer's instructions to generate unidirectional 5′ SalI and 3′ NotI inserts. The number of clones of the library (transformed into Escherichia coli EMDH10B) was about 3 × 106 CFU, with an average insert size of 1.6 kb and 100% of clones having inserts larger than 200 bp; 10,000 individual colonies were transferred to 110 96-well microtiter plates for storage at −80°C.
Real-time PCRs, plasmid DNA extraction, and sequencing.
All the PCRs and reverse transcription-PCRs (RT-PCRs) were performed using an ABI Prism 7700 sequence detection system (Perkin-Elmer Applied Biosystems). TaqMan EZ RT-PCR kits (Applied Biosystems) were used for RT-PCRs, and the TaqMan Universal PCR Master Mix kit was used for PCRs. The thermal cycling conditions comprised an initial step at 50°C for 2 min, followed by 10 min at 95°C and 40 cycles of 95°C for 15 s and 60°C for 1 min. The reactions and calculations were performed as described previously (65). Table S2 in the supplemental material describes the primers and Lux fluorescent probes (Invitrogen) used in this work. Plasmid DNA was prepared by standard alkaline lysis procedures and also by the boiling method described previously (48). Single-run sequencing was done with the dideoxy chain termination method and dye termination chemistry (Applied Biosystems) using universal M13 primers. The sequence analysis was performed on ABI 377 and 3100 fluorescence automated sequencers. A pipeline was built to analyze and assemble the A. nidulans EST sequences (the EST project is available at http://143.107.203.68/camptothean/nidulans.html).
Construction of the A. nidulans macroarrays.
The macroarray consisted of 3,619 elements that are representative cDNA clones from partial sequences of 2,787 A. nidulans ORFs. Twenty microliters of the plasmid DNA was dried to completion and then resuspended in 40 μl of 50% dimethyl sulfoxide to a final concentration ranging from 150 to 250 ng/μl. The purity and quality of all plasmid DNAs were checked by agarose gel electrophoresis. Positively charged nylon membranes (222 mm by 222 mm; Genetix, United Kingdom) were placed into denaturing solution (NaCl, 1.5 M; NaOH, 0.5 M) and onto a Whatman no. 1 paper prewetted in the same denaturing solution. The plasmid DNAs were spotted in duplicate onto the nylon membranes by using the “Q”-bot robot (Genetix, United Kingdom) in a 5-by-5 array configuration, with spacing of 900 μm between each spot. After arraying, the membranes were denatured by the same procedure described above and placed in the neutralizing solution (NaCl, 1.5 M; Tris-HCl, 1 M; pH 7.0) twice for 10 min each time. The DNA was fixed to filters by UV light (160.000 μJ/cm2) and stored at room temperature until used for hybridizations.
Probe preparation and array hybridization.
About 1 × 107 conidia/ml of the A. nidulans wild-type strain GR5 were grown for 16 h in a reciprocal shaker (250 rpm) at 37°C. Mycelia were aseptically transferred to fresh YUU medium in the presence or absence of 25 μM CPT. Samples were taken after 30, 60, and 120 min of CPT treatment. Total RNA was isolated from CPT-treated and untreated mycelia as described above. cDNA probes were produced as described previously (64). Briefly, 30 μg of total RNA was reverse transcribed with Superscript II RNase H (Invitrogen), using oligo-dT18V (156 pmol) with 50 μCi [α-33P]dCTP (2,500 Ci/mol) and unlabeled dATP, dGTP, and dTTP (1 mM each). After an initial 20 min of incubation, unlabeled dCTP was added to a final concentration of 1 mM and the reaction was continued for additional 40 min at 42°C. Unincorporated nucleotides were separated from the probe by passage through a Sephadex G50 column (Amersham Bioscience). The radioactive signal was measured for each set of probes, and about equal cpm of each probe were added to the hybridization mixtures. Variations in the amount of DNA in the spots were estimated by hybridizing the membranes with an oligonucleotide probe that recognized the sequence of the Ampr gene of the pSPORT1 vector (overgo probe). This probe was synthesized with the primers OSG208 (5′ GTGGTCCTGCAACTTTATCCGC 3′) and OGA243 (5′ TAGACTGGATGGAGGCGGATAA 3′) in the presence of [α-33P]dCTP according to the protocol described by J. D. McPherson (http://www.tree.caltech.edu/protocols/overgo.html). Filters were first hybridized with the oligonucleotide vector probe for 18 h at 58°C (for details, see http://www.tree.caltech.edu/protocols/overgo.html). After hybridization and washing, the filters were exposed for 96 h to a storage phosphor screen (Imaging Plates; Fujifilm, Tokyo, Japan) and then scanned in a Fuji FLA3000 phosphorimager (Fujifilm, Tokyo, Japan) for image acquisition. The vector probe was removed by pouring a boiling solution of 0.1% (wt/vol) sodium dodecyl sulfate (SDS) over the filters and incubating them for 5 min at room temperature. The probe stripping for the overgo probes was performed twice, and the efficiency of probe removal was evaluated by phosphorimager scanning after 48 h of exposure in the image plates. After stripping, the filters were prehybridized with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10× Denhardt's solution, 20 mM Tris-HCl (pH 7.5), 1% SDS, 50% formamide, and 100 μg/ml of salmon sperm DNA for 4 h at 42°C. An α-33P-labeled cDNA probe was heat denatured for 5 min at 100°C, quickly cooled on ice, and added to a prewarmed hybridization mixture that consisted of 5× SSC, 2× Denhardt's solution, 20 mM Tris-HCl (pH 7.5), 1% SDS, 50% formamide, 5% dextran sulfate, and 100 μg/ml of salmon sperm DNA. Hybridization was carried out for 24 h at 42°C. The filters were washed twice in 2× SSC and 0.1% SDS for 15 min at 65°C, once in 1× SSC and 0.1% SDS for 15 min at 65°C, and twice in 0.1× SSC and 0.1% SDS for 15 min at 65°C. After the last wash, the filters were exposed to image plates for 96 h and scanned, as described above. Radiolabeled cDNA probes were completely stripped from the filters by washing them twice with a solution containing 0.4 M NaOH and 0.1% SDS at 65°C following two neutralizing washes using a solution of 2 M Tris-HCl (pH 8) and 0.1% SDS at room temperature. The filters were then reexposed to the image plates for 48 h in order to assess the residual labeled probe on each spot. If a residual signal was detected, the filters were rewashed as described above and reexposed for an additional 48 h. The result of the additional wash was determined. No more than two washes were ever required to completely remove the hybridization signal. Duplicate membranes were probed four times without detectable loss of signal. No cross hybridization signals were observed.
Macroarray analysis.
Phosphorimages were analyzed using Arrayvision software (Imaging Research, Canada). For each membrane, the grids were predefined and manually adjusted to obtain maximum spot recognition, and the spots were then individually quantified. The local background was automatically subtracted from each spot by taking the average intensity of the area surrounding each spot and subtracting it from the intensity value of each spot (sVol value). The intensity values generated by the probe vector hybridizations were evaluated to identify duplicated spots in which the amounts of DNA deposited onto the membrane were unequal. The coefficients of variation for the duplicated spots in the filters were calculated, and only spots presenting a coefficient of variation of ≤10% were used for further analysis. The macroarray data were pooled by averaging the two signal intensity values of the duplicated DNA spots and used to calculate the expression ratios between CPT-treated and untreated samples for the 30, 60, and 120 min of drug exposure. Each replicate membrane filter was treated as a separate hybridization but with the same biological material (variability between replicates was about 10%). To reduce the fluctuations among replica filters due to differences in the experimental conditions, the sign from every spot was normalized using the median of all signals on that membrane; i.e., the median was set to 1 (47, 64), generating the nVol value. Genes with expression levels close to the background value were discarded. In order to determine whether results were reproducible, correlation coefficients for duplicate hybridizations were calculated, and these were greater than 0.91 for all hybridizations. Differentially expressed genes were identified by Student's t test by using the log2s of normalized values from the two replicated experiments, with a P value of <0.05. The data sets were then imported into Microsoft Excel spreadsheets for further calculations. Means (M) of the normalized signals for the genes that showed up-regulation at at least one of the three times of CPT treatment (30, 60, or 120 min) were then calculated. The fold increase for each ORF was calculated as M(nVol)CPT-treated/M(nVol)untreated 120 min. The transcripts were clustered using the TIGR Multi Experiment Viewer 3.1 program (http://tigr.org/software/tm4/), using agglomerative hierarchical average linkage based on the Euclidean distance metric.
RESULTS
Establishment of the macroarray.
The EST approach is a rapid and relatively efficient method for quick gene discovery and also for the establishment of arrays, because the cDNA clones or their inserts can be spotted on nylon membranes or glass slides. Since we are interested in investigating the effect of DNA damage caused by the anti-topoisomerase I drug CPT on the large-scale gene expression of A. nidulans, we generated a macroarray for this organism based on this approach. Thus, a unidirectional cDNA library was constructed for the identification of genes expressed when A. nidulans was exposed to 25 μM CPT for 60 min. As previously demonstrated (22), by using these conditions the uvsCRAD51 (the homologue of the RAD51 gene) mRNA accumulation is increased (see Fig. 4F) and the DNA damage response is active in A. nidulans. We obtained 8,745 ESTs with a minimum length of 300 bases and a Phred quality value of at least 20 (2, 20, 21, 29, 31, 73). We sequenced from the 5′ end 8,575, ESTs while 170 were sequenced from the 3′ end. Clustering by the CAP3 program (34) identified 1,214 singlets and 2,405 contigs. Comparison of these ESTs with the assigned A. nidulans ORFs showed us that these 3,619 ESTs corresponded to 2,787 A. nidulans ORFs (http://www-genome.wi.mit.edu/annotation/fungi/aspergillus/).
FIG. 4.
