Abstract
The eukaryotic genome is packaged together with histone proteins into chromatin following DNA replication. Recent studies have shown that histones can also be assembled into chromatin independently of DNA replication and that this dynamic exchange of histones may be biased toward sites undergoing transcription. Here we show that epitope-tagged histone H4 can be incorporated into nucleosomes throughout the budding yeast (Saccharomyces cerevisiae) genome regardless of the phase of the cell cycle, the transcriptional status, or silencing of the region. Direct comparisons reveal that the amount of histone incorporation that occurs in G1-arrested cells is similar to that occurring in cells undergoing DNA replication. Additionally, we show that this histone incorporation is not dependent on the histone H3/H4 chaperones CAF-1, Asf1, and Hir1 individually. This study demonstrates that DNA replication and transcription are not necessary prerequisites for histone exchange in budding yeast, indicating that chromatin is more dynamic than previously thought.
The nucleosome is the basic repeating unit of chromatin and comprises 147 bp of DNA wrapped around two molecules each of histones H2A, H2B, H3, and H4 (23). Chromatin is assembled following DNA replication, when two new daughter DNA strands are created (26, 41). However, it is becoming increasingly apparent that chromatin can be assembled at times other than during DNA replication; this is termed replication-independent chromatin assembly. The clearest evidence for replication-independent chromatin assembly is provided by the observation that the Drosophila histone variant H3.3 is incorporated into sites undergoing transcription (3, 27, 36, 43). As yeast (Saccharomyces cerevisiae) lacks H3.1, the histone variant assembled during DNA replication in higher eukaryotes, the sole copy of noncentromeric H3 is the equivalent of H3.3. Accordingly, it has been shown that histones are disassembled from the promoters and open reading frames (ORFs) of highly transcribed yeast genes during transcription and that they rapidly reassemble following transcriptional repression (1, 2, 5, 31, 34, 45).
Chromatin disassembly and assembly are mediated by histone chaperones. The histone H3/H4 chaperones antisilencing function 1 (Asf1) and histone regulatory 1 (Hir1) mediate replication-independent chromatin assembly in vitro (9, 39). Similarly, Asf1 and chromatin assembly factor 1 (CAF-1) work together to mediate DNA synthesis-coupled chromatin assembly in vitro (8, 42) and in vivo in higher eukaryotes (29, 33).
Although it is clear that histones are disassembled and reassembled within transcription units in a replication-independent manner, whether replication-independent histone exchange occurs in the remainder of the genome is unclear. In this study, we show that an inducible epitope-tagged histone H4 can be readily incorporated into nucleosomes broadly throughout the budding yeast genome, irrespective of cell cycle phase, transcriptional state, or chromatin silencing. Furthermore, the histone chaperones Asf1, CAF-1, and Hir1 do not make major individual contributions to either replication-independent or replication-dependent histone exchange in yeast. This work indicates that chromatin is much more dynamic and fluid than previously assumed.
MATERIALS AND METHODS
Yeast strains, media, and plasmids.
All strains used in this study are haploid derivatives of W303-1a (40): JKT0010, MATa his3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 (30); NWY0008, MATa his3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 cac2::HPH (21); JLY091, MATa his3-11 leu2-3,112 lys2 trp ura3-1 bar1::LEU2 hir1::KAN (this study); and JKT0018, MATa his3-11 leu2-3,112 lys2 trp1-1 ura3-1 bar1::LEU2 asf1::HIS5 (this study). Deletion mutants were created by one-step PCR-mediated integration, as described previously (22). Yeast cells were grown in synthetic complete medium lacking uracil and supplemented with 2% raffinose, galactose, or glucose. G1 arrest was achieved by adding α-factor directly to the growth medium to 150 ng/ml for 3 to 4 h and monitoring arrest microscopically. For G2/M arrest, α-factor-arrested cells were washed three times, and then pronase (1 μg/ml, final concentration) and nocodazole (15 μg/ml) were added to the medium. The 3HA tag (three repeats of the influenza virus hemagglutinin epitope) was incorporated onto