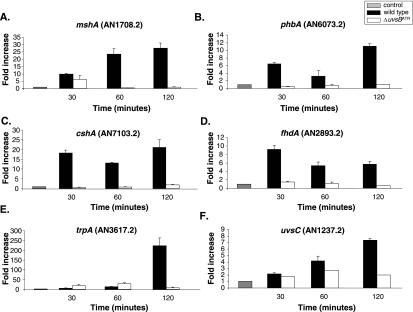
Expression of phbA (A), mshA (B), cshA (C), fhdA (D), tprA (E), and uvsC (F) mRNAs is dependent on the uvsBATR gene when A. nidulans is grown in the presence of CPT. The wild-type and ΔuvsB strains were grown in YG medium for 16 h at 37°C and then transferred to YG or YG plus 25 μM of CPT for 30, 60, and 120 min. RNA was extracted and DNase treated, and RT-PCRs were run. The measured quantity of each gene mRNA in each of the treated samples was normalized using the cycle threshold values obtained for the tubC RNA amplifications run in the same plate. The relative quantitation of each gene and tubulin gene expression was determined by a standard curve (i.e., cycle threshold values plotted against logarithm of the DNA copy number). Results of four sets of experiments were combined for each determination; means ± standard deviations are shown. The values represent the number of times that the genes are expressed compared to that for the wild type or the ΔuvsB mutant grown without any drug (represented absolutely as 1.00).
Macroarray analysis of the CPT transcriptional response in the wild-type strain.
To assess A. nidulans gene expression on a large scale, we constructed a 3,619-element cDNA macroarray by spotting cDNA clones representative of partial sequences of 2,787 nonredundant expressed genes from A. nidulans. There are two independent copies of each element in our array. These cDNA clones contained an average length of about 1.0 kb. The annotation information provided by the A. nidulans genome project was used for the designation of genes in this array (http://www-genome.wi.mit.edu/annotation/fungi/aspergillus/) (27).
To identify A. nidulans genes that display differential expression when the mycelia are exposed to CPT 25 μM during growth, we assessed gene expression at 30, 60, and 120 min in the presence and absence of CPT. Time course experiments were performed twice for each time point, yielding 12 independent hybridizations. For each experiment, samples were collected and total RNA was prepared. The cDNAs were generated from the total RNA samples and labeled with [α-33P]dCTP. Equal amounts of radioactively labeled cDNA probes were separately hybridized to duplicate nylon membranes. The signals were converted to relative intensity values, quantified, and statistically analyzed. Statistical analysis of the data generated by these hybridization experiments (Student's t test; P < 0.05) showed 1,512 genes that displayed modulation in their expression at at least one experimental time point. We were able to observe several genes involved with a variety of cellular processes, and their specific modulation is likely to be implicated in the A. nidulans response to CPT (Table 2). These genes were classified into broad functional categories (DNA repair, DNA metabolism, proteasome, signal transduction, and cell cycle). The data for the time course experiments as well as the complete list of regulated genes are available online at http://143.107.203.68/camptothean/html_iran_pub/.
TABLE 2.
CPT-up-regulated A. nidulans genes revealed by transcript profiling in the wild-type strain
| Functional category and ORF | Homologue (NCBI protein accession no.) | BLAST results (species and E value/ % identity/% similarity) | Induction time(s) (min) |
|---|---|---|---|
| Chromatin/chromosome | |||
| AN1286.2 | Putative hira protein; histone transcription regulator (NP596575) | Schizosaccharomyces pombe e−156/34/51 | 60, 120 |
| AN2770.2 | Putative histone promoter control protein (NP595268) | S. pombe 1e−18/37/51 | 120 |
| AN2765.2 | Histone H1(AAF16011) | Ascobolus immersus 4e−17/55/76 | 60 |
| AN0734.2 | Histone H4 (XP_328073) | Neurospora crassa 6e−44/100/100 | 30, 60, 120 |
| AN3468.2 | Histone H2A (CAA75581) | Aspergillus niger 3e−48/75/76 | 30, 60 |
| AN8039.2 | Histone H2A variant (NP595630) | S. pombe 7e−43/83/90 | 30 |
| AN3469.2 | Histone H2B (AAP69672) | Actinobacillus capsulatus 3e−42/98/98 | 60 |
| AN3071.2 | Histone acetyltransferase (AAF72665) | Homo sapiens 3e−/42/60 | 120 |
| AN8863.2 | Probable nucleosome assembly protein I (CAD70974) | N. crassa e−124/59/66 | 30, 120 |
| AN8851.2 | Centromere/microtubule binding protein CBF5 (NP013276) | S. cerevisiae 1e−71/70/80 | 30, 60 |
| AN0253.2 | Topoisomerase I (AAB39507) | Candida albicans 0.0/50/65 | 30 |
| AN2751.2 | Topoisomerase II-associated protein PAT1 homolog (NP595976) | S. pombe 3e−90/29/45 | 30 |
| AN6364.2 | Putative chromosome-associated protein (NP593260) | S. pombe 0.0/46/64 | 30, 60 |
| AN1698.2 | Putative involvement in chromatin structure (NP593191) | S. pombe e−180/43/59 | 30 |
| AN8742.2 | Structural maintenance of chromosome protein (AAL82734) | A. fumigatus 0.0/57/71 | 30 |
| DNA repair | |||
| AN3129.2 | Related to NAD+ ADP-ribosyltransferase (PARP) (CAD21266) | N. crassa 1e−127/43/67 | 30, 60, 120 |
| AN7309.2 | Postreplication repair protein uvs-2 (P33288) | N. crassa 1e−75/42/56 | 120 |
| AN0076.2 | Putative excision-repair protein (NP593167) | S. pombe 1e−38/25/44 | 60 |
| AN5344.2 | DNA repair protein mus-8 (P52493) | N. crassa 3e−63/79/83 | 30 |
| AN7103.2 | Similar to excision repair protein ERCC-6 (Cockayne syndrome protein CSB) (XP224627) | Rattus norvegicus e−124/51/65 | 30, 120 |
| AN1708.2 | MutS family DNA mismatch repair protein Msh6 (NP588344) | S. pombe 0.0/50/69 | 30 |
| AN2764.2 | DNA repair protein Rad2 (NP594972) | S. pombe e−124/60/75 | 120 |
| AN4407.2 | DNA repair and recombination protein Rad22 (RadC A. nidulans) (NP593207) | S. pombe 2e−53/33/45 | 60 |
| AN4365.2 | Related to DNA mismatch repair homologue (HPMS2) (XP_325240) | N. crassa 4e−86/31/46 | 120 |
| AN1237.2 | UvsC (Rad51 homologue) (EAL90369.1) | A. fumigatus 0.0/98/100 | 30, 60, 120 |
| DNA metabolism | |||
| AN6303.2 | Related to replication factor C protein (RP-C) (CAB91757) | N. crassa 0.0/46/59 | 60, 120 |
| AN7423.2 | Replication protein A (RP-A) (NP595092) | S. pombe e−150 46/63 | 60 |
| AN0415.2 | Proliferating cell nuclear antigen (PCNA) (NP596504) | S. pombe 1e−92/64/82 | 120 |
| AN8201.2 | DNA helicase, homologue of human XPBC (NP012123) | S. cerevisiae 0.0/64/75 | 30, 60, 120 |
| AN0067.2 | Ribonucleotide reductase small subunit (BAB13815) | Lentinula edodes 1e−138/61/71 | 30, 60 |
| AN4380.2 | Ribonucleotide-diphosphate reductase large chain (XP322797) | N. crassa 0.0/80/87 | 60, 120 |
| AN0687.2 | Spermidine synthase (XP327013) | N. crassa e−131/76/85 | 30, 60, 120 |
| AN8216.2 | Nucleoside diphosphate kinase (AAP85295) | A. fumigatus 1e−67/79/90 | 30 |
| AN0271.2 | Deoxyuridine-5′-triphosphate nucleotide (NP593873) | S. pombe 4e−46/69/78 | 30, 120 |
| AN8185.2 | Exonuclease II (NP593482) | S. pombe 0.0/48/64 | 30, 120 |
| AN2868.2 | Small fragment nuclease (NP077195) | Mus musculus 4e−37/48/65 | 30, 120 |
| AN5932.2 | DNA-directed DNA polymerase (CAB57881) | S. pombe 0.0/43/60 | 30 |
| AN2285.2 | DNA helicase (NP594979) | S. pombe e−159/39/52 | 120 |
| AN2739.2 | DNA polymerase theta isoform 2 (NP955452) | H. sapiens e−101/31/48 | 60, 120 |
| Protein degradation | |||
| AN2761.2 | Ubiquitin-conjugating enzyme E2, 16 kDa (ubiquitin-protein ligase) | Glomerella cingulata 6e−82/95/96 | 30, 120 |
| AN3716.2 | 19S proteasome regulatory subunit (NP593594) | S. pombe 9e−91/46/66 | 30 |
| AN7422.2 | Ubiquitin carboxyl-terminal hydrolase (NP596085) | S. pombe e−130/37/53 | 120 |
| AN6354.2 | Ubiquitin carboxyl-terminal hydrolase (NP588530) | S. pombe e−118/35/53 | 30, 120 |
| AN4236.2 | Probable 26S proteasome subunit and member of the CDC48/PAS1/SEC18 family of ATPases (NP014760) | S. cerevisiae e−168/73/85 | 60, 120 |
| AN0909.2 | Hypothetical protein (ubiquitin domain profile) (EAA65938) | A. nidulans 0.0/92/92 | 30, 120 |
| AN5981.2 | Hypothetical protein (ubiquitin domain profile) (EAA57730) | A. nidulans 0.0/77/77 | 60, 120 |
| AN2085.2 | Putative proteasome subunit PUP1 (NP014800) | S. cerevisiae 7e−93/66/79 | 60, 120 |
| AN4225.2 | 26S proteasome regulatory subunit (NP593111) | S. pombe e−108/50/68 | 120 |
| AN2000.2 | Polyubiquitin (AAK19308) | Tuber borchii e−166/98/100 | 30, 60, 120 |
| AN7254.2 | Cdc48 (AAM08677) | A. fumigatus 0.0/90/93 | 30, 60, 120 |
| AN4236.2 | Probable 26S protease subunit and member of the CDC48/PAS1/SEC18 family of ATPases (NP014760) | S. cerevisiae e−168/73/85 | 60, 120 |
| AN2212.2 | Ubiquitin-conjugating enzyme (NP596239) | S. pombe 1e−34/49/65 | 30, 120 |
| AN5793.2 | 20S proteasome beta-type subunit (NP015007) | S. cerevisiae 1e−75/59/69 | 30, 60, 120 |
| AN4869.2 | 20S proteasome component (NP036098) | M. musculus 7e−79/57/75 | 60, 120 |
| AN2416.2 | Ubiquitin-activating enzyme E1C isoform 1; Nedd8-activating enzyme (NP003959) | H. sapiens 4e−81/38/56 | 120 |
| AN5705.2 | Ubiquitin carboxyl terminus (T38954) | S. pombe 1e−90/62/90 | 60, 120 |
| AN1700.2 | 26S proteasome regulatory subunit (NP596381) | S. pombe 0.0/43/62 | 60 |
| AN4872.2 | Monoubiquitin/carboxy extension protein fusion (AAC24705) | Botryotinia fuckeliana 6e−60/73/79 | 30 |
| AN4016.2 | Ubiquitin fusion protein (CAB50892) | Kluyveromyces lactis 8e−66/96/100 | 30, 60, 120 |
| AN2174.2 | Ubiquitin-activating enzyme E1 (O94609) | S. pombe 0.0/58/72 | 30, 60, 120 |
| AN5783.2 | 20S proteasome component (beta 7) (NP595308) | S. pombe 9e−66/51/69 | 120 |
| AN6988.2 | Probable 26S proteasome subunit protein 8 homolog (NP595870) | S. pombe e−165/77/86 | 120 |
| AN2416.2 | Ubiquitin-activating enzyme E1C isoform 1; Nedd8-activating enzyme (NP003959) | H. sapiens 4e−81/38/56 | 120 |
| AN1191.2 | Ubiquitin-like modifier (NP596035) | S. pombe 8e−20/50/64 | 120 |
| Signal transduction, cell cycle | |||
| AN1019.2 | Putative Scf complex protein (CAD28438) | A. fumigatus 0.0/79/85 | 30, 60, 120 |
| AN6044.2 | Similar to cell division cycle 2 homologue protein kinase (NPKA) (XP235722) | S. pombe 2e−77/58/80 | 60 |
| AN2412.2 | Ca/CaM-dependent kinase-1 (AAL14118) | N. crassa 1e−152/71/81 | 60, 120 |
| AN6508.2 | Protein kinase Skp1 (NP593134) | S. pombe e−154/70/84 | 120 |
| AN4182.2 | Cell division control protein 2 CDC2 (cyclin-dependent protein kinase) (XP330428) | N. crassa 1e−140/79/85 | 30, 120 |
| AN5100.2 | Similar to yeast Cdc50 (NP595126) | S. pombe 9e−82/44/61 | 30, 60 |
| AN5102.2 | Cell division control protein 68 CDC68 gene product (Q00976) | K. lactis 0.0/42/60 | 30, 60, 120 |
| AN5744.2 | 14-3-3-like protein (AAP22960) | Paracoccidioides brasiliensis e−124/92/97 | 30, 120 |
| AN1560.2 | Polo-like kinase (BAB18588) | Hemicentrotus pulcherrimus 1e−59/42/61 | 30, 120 |
| AN2047.2 | Calmodulin (AAC96324) | Magnaporthe grisea 1e−76/97/100 | 120 |
| AN4583.2 | Peptidylprolyl isomerase D (cyclophilin D) (NP080628) | M. musculus 1e−86/48/59 | 60, 120 |
| Miscellaneous | |||
| AN6073.2 | Putative prohibitin (NP588144) | S. pombe 4e−88/68/78 | 30, 60, 120 |
| AN2893.2 | Smad nuclear interacting protein (35.8 kDa) (NP451217) | Caenorhabditis elegans 3e−29/43/63 | 120 |
| AN7374.2 | Hypothetical protein (EAA61745) | A. nidulans e−119/100/100 | 30, 60, 120 |
| AN3617.2 | Tetratrico peptide repeat (ZP00109827) | Nostoc punctiforme 3e−2/43/64 | 30, 60, 120 |
| AN8562.2 | Hypothetical protein (ankirin repeats) (EAA66987) | A. nidulans 0.0/95/95 | 60, 120 |
| AN1556.2 | WD-40 repeat protein family (NP564469) | Arabidopsis thaliana 4e−39/29/48 | 120 |
| AN7170.2 | Hypothetical protein (Myc type, helix-loop-helix) (EAA61422) | A. nidulans e−101/70/70 | 30, 60, 120 |
| AN4119.2 | Major facilitator superfamily (AAO49453) | Leptosphaeria maculans e−133/48/64 | 120 |
| AN8489.2 | ABC transporter PMR5 (BAB59028) | Penicillium digitatum 0.0/71/83 | 120 |
| AN2210.2 | Probable ABC-transporter (NP200887) | A. thaliana e−170/55/72 | 30, 60, 120 |
| AN8746.2 | Transcription factor Bft3 homologue (NP594757) | S. pombe 4e−37/52/68 | 60, 120 |
| AN1217.2 | LIM/homeobox transcription factor (Q25132) | Halocynthia roretzi 3e−07/38/58 | 30, 60, 120 |
| AN2919.2 | Zinc finger transcription factor ACE I (AAL69549) | Talaromyces emersonii e−131/43/56 | 30, 60, 120 |
| AN0273.2 | Transcription factor that activates expression of early G1-specific genes (NP013232) | S. cerevisiae 5e−11/28/42 | 30, 60, 120 |
Cluster analysis of transcriptionally induced genes.
The 1,512 modulated elements from wild-type A. nidulans have also been analyzed with the aid of a K-means algorithm, in an attempt to cluster genes according to the similarities in their expression profiles. Their distribution into 60 distinct clusters shows a large number of genes with minor alterations in their expression levels, while others were dramatically up- or down-regulated at one or more steps of exposure to CPT (Fig. 1). We focused our attention in the two clusters that seemed to contain genes with the most intense and consistent up-regulation profiles (counting from the top left to right, clusters 35 and 49 in Fig. 1). In cluster 35, genes were highly expressed upon induction with CPT for 60 and 120 min, while cluster 49 contained genes that were overexpressed at all the time points. Most of the genes which are down-regulated, which are present in the clusters 16, 53, 54, and 55, encode hypothetical proteins (http://143.107.203.68/camptothean/html_iran_pub/).
FIG. 1.
Clusters of gene expression generated by the K-means algorithm. The 1,512 genes that showed modulation in expression during the exposure of the A. nidulans wild-type strain to CPT were evaluated by a figure-of-merit algorithm. The results obtained supported their subdivision into 60 clusters, which was achieved with the aid of a K-means algorithm. Groups of genes with similar modulation of gene expression during exposure to CPT are located in each cluster. The figure shows, on the y axis, the variation in the log2(Cy5/Cy3) ratios (from −4 to 4) and, on the x axis, the different time points of the exposure to CPT, taking as a reference their respective expression levels at the corresponding time in the absence of CPT. Clusters 35 and 49, containing genes that displayed the most intense and consistent up-regulation profiles, are indicated by boldface and are shown in more detail in Fig. 5.
In cluster 35, we have observed eight genes encoding hypothetical proteins (AN1066.2, AN4049.2, AN5352.2, AN9412.2, AN2029.2 [contains an F-box domain], AN7374.2, AN8462.2, and AN8562.2 [contains ankirin repeats]) and four other genes encoding a putative ATP-dependent helicase (AN7753.2), a fumarate hydratase (AN8707.2), a 6-phosphogluconate dehydrogenase (AN3954.2), and a forkhead-associated domain protein (AN2893.2). Cluster 49 shows the up-regulation of three genes encoding the poly(ADP-ribose) polymerase (PARP) (AN3129.2; named prpA previously [67]), a hypothetical protein that contains a tetratrico peptide repeat (AN3617.2), and a multidrug ABC transporter (AN8489.2).
To assess the reliability of the macroarray results and validate the expression of some of these genes, we chose three genes from clusters 35 and 49, manually selected 20 genes from our significant-gene list (Table 2) of more expressed genes, designed Lux probes, and used real-time RT-PCR analysis to quantify their expression in the presence of CPT (see Fig. S1 in the supplemental material). The results were expressed as the relative A. nidulans growth in the presence of CPT divided by the growth in the absence of CPT transcripts, where the growth in the absence of CPT transcript copy number was given a value of 1 (see Fig. S1 in the supplemental material). We also compared the gene expression variation estimated by these two methodologies by calculating both Pearson's (RP) and Spearman's (RS) correlation coefficients for the log2 ratios obtained by the two approaches. As shown in Table 3, the value of either RP or RS was above 0.50 (indicating moderate to strong correlation) for 19 out of 23 genes (about 80% of the cases). Thus, although we were able to detect some discrepancies between the two methodologies, it seems that our macroarray hybridization approach is capable of providing information about A. nidulans gene expression modulation with a considerably high level of confidence. Our results strongly indicate that the genes described here might have their mRNA expression increased when A. nidulans is growing in the presence of CPT and possibly are involved in the DNA damage response.
TABLE 3.
Comparison of gene expression values obtained with macroarray hybridization and real-time RT-PCR
| Gene product (ORF) | Log2 ratio between value after the indicated time (min) of CPT treatment and reference point value (array/RT-PCR)
|
Correlation coefficienta (RP/RS) | ||
|---|---|---|---|---|
| 30 | 60 | 120 | ||
| Ubiquitin carboxyl-terminal hydrolase (AN7422.2) | 0.17/0.32 | 0.15/2.12 | 0.84/3.59 | 0.82/0.46 |
| Ubiquitin-like modifier (AN1191.2) | −0.22/1.48 | −0.12/1.90 | 0.39/3.24 | 0.99/0.94 |
| Histone acetyltransferase (AN3071.2) | 0.45/0.06 | 0.52/0.43 | 0.95/1.35 | 0.99/0.89 |
| Ubiquitin-activating enzyme Ned8 (AN2416.2) | −0.14/1.28 | −0.25/1.23 | 0.49/1.39 | 0.99/0.94 |
| Histone H2A variant (AN8039.2) | 0.25/1.72 | −0.13/1.04 | −0.54/0.78 | 0.96/0.89 |
| Rad52 (AN4407.2) | 0.27/1.29 | 2.07/3.35 | 0.23/2.83 | 0.68/0.49 |
| PARP (AN3129.2) | 1.17/3025 | 3.06/3.88 | 3.39/4.46 | 0.94/0.94 |
| Rad2 (AN2764.2) | −1.10/1.65 | −0.12/1.08 | 0.37/0.63 | −0.99/−0.98 |
| Rad26 (AN7103.2) | 0.36/2.18 | −0.08/3.71 | 0.68/4.38 | 0.20/0.50 |
| Rad18 (uvsH) (AN7309.2) | 0.60/1.57 | −0.55/2.37 | 0.69/3.76 | 0.22/0.43 |
| MutS (AN1708.2) | 0.78/1.17 | 0.79/4.55 | −0.06/4.30 | −0.06/0.43 |
| Smad (AN2893.2) | 0.07/1.33 | 1.76/2.41 | 2.66/2.65 | 0.98/0.89 |
| Prohibitin (AN6073.2) | 1.21/1.06 | 0.34/1.63 | 0.74/3.47 | −0.27/−0.50 |
| Sulfur metabolite repression control protein SconB (AN6359.2) | 0.69/0.97 | 0.12/2.01 | 0.49/2.38 | −0.58/−0.37 |
| Helicase (AN2285.2) | −1.09/0.19 | −0.91/2.91 | 1.91/3.76 | 0.72/0.89 |
| Topoisomerase I (AN0253.2) | 0.96/2.06 | −0.05/1.50 | −0.21/2.12 | 0.30/−0.41 |
| UvsC (Rad51 homologue) (AN1237.2) | 0.43/1.43 | 0.67/1.98 | 1.20/0.33 | −0.17/0.08 |
| Ribonucleotide reductase small subunit (AN0067.2) | 1.08/0.09 | 1.23/0.92 | 0.39/2.12 | −0.83/−0.49 |
| Ribonucleotide-diphosphate reductase large chain (AN4380.2) | 0.43/1.52 | 0.94/1.97 | 1.17/3.31 | 0.91/0.89 |
| Myb-like DNA binding protein Myb-1 (AN0279.2) | 0.76/0.32 | 0.69/1.14 | −0.69/1.19 | −0.58/−0.99 |
| Tetratrico peptide repeat (AN3617.2) | 2.75/2.26 | 3.28/3.64 | 4.23/7.78 | 0.99/0.87 |
| HpmS2 (AN4365.2) | 0.73/0.99 | 1.58/2.03 | 1.22/2.55 | 0.72/0.50 |
| Cell division cycle 2 homolog protein kinase (NPKA) (AN6044.2) | −0.22/1.07 | 0.74/0.97 | −0.29/3.30 | −0.58/0.94 |
Correlation coefficients calculated after comparison of the array and RT-PCR data for each gene. We calculated both Pearson's (RP) and Spearman's (RS) correlation coefficients for each pair of curves. The former is more adequate for comparisons in which the data assume a normal distribution, while the latter is more appropriate for data sets which do not follow a normal distribution. The highest R value is in boldface.
Macroarray analysis of the CPT transcriptional response in the uvsBATR strain.
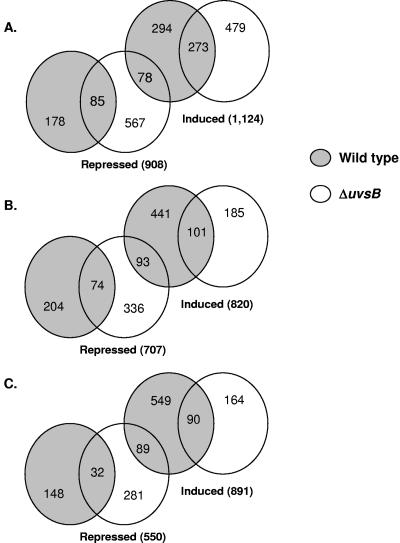
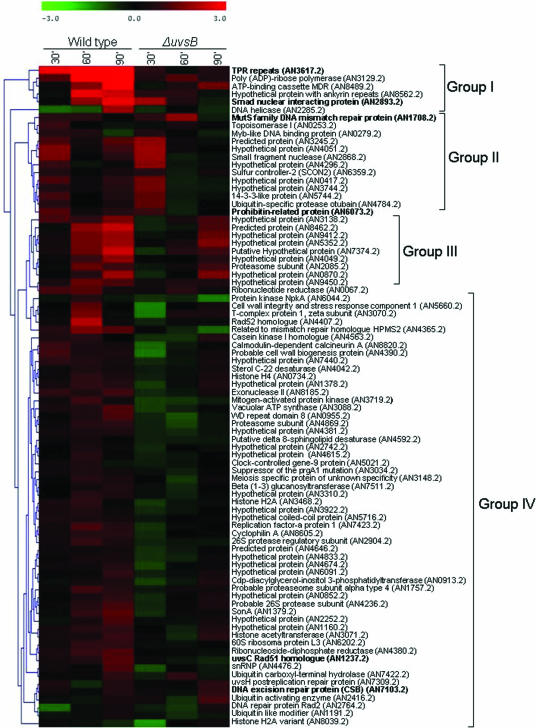
In several eukaryotic organisms, DNA damage checkpoint activation is controlled by the conserved family of ATM/ATR kinases (55, 68), which includes A. nidulans UvsBATR and AtmAATM (14, 46). It has already been shown that ATR is important in responding to the replication-associated DNA damage from topoisomerase poisons (12). Thus, to identify genes that have their expression dependent on uvsBATR (the homologue of the ATR gene), we once more assessed gene expression when the uvsBATR mutant strain was grown for 30, 60, and 120 min in the presence and absence of CPT. The same experimental design previously described for the wild-type strain was again followed for the uvsBATR mutant strain; i.e., time course experiments were performed twice for each time point, yielding 12 independent hybridizations. It is important to note that we did not detect any significant difference in survival with CPT treatment when the uvsBATR strain was compared to the wild-type strain (data not shown), so the expression data corresponded mainly to differences due to the presence or absence of this gene. Statistical analysis of the data generated by these hybridization experiments (Student's t test; P < 0.05) showed about 1,700 genes that displayed modulation in their expression at at least one experimental time point. These data were compared to the corresponding data for wild-type expression during growth in the presence of camptothecin (Fig. 2 and 3). Altogether, there are 1,124, 820, and 891 genes that are up-regulated and 908, 707, and 550 genes that are down-regulated when both strains are exposed to CPT for 30, 60, and 120 min (Fig. 2). Surprisingly, there is only one gene (AN5014.2, encoding a homologue of the 60S ribosomal protein L22) that showed overlap in the Venn diagram between the three time points for increased and decreased mRNA expression in the wild-type and the ΔuvsB mutant strain, respectively. In order to have a comparative view of these alterations, we arbitrarily and manually selected 84 genes from our list of significant genes and hierarchically clustered them (Fig. 3). (The complete list of gene expression for the wild-type and ΔuvsB mutant strains is available in Table S3 in the supplemental material and at http://143.107.203.68/camptothean/html_iran_pub/.) This hierarchical clustering could be divided into four groups (I to IV). We characterized in more detail some representative genes belonging to the different groups. In group I, we chose ORFs AN2893.2 and AN3617.2, which correspond to genes encoding a forkhead-associated domain protein (named fhdA) and a hypothetical protein that contains a tetratrico peptide repeat (named tprA), respectively. In group II, two genes were selected: ORFs AN1708.2, corresponding to a gene encoding a MutS homologue (mshA) involved in mismatch repair, and AN6073.2, a prohibitin gene homologue (phbA) which encodes a protein localized to the mitochondria, where it might have a role in the maintenance of mitochondrial function and protection against senescence (51). In group IV, uvsCRAD51 (AN1237.2), and ORF AN7103.2 (cshA), which corresponds to a gene that encodes a homologue of the excision repair protein CSB (Cockayne's syndrome protein; more highly expressed at 30 and 120 min [Table 2]), were selected for further studies. Additionally, we have recognized the npkA gene (AN6044.2) in group III, which encodes a cdc2-related kinase that we had previously shown to interact with uvsBATR (23).
FIG. 2.
Venn diagram representation of transcripts identified as exposed to CPT by 30 (A), 60 (B), and 120 (C) minutes. Wild-type and uvsBATR deletion mutant strains are identified as gray and white circles, respectively.
FIG. 3.
Hierarchical clustering showing the pattern of expression of arbitrarily selected genes from A. nidulans wild-type and uvsBATR deletion mutant strains when exposed to CPT. The color code displays the log2 ratio for each time point, having as the reference value the treatment without CPT for each time point. The genes highlighted in boldface were deleted in this work. Note that all the genes from clusters 35 (except AN1066.2, AN2029.2, AN7753.2, AN8707.2, and AN3954.2) and 49 (identified in Fig. 2) are present in this list of selected genes.
First, we checked whether the mRNA expression of phbA, mshA, cshA, fhdA, tprA, and uvsC was dependent on the uvsBATR deletion mutant background. Thus, the uvsBATR deletion mutant was grown in the absence of any drug and transferred to 25 μM of CPT for 30, 60, and 120 min. Then, mRNA was isolated, and real-time RT-PCR was performed. As previously shown (see Fig. S1 in the supplemental material), all these genes are induced at the mRNA level when the wild type is exposed to CPT; however, when the ΔuvsBATR mutant is exposed to CPT, the mRNA expression of all these genes is broadly down-regulated (Fig. 4). These results show that the induced transcript levels of the phbA, mshA, cshA, fhdA, trpA, and uvsC genes in the presence of CPT require uvsBATR.
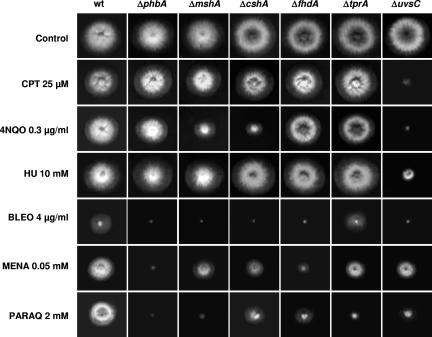
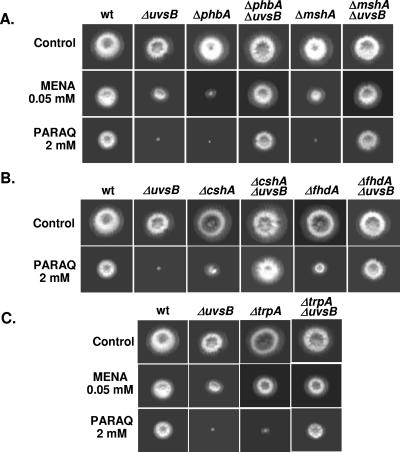
To have more information about the function of some of the genes identified as being more expressed in the wild type and repressed in the ΔuvsBATR mutant when both strains are exposed to camptothecin, we inactivated the phbA, mshA, cshA, fhdA, and tprA genes by PCR-mediated deletion (see Fig. S2 in the supplemental material). The uvsCRAD51 gene has been previously deleted (35). These deletion mutants and the corresponding parental strain were grown in the presence of different DNA-damaging and oxidative stress agents. Surprisingly, only the ΔuvsC mutant strain was sensitive to CPT; in addition, this strain was also sensitive to 4-nitroquinoline-1-oxide (4-NQO), HU, bleomycin (BLEO), and paraquat (PARAQ) but was as sensitive as the wild type to menadione (MENA) (Fig. 5). However, all the mutant strains except the ΔtprA mutant are more BLEO sensitive (Fig. 5). The ΔcshA and ΔmshA mutant strains showed slight sensitivity to 4-NQO (Fig. 5). The ΔphbA and ΔfhdA mutants showed MENA sensitivity, while all mutant strains displayed PARAQ sensitivity, except the ΔcshA and ΔfhdA mutants, which are as sensitive to PARAQ as the wild type. No significant difference in sensitivity to UV light was observed between these null mutants and the corresponding wild-type strain (data not shown). Taken together, these results indicate that the selected genes when inactivated displayed very complex and heterogeneous phenotypes of sensitivity during growth in the presence of agents that directly or indirectly cause DNA damaging. All these phenotypes are identical in media either supplemented with UU or not. Furthermore, since most of these inactivation mutants are not sensitive to CPT, the data also indicated that the transcriptional response of A. nidulans to CPT does not necessarily identify genes that directly protect against this genotoxic agent.
FIG. 5.
Growth phenotypes of the inactivation mutants. Wild-type (wt) (UI224), ΔphbA (IM70), ΔmshA(IM71), ΔcshA (IM72-3), ΔfhdA (IM73-104), ΔtprA (IM74-10), and ΔuvsC (ATr60cd1-14) strains were grown for 72 h at 37°C in YUU, YUU plus CPT (25 μM), YUU plus 4-NQO (0.3 μg/ml), YUU plus HU (10 mM), YUU plus BLEO (4 μg/ml), YUU plus MENA (0.05 mM), and YUU plus PARAQ (2 mM).
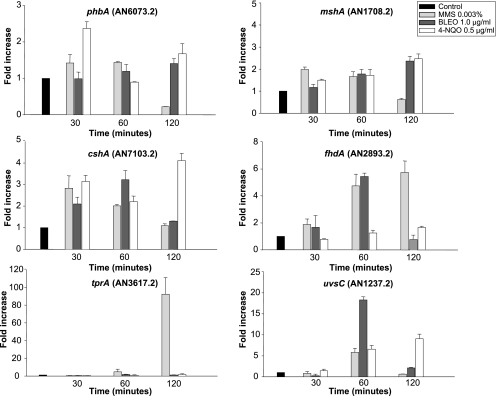
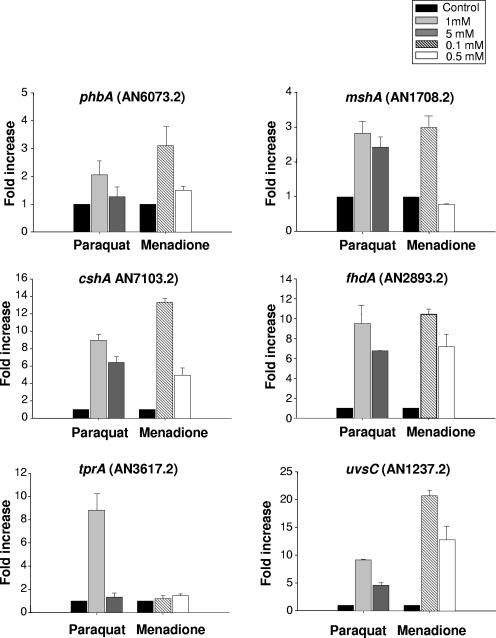
To learn more about the relationship between sensitivity to growth in the presence of these agents and the transcriptional response of the selected genes to them, we exposed the wild-type strain to MMS, BLEO, and 4-NQO (Fig. 6) and MENA and PARAQ (Fig. 7) and verified the mRNA expression of these genes. The mshA and cshA genes showed a low level of mRNA increase in the presence of MMS, BLEO, and 4-NQO (2- to 3-fold), while phbA responded (1.2- to 2-fold induction) to MMS and 4-NQO and fhdA responded (about 5-fold at 60 min) to MMS and BLEO. The uvsC gene showed about 6-, 17-, and 7-fold mRNA increases when the wild-type strain was exposed to MMS, BLEO, and 4-NQO for 60 min and a 9-fold increase when exposed to 4-NQO for 120 min. The tprA gene showed the highest expression (about 80-fold) after 120 min in the presence of MMS. When the wild-type strain was exposed to different concentrations of PARAQ and MENA, the mshA and phbA genes showed low levels of mRNA increase (about 1.2- to 3-fold) (Fig. 7). In contrast, the fhdA, cshA, and uvsC genes displayed high levels of mRNA expression when the wild-type strain was exposed to different concentrations of PARAQ (about 9-fold) and MENA (10- to 12-fold) (Fig. 7). The tprA gene showed increased mRNA expression only in the presence of 1 mM PARAQ (Fig. 7). These data suggest a very complex transcriptional behavior of these genes when A. nidulans is exposed to different DNA-damaging and oxidative stress agents.
FIG. 6.
Expression of phbA, mshA, cshA, fhdA, tprA, and uvsC mRNAs when A. nidulans is exposed to different DNA-damaging agents. The wild-type strain was grown in YG medium for 16 h at 37°C and then transferred to YG, YG plus 0.003% MMS, YG plus 1.0 μg/ml BLEO, or YG plus 0.5 μg/ml 4-NQO for 30, 60, and 120 min. RNA was extracted and DNase treated, and RT-PCRs were run. The measured quantity of each gene mRNA in each of the treated samples was normalized using the cycle threshold values obtained for the tubC RNA amplifications run in the same plate. The relative quantitation of each gene and tubulin gene expression was determined by a standard curve (i.e., cycle threshold values plotted against logarithm of the DNA copy number). Results of four sets of experiments were combined for each determination; means ± standard deviations are shown. The values represent the number of times that the genes are expressed compared to that for the wild type or the ΔuvsB mutant grown without any drug (represented absolutely as 1.00).
FIG. 7.
Expression of phbA, mshA, cshA, fhdA, tprA, and uvsC mRNAs when A. nidulans is exposed to different oxidative stress agents. The wild-type strain was grown in YG medium for 16 h at 37°C and then transferred to YG, YG plus PARAQ (1 or 5 mM), or YG plus MENA (0.1 or 0.5 mM) for 60 min. RNA was extracted and DNase treated, and RT-PCRs were run. The measured quantity of each gene mRNA in each of the treated samples was normalized using the cycle threshold values obtained for the tubC RNA amplifications run in the same plate. The relative quantitation of each gene and tubulin gene expression was determined by a standard curve (i.e., cycle threshold values plotted against logarithm of the DNA copy number). Results of four sets of experiments were combined for each determination; means ± standard deviations are shown. The values represent the number of times that the genes are expressed compared to that for the wild type or the ΔuvsB mutant grown without any drug (represented absolutely as 1.00).
Interestingly, we have also observed during the growth of the ΔmshA mutant strain the appearance of sectors, which could be a hallmark of genetic instability for this strain. Thus, we investigated this phenotype by growing the wild-type and ΔmshA mutant strains in the absence and presence of 25 μM of CPT and 0.04 mM of MENA. The ΔmshA mutant strain grew about 25% faster than the wild-type strain in the absence and presence of CPT; however, it grew at about the same rate as the wild type in the presence of MENA. The number of sectors in the ΔmshA mutant strain was comparable in the absence of any drug treatment and in the presence of MENA, but the number of sectors was increased in the presence of CPT (data not shown), confirming that the mshA gene is an important component in the pathways that contribute to A. nidulans genetic stability.
We constructed double-inactivation mutants with uvsBATR and these deleted genes (phbA, mshA, cshA, fhdA, tprA, and uvsCRAD51) to screen for possible genetic interactions. Surprisingly, in most cases, the deletion mutants exhibited cosuppression with uvsBATR (Fig. 8). For example, whereas both the uvsB and phbA mutants were extremely sensitive to PARAQ, the double mutant grew almost as well as the wild type. These observations suggest that a complex network of functions controls the response to oxidative inducing agents such as MENA and PARAQ. Furthermore, many of these functions appear to act in a manner that is antagonistic to UvsB, which could account for the observed cosuppression.
FIG. 8.
The double-inactivation mutants are more sensitive to oxidative stress agents. Wild-type (UI224), ΔuvsBATR, ΔphbA (IM70), ΔphbA ΔuvsB (IM70-14), ΔmshA (IM71), ΔmshA ΔuvsB (IM71-14), ΔcshA (IM72-3), ΔcshA ΔuvsB (IM72-2.14), ΔfhdA (IM73-104), ΔfhdA ΔuvsB (IM73-104.14A), ΔtprA (IM74-10), and ΔtrpA ΔuvsB (IM74-10.14A) strains were grown for 72 h at 37°C in YUU, YUU plus MENA (0.05 mM), and YUU plus PARAQ (2 mM).
DISCUSSION
Topoisomerases are enzymes that modify and regulate the topological state of DNA (72). Since they are required for replication, transcription, recombination, and chromosome segregation, they have an important role in the maintenance of genome integrity (6, 43). During the cleavage reaction, topoisomerases covalently attach to newly generated DNA 5′ phosphotyrosyl bonds (members of the type IA subfamily, including E. coli topoisomerases I and III, eukaryotic topoisomerase III, and archaeal reverse gyrase) or 3′ phosphotyrosyl bonds (members of the type IB subfamily, including eukaryotic topoisomerase I, archaeal topoisomerase V, and the poxvirus type I topoisomerases) (26, 33, 43, 44, 70). Under normal circumstances, these covalent enzyme-DNA cleavage complexes are fleeting catalytic intermediates and are present at low steady-state concentrations that are tolerated by the cell. However, conditions that significantly increase the physiological concentration or lifetime of these breaks, such as exposure to the anti-topoisomerase I drug CPT, cause several deleterious side effects, including mutations, insertions, deletions, and chromosomal aberrations. The basic mechanism of action for CPT is well characterized (26). Briefly, CPT generates replication-mediated DSBs, which in turn induce reversible or permanent cell cycle arrest in G2-M. The repair of a topoisomerase lesion presents special problems because the strand break is entangled with a covalently bound polypeptide. To restore the integrity of the chromosomal DNA, this polypeptide must be removed. Mutants defective in DNA repair show increased sensitivity to topoisomerase poisons, and it has already been reported that in Saccharomyces cerevisiae multiple pathways could repair topoisomerase damage (45, 69). Thus, to examine gene expression during DNA damage caused by CPT, we have performed a high-throughput macroarray hybridization analysis of A. nidulans.
We evaluated gene expression in both A. nidulans wild-type and uvsBATR deletion mutant strains in a time course exposure to CPT. We concentrated our analysis on the genes that are transcriptionally induced during the exposure of the A. nidulans wild-type strain to CPT and analyzed whether the mRNA expression of these genes was different in the uvsBATR deletion mutant background compared to the wild-type strain. Some of these genes were investigated in more detail by generating deletion mutants and observing their mRNA expression when the wild-type and uvsBATR deletion mutant strains were exposed to several DNA-damaging and oxidative stress agents. Furthermore, we also observed possible genetic interactions with the uvsBATR deletion mutant by constructing double mutants with these deletion strains. Surprisingly, we have shown that among six genes induced by CPT at the mRNA level, only one (uvsCRAD51) displayed CPT sensitivity when inactivated, indicating that their roles in processing DNA lesions produced by DNA-damaging agents cannot easily be inferred from gene expression profiling. It has already been shown that the transcriptional response of S. cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents (7). However, in contrast to those authors, we have observed a much more complex behavior and relationship between mRNA expression and protection against these agents. Actually, this behavior was observed not only for CPT but also for other DNA-damaging and oxidative stress agents. For instance, most of the genes when inactivated caused BLEO sensitivity, but they were either not induced by this drug or showed only a slight mRNA increase. Furthermore, uvsCRAD51 was highly induced by MENA and PARAQ, but the uvsCRAD51 deletion mutant strain is not very sensitive to these agents.
Genes induced during DNA damage caused by CPT.
Transcriptional and translational changes after DNA damage can be either primary responses to the damage or secondary to cellular processes responding to the damage. Identification of changes in gene expression levels can provide a useful link between DNA damage and CPT sensitivity. The transcriptional regulation of genes by CPT-induced DNA damage could reflect resistance mechanisms, and resistance to this drug could be the result of (i) alterations in topoisomerase I that confer resistance to CPT, (ii) cellular accumulation and transport of CPT, and (iii) alterations in the ternary complex formation (60). We have observed several transporters from the ABC family and major facilitator superfamily as being more expressed during exposure to CPT. These genes could be involved in the detoxification of CPT by pumping this drug out of the cytoplasm. In yeast, mutations in the ABC protein Snq2 result in CPT resistance (61). In mammalian cells, MDR1 overexpression confers resistance to CPT (11), while inhibition of MRP2 gene expression can increase cellular sensitivity to CPT derivatives (41).
There is relatively little known about the pathways downstream from CPT-topoisomerase I-DNA ternary complex formation that lead to repair of DNA damage or cell death. Several DNA replication, DNA damage checkpoint, and DNA repair proteins have been involved in the response to cleavage complex formation (for a review, see reference 60). Taking into consideration the repair of CPT-induced DNA damage, both mismatch repair and base excision repair systems are implicated. Accordingly, we have observed several genes involved in these pathways as being induced upon CPT exposure (Table 2). In addition to DNA repair genes, we have also detected genes involved in (i) chromatin and DNA metabolism, such as histone and ribonucleotide reductase genes; (ii) protein degradation, such as genes encoding ubiquitin and subunits of the proteasome; and (iii) signal transduction and cell cycle, such as polo-like kinase and calmodulin genes. Some of the genes observed to be more induced in the presence of CPT were also seen to be expressed in other studies addressing the global transcriptional regulation of genes by CPT-induced DNA damage (13, 32, 50, 52, 77). Genes that act in the ubiquitin/proteasome-dependent degradation pathway (77), in DNA repair (such as RAD51 and PARP) (52), and in chromatin architecture (such as the histone H2A gene) (50) were also detected as being more induced by CPT. The participation of these genes in the CPT DNA damage transcriptional response remains to be determined.
We have observed that the genes encoding fumarate hydratase, 6-phosphogluconate dehydrogenase, and PARP are the most expressed genes in our macroarray hybridizations. The fumarate hydratase is a nucleus-encoded mitochondrial protein and an enzyme of the tricarboxylic acid cycle that has been implicated in tumor susceptibility (30). Mutations in mitochondrial tumor suppressor genes can contribute to tumor formation through redox stress resulting from increased production of reactive oxygen species (ROS) in mitochondria (30). Increased production of this gene could contribute to avoidance of DNA damage caused by ROS production. The 6-phosphogluconate dehydrogenase is an enzyme of the pentose phosphate pathway that produces important precursors for DNA biosynthesis and repair. Zhang et al. (75) have shown that in the highly resistant bacterium Deinococcus radiodurans, the pentose phosphate pathway augmented the excision repair system by providing cells with adequate metabolites. PARP consumes NAD+ to catalyze the formation of ADP-ribosyl groups on target proteins. Modification of a target by PARP is a rapid response capable of altering protein activity and/or stability (reviewed in references 36 and 40). Accordingly, PARP plays an integral role in the cellular response to a variety of stresses, most notably DNA damage (for a review, see reference 3). Recently, we characterized a putative PARP homologue (PrpA) in A. nidulans (67). The genetic analysis of prpA demonstrates that it is an essential gene whose role in the DNA damage response is sensitive to gene dosage. Notably, temporal patterns of prpA expression and PrpA-GFP nuclear localization suggest that PrpA acts early in the A. nidulans DNA damage response. Interestingly, the prpA gene is highly expressed at all time points in our array, highlighting its importance upon DNA damage in A. nidulans.
We have further investigated the participation of inactivated phbA, mshA, tprA, chsA, fhdA, and uvsCRAD51 genes by growing null mutant strains in the presence of several DNA-damaging and oxidative stress agents. Most of these deletion strains were more sensitive to BLEO than the corresponding wild-type strain. Furthermore, the ΔphbA, ΔmshA, and ΔcshA mutant strains were more sensitive to oxidative stress agents, such as MENA and PARAQ. The phbA, mshA, and cshA genes are homologues of the prohibitin, MutS, and CSB genes, respectively, while tprA and fhdA encode hypothetical proteins that have tetrarico repeats and a forkhead-associated domain, respectively. Two prohibitins (PHB and PHB2 products) together can form a high-molecular-weight complex, and these complexes have been identified in both the mitochondria and the plasma membrane (51). The least-controversial and best-described function of the prohibitins is as chaperone proteins in the mitochondria in the stabilization of newly synthesized subunits of mitochondrial respiratory enzymes (51). However, prohibitins have potential roles as tumor suppressors, as antiproliferative proteins, as regulators of cell cycle progression, in the yeast replicative life span, and in apoptosis (37, 38, 51). Besides the phbA gene (AN6073.2), A. nidulans has another prohibitin homologue, phbB (AN6861.2 [product 51% identical and 72% similar to PhbA; E value of 2e−64]), and it remains to be determined if the proteins encoded by these two genes also interact to form a complex. Our results emphasize a possible role played by phbA in the DNA damage and oxidative stress response.
We have observed that A. nidulans ΔmshA and ΔcshA mutant strains are more sensitive to 4-NQO, MENA, and PARAQ. The mshA product is the homologue of MutS, and several studies on prokaryotic and eukaryotic MutS homologues also demonstrated their involvement in the repair of oxidative DNA damage (4, 9, 17, 18, 19, 49, 54, 72). Interestingly, we observed a very high genetic instability in the ΔmshA mutant strain that was translated by the presence of several sectors when a colony of this strain was exposed to CPT or MENA. The cshA gene is the homologue of CSB, which is involved in the transcription-coupled repair subpathway of nucleotide excision repair, providing the cell with a mechanism to remove transcription-blocking lesions from the transcribed strands of actively transcribed genes (16). Mutations in human CSA and CSB cause Cockayne's syndrome, a rare inherited disorder characterized by UV sensitivity, severe neurological abnormalities, and progeriod symptoms (15). Using a mouse model for CSB, it has been shown that CSB-deficient cells and animals are sensitive to oxidative DNA damage. In contrast to CSB−/− mouse embryonic fibroblasts, CSA−/− mouse embryonic fibroblasts are not hypersensitive to paraquat (16). These differences in the DNA damage response between human CSA and CSB and A. nidulans cshA not only uncover a clear difference in oxidative DNA damage sensitivity between CSA- and CSB-deficient cell lines and mice but also show differences in this system in the two species. Our data confirm that in A. nidulans the CSB homologue (cshA) is also involved in the repair of oxidative DNA damage. A CSA homologue has also been identified in this organism (AN5235.2), but its function has not been investigated yet.
Our results indicate that CPT is able to induce the expression of several genes in different pathways, including DNA repair, which are also involved in DNA damage caused by oxidative stress.
The uvsBATR gene is important for the response to DNA damage induced by CPT and oxidative stress.
It has been shown that ATR is important in responding to the replication-associated DNA damage caused by camptothecin (12, 25). Down-regulation of ATR or Chk1 sensitized cells to 7-ethyl-10-hydroxycamptothecin (SN-38) and camptothecin, but in contrast, down-regulation of ATM and Chk2 had a minimal effect on sensitivity to these drugs (25). Interestingly, besides the known sensitivity to other DNA-damaging agents, such as CPT (24), the uvsBATR deletion mutant was shown to be more sensitive to oxidative stress agents, such as MENA and PARAQ.
We have observed that the mRNA expression of a great number of genes was affected by the absence of uvsBATR. Furthermore, not only is the expression of some of these genes dependent on uvsBATR, but when some of them are inactivated, such as phbA, mshA, tprA, chsA and fhdA, they genetically interact with the uvsBATR deletion mutation. Curiously, all these genetic interactions partially suppressed growth inactivation in the presence of MENA and PARAQ but not DNA-damaging agents. These results suggest that UvsBATR probably monitors DNA damage caused by oxidative stress agents and that PhbA, MshA, TprA, CshA, and FhdA genetically interact with the UvsBATR complex during oxidative stress damage caused by these agents. It has been reported that during DNA damage caused by the DNA-methylating agent N-methyl-N′-nitro-N-nitrosoguanidine, the MSH2 (MutS homologue 2) protein interacts with the ATR kinase to form a signaling module and regulate the phosphorylation of Chk1 and SMC1 (structure maintenance of chromosome 1) (71). The interactions with phbA and chsA inactivation mutants are novel and emphasize the role played by these genes in the DNA damage response mediated by uvsBATR.
In summary, we have provided information about genes which are regulated at the mRNA level in the presence of CPT in A. nidulans and have evaluated the influence of the mutation ΔuvsBATR on the mRNA expression of these genes in the presence of CPT. Our results emphasize the complex network of interactions involved in DNA damage. The response to CPT may be robust in that it involves multiple redundant pathways. By contrast, the responses to BLEO and ROS may not be so robust and can be easily perturbed by a single mutation. In addition, multiple functions contribute to ROS-induced lethality in ATR mutants, because mutations in several functionally unrelated genes can all suppress ΔuvsBATR.
Supplementary Material
Acknowledgments
We thank the Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil, for financial support for our research.
We thank Davidson Custódio Duarte Ribeiro for the bioinformatics analysis and Everaldo dos Reis Marques for constructing the cDNA library. We also thank Y. Itoh and B. R. Oakley for providing the ΔuvsC and ΔnkuA mutant strains, respectively, and the three anonymous reviewers for their suggestions.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped blast and Psi-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ame, J. C., C. Spenlehauer, and G. de Murcia. 2004. The PARP superfamily. Bioessays 26:882-893. [DOI] [PubMed] [Google Scholar]
- 4.Barzilai, A., and K-I. Yamamoto. 2004. DNA damage responses to oxidative stress. DNA Repair 3:1109-1115. [DOI] [PubMed] [Google Scholar]
- 5.Bergen, L. G., and N. R. Morris. 1983. Kinetics of the nuclear division cycle of Aspergillus nidulans. J. Bacteriol. 156:155-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger, J. M. 1998. Structure of DNA topoisomerases. Biochim. Biophys. Acta 1400:3-18. [DOI] [PubMed] [Google Scholar]
- 7.Birrell, G. W., J. A. Brown, H. I. Wu, G. Giaever, A. M. Chu, R. W. Davids, and J. M. Brown. 2002. Transcriptional response of Saccharomyces cerevisiae to DNA-damaging agents does not identify the genes that protect against these agents. Proc. Natl. Acad. Sci. USA 99:8778-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruschi, G. C. M., C. C. de Souza, M. R. Z. K. Fagundes, M. A. C. Dani, M. H. S. Goldman, N. R. Morris, L. Liu, and G. H. Goldman. 2001. Sensitivity to camptothecin in Aspergillus nidulans identifies a novel gene, scaA, related to the cellular DNA damage response. Mol. Genet. Genomics 265:264-275. [DOI] [PubMed] [Google Scholar]
- 9.Chang, C. L., G. Marra, D. P. Chauhan, H. T. Ha, D. K. Chang, L. Ricciardiello, A. Randolph, J. M. Carethers, and R. Boland. 2002. Oxidative stress inactivates the human DNA mismatch repair system. Am. J. Physiol. Cell Physiol. 283:C148-C154. [DOI] [PubMed] [Google Scholar]
- 10.Chaveroche, M. K., J. M. Ghigo and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97-E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, A. Y., C. Yu, M. Potmesil, M. E. Wall, M. C. Wani, and L. F. Liu. 1991. Camptothecin overcomes MDR1-mediated resistance in human KB carcinoma cells. Cancer Res. 51:6039-6044. [PubMed] [Google Scholar]
- 12.Cliby, W. A., K. A. Lewis, K. K. Lilly, and S. H. Kaufmann. 2002. S phase and G2 arrests induced by topoisomerase I poisons are dependent on ATR kinase function. J. Biol. Chem. 277:1599-1606. [DOI] [PubMed] [Google Scholar]
- 13.Daoud, S. S., P. J. Munson, W. Reinhold, L. Young, V. V. Prabhu, Q. Yu, J. LaRose, K. W. Kohn, J. N. Weinstein, and Y. Pommier. 2003. Impact of p53 and knockout and topotecan treatment on gene expression profiles in human colon carcinoma cells: a pharmacogenomic study. Cancer Res. 63:2782-2793. [PubMed] [Google Scholar]
- 14.De Souza, C. P., X. S. Ye, and S. A. Osmani. 1999. Checkpoint defects leading to premature mitosis also cause endoreplication of DNA in Aspergillus nidulans. Mol. Biol. Cell 10:3661-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Waard, H., J. de Wit, T. G. Gorgels, G. Van van den Aardweg, J. O. Andressoo, M. Vemeij, H. Van Steeg, J. H. Hoeimakers, and G. T. J. Van der Horst. 2003. Cell type-specificity hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair 2:13-25. [DOI] [PubMed] [Google Scholar]
- 16.De Waard, H., J de Wit, J.-O. Andressoo, C. T. M. van Oostrom, B. Riis, A. Weimann, H. E. Poulsen, H. Van Steeg, J. H. J. Hoeimakers, and G. T. J. Van der Horst. 2004. Different effects of CSA and CSB deficiency on sensitivity to oxidative DNA damage. Mol. Cell. Biol. 24:7941-7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeWeese, T. L., J. M. Shipman, N. A. Larrier, N. M. Buckley, L. R. Kidd, J. D. Groopman, R. G. Cutler, H. te Riele, and W. G. Nelson. 1998. Mouse embryonic stem cells carrying one or two defective Mhs2 alleles respond abnormally to oxidative stress inflicted by low-level radiation. Proc. Natl. Acad. Sci. USA 95:11915-11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dzierzbicki, P., P. Koprowski, M. U. Fikus, E. Malc, and Z. Ciesla. 2004. Repair of oxidative damage in mitochondrial DNA of Saccharomyces cerevisiae: involvement of the MHS-1 dependent pathway. DNA Repair 3:403-411. [DOI] [PubMed] [Google Scholar]
- 19.Earley, M. C., and G. F. Crouse. 1998. The role of mismatch repair in the prevention of base pair mutations in Saccharomyces cerevisae. Proc. Natl. Acad. Sci. USA 95:15487-15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 21.Ewing, B., and P. Green. 1998. Base calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 22.Fagundes, M. R. Z. K., L. Fernandes, M. Savoldi, S. D. Harris, M. H. S. Goldman, and G. H. Goldman. 2003. Identification of a topoisomerase I mutant, scsA1, as an extragenic suppressor of a mutation in scaANBS1, the apparent homolog of human nibrin in Aspergillus nidulans. Genetics. 164:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fagundes, M. R. Z. K., J. F. Lima, M. Savoldi, I. Malavazi, R. E. Larson, M. H. S. Goldman, and G. H. Goldman. 2004. The Aspergillus nidulans npkA gene encodes a Cdc2-related kinase that genetically interacts with the UvsBATR kinase. Genetics 167:1629-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fagundes M. R., C. P. Semighini, I. Malavazi, M. Savoldi, J. F. de Lima, M. H. S. Goldman, S. D. Harris, and G. H. Goldman. 2005. Aspergillus nidulans uvsBATR and scaANBS1 genes show genetic interactions during recovery from replication stress and DNA damage. Eukaryot. Cell 4:1239-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flatten, K., N. T. Dai, B. T. Vroman, D. Loegering, C. Erlichman, L. M. Karnitz, and S. H. Kaufmann. 2005. The role of checkpoint kinase 1 in sensitivity to topoisomerase I poisons. J. Biol. Chem. 280:14349-14355. [DOI] [PubMed] [Google Scholar]
- 26.Froelich-Ammon, S. J., and N. Osheroff. 1995. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J. Biol. Chem. 270:21429-21432. [DOI] [PubMed] [Google Scholar]
- 27.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae, Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 28.Goldman, G. H., and E. Kafer. 2004. Aspergillus nidulans as a model system to characterize the DNA damage response in eukaryotes. Fungal Genet. Biol. 41:428-442. [DOI] [PubMed] [Google Scholar]
- 29.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 30.Gottlieb, E., and P. M. Tomlinson. 2005. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat. Rev. Cancer 5:857-866. [DOI] [PubMed] [Google Scholar]
- 31.Green, P. 1996. http://bozeman.mbt.washington.edu/phrap.html.
- 32.Guo, X. Q., J. Zhang, X. Fu, Q. Wei, Y. Lu, Y. Li, G. Yin, Y. Mao, Y. Xie, Y. Rui, and K. Ying. 2006. Analysis of common gene expression patterns in four human tumor cell lines exposed to camptothecin using cDNA microarray: identification of topoisomerase-mediated DNA damage response pathways. Front. Biosci. 11:1924-1931. [DOI] [PubMed] [Google Scholar]
- 33.Gupta, M., A. Fujimori, and Y. Pommier. 1995. Eukaryotic DNA topoisomerases I. Biochim. Biophys. Acta 1262:1-14. [DOI] [PubMed] [Google Scholar]
- 34.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9:868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ichioka, D., T. Itoh, and Y. Itoh. 2001. An Aspergillus nidulans uvsC null mutant is deficient in homologous DNA integration. Mol. Gen. Genet. 264:709-715. [DOI] [PubMed] [Google Scholar]
- 36.Jagtap, P., and C. Szabo. 2005. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev. Drug Discov. 4:421-440. [DOI] [PubMed] [Google Scholar]
- 37.Jazwinski, S. M. 2004. Yeast replicative life span—the mitochondrial connection. FEMS Yeast Res. 5:119-125. [DOI] [PubMed] [Google Scholar]
- 38.Joshi, B., D. Ko, D. Ordonez-Ercan, and S. P. Chellappan. 2003. A putative coiled-coil domain of prohibitin is sufficient to repress E2F1-mediated transcription and induce apoptosis. Biochem. Biophys. Res. Commun. 312:459-466. [DOI] [PubMed] [Google Scholar]
- 39.Kafer, E. 1977. Meiotic and mitotic recombination in Aspergilllus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 40.Kim, M. Y., T. Zhang, and W. L. Kraus. 2005. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 19:1951-1967. [DOI] [PubMed] [Google Scholar]
- 41.Koike, K., T. Kawabe, T. Tanaka, S. Toh, T. Uchiumi, M. Wada, S. Akiyama, M. Ono, and M. Kuwano. 1997. A canalicular multispecific organic anion transporter (cMOAT) antisense cDNA enhances drug sensitivity in human hepatic cancer cells. Cancer Res. 57:5475-5479. [PubMed] [Google Scholar]
- 42.Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushihara, and Y. Tanaka. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu, L. F. 1989. DNA topoisomerase poisons as antitumor drugs. Annu. Rev. Biochem. 58:351-375. [DOI] [PubMed] [Google Scholar]
- 44.Liu, L. F. 1994. DNA topoisomerases. Topoisomerase-targeting drugs. Advances in pharmacology, vol. 29B. Academic Press, San Diego, Calif.
- 45.Liu, C., J. J. Pouliot, and H. A. Nash. 2002. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. USA 99:14970-14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malavazi, I., C. P. Semighini, M. R. Zeska Kress, S. D. Harris, and G. H. Goldman. 2006. Regulation of hyphal morphogenesis and the DNA damage response by the Aspergillus nidulans ATM homologue, AtmA. Genetics 173:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marques, E. R., M. E. S. Ferreira, R. D. Drummond, J. M. Felix, M. Menossi, M. Savoldi, L. R. Travassos, R. Puccia, W. L. Batista, K. C. Carvalho, M. H. S. Goldman, and G. H. Goldman. 2004. Identification of genes preferentially expressed in the pathogenic yeast phase of Paracoccidioides brasiliensis using suppression subtraction hybridization and macroarray differential analysis. Mol. Gen. Genomics 271:667-677. [DOI] [PubMed] [Google Scholar]
- 48.Marra, M. A., T. A. Kucaba, L. W. Hillier, and R. H. Waterston. 1999. High throughput plasmid DNA purification for 3 cents per sample. Nucleic Acids Res. 27:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazurek, A., M. Berardini, and R. Fishel. 2002. Activation of human MutS homologs by 8-oxo-guanine DNA damage. J. Biol. Chem. 277:8260-8266. [DOI] [PubMed] [Google Scholar]
- 50.Minderman, H., J. M. Conroy, K. L. O'Loughlin, D. McQuaid, P. Quinn, S. Li, L. Pendyala, N. J. Nowak, and M. R. Baer. 2005. In vitro and in vivo irinotecan-induced changes in expression profiles of cell cycle and apoptosis-associated genes in acute myeloid leukemia cells. Mol. Cancer Ther. 4:885-900. [DOI] [PubMed] [Google Scholar]
- 51.Mishra, S., L. C. Murphy, B. L. Gregoire Nyomba, and L. J. Murphy. 2005. Prohibitin: a potential target for new therapeutics. Trends Mol. Med. 11:192-197. [DOI] [PubMed] [Google Scholar]
- 52.Morandi, E., C. Zingaretti, D. Chiozzotto, C. Severini, A. Semeria, W. Horn, M. Vaccari, R. Serra, P. Silingardi, and A. Colacci. 2006. A cDNA-microarray analysis of camptothecin resistance in glioblastoma cell lines. Cancer Lett. 231:74-86. [DOI] [PubMed] [Google Scholar]
- 53.Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil, S. L. Murray, M. J. Hynes, S. A. Osmani, and B. R. Oakley. 2006. A versatile and efficient gene targeting system for Aspergillus nidulans. Genetics 172:1557-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ni, T. T., G. T. Marsischky, and R. D. Kolodner. 1999. MSH2 and MSH6 are required for removal of adenine misincorporated opposite to 8-oxo-guanine in S. cerevisiae. Mol. Cell 4:439-444. [DOI] [PubMed] [Google Scholar]
- 55.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 56.Osborn, A. J., S. J. Elledge, and L. Zou. 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12:509-516. [DOI] [PubMed] [Google Scholar]
- 57.Osmani, S. A., G. S. May, and N. R. Morris. 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 104:1495-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osmani, S. A., and P. M. Mirabito. 2004. The early impact of genetics on our understanding of cell cycle regulation in Aspergillus nidulans. Fungal Genet. Biol. 41:401-410. [DOI] [PubMed] [Google Scholar]
- 59.Qin, J., and L. Li. 2003. Molecular anatomy of the DNA damage and replication checkpoints. Radiat. Res. 159:139-148. [DOI] [PubMed] [Google Scholar]
- 60.Rasheed, Z. A., and E. H. Rubin. 2003. Mechanisms of resistance to topoisomerase I-targeting drugs. Oncogene 22:7296-7304. [DOI] [PubMed] [Google Scholar]
- 61.Reid, R. J., E. A. Kauh, and M. A. Bjornsti. 1997. Camptothecin sensitivity is mediated by the pleiotropic drug resistance network in yeast. J. Biol. Chem. 272:12091-12099. [DOI] [PubMed] [Google Scholar]
- 62.Rotman, G., and Y. Shiloh. 1998. ATM: from gene to function. Hum. Mol. Genet. 7:1555-1563. [DOI] [PubMed] [Google Scholar]
- 63.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 64.Schummer, M., V. L. V. Ng, R. E. Baumgarner, P. S. Nelson, B. Schummer, D. W. Bednarski, L. Hassell, R. L. Baldwin, B. Y. Karlan, and L. Hood. 1999. Comparative hybridization of an array of 21 500 ovarian cDNAS for the discovery of genes overexpressed in ovarian carcinomas. Gene 238:375-385. [DOI] [PubMed] [Google Scholar]
- 65.Semighini, C. P., M. Marins, M. H. S. Goldman, and G. H. Goldman. 2002. Quantitative analysis of the relative transcript levels of ABC transporter Atr genes in Aspergillus nidulans by real-time reverse transcripition-PCR assay. Appl. Environ. Microbiol. 68:1351-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Semighini, C. P., M. R. Z. K. Fagundes, J. C. Ferreira, R. C. Pascon, M. H. S. Goldman, and G. H. Goldman. 2003. Different roles of the Mre11 complex in the DNA damage response in Aspergillus nidulans. Mol. Microbiol. 48:1693-1709. [DOI] [PubMed] [Google Scholar]
- 67.Semighini, C. P., M. Savoldi, G. H. Goldman, and S. D. Harris. 2006. Functional characterization of the putative Aspergillus nidulans poly(ADP-ribose) polymerase homologue, PrpA. Genetics 173:87-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shiloh, Y. 1997. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu. Rev. Genet. 31:635-662. [DOI] [PubMed] [Google Scholar]
- 69.Vance, J. R., and T. E. Wilson. 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA 99:13669-13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]
- 71.Wang, Y., and J. Qin. 2003. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl. Acad. Sci. USA 100:15387-15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, G., P. Alamuri, M. Zafri Humayun, D. E. Taylor, and R. J. Maler. 2005. The Helicobacter pylori MutS protein confers protection from oxidative DNA damage. Mol. Microbiol. 58:166-176. [DOI] [PubMed] [Google Scholar]
- 73.White, O., and A. R. Kerlavage. 1996. TDB: new databases for biological discovery. Methods Enzymol. 266:27-40. [DOI] [PubMed] [Google Scholar]
- 74.Yang, J., Y. Yu, H. E. Hamrick, and P. J. Duerksen-Hugues. 2003. ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinogen 24:1571-1580. [DOI] [PubMed] [Google Scholar]
- 75.Zhang, Y.-M., J.-K. Liu, and T.-Y. Wong. 2003. The DNA excision repair system of the highly radioresistant bacterium Deinococcus radiodurans is facilitated by the pentose phosphate pathway. Mol. Microbiol. 48:1317-1323. [DOI] [PubMed] [Google Scholar]
- 76.Zhou, B. B., and S. J. Elledge. 2002. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]
- 77.Zhou, Y., F. G. Gwadry, W. C. Reinhold, L. D. Miller, L. H. Smith, U. Scherf, E. T. Liu, K. W. Kohn, Y. Pommier, and J. N. Weinstein. 2002. Transcriptional regulation of mitotic genes by camptothecin-induced DNA damage: microarray analysis of dose- and time-dependent effects. Cancer Res. 62:1688-1695. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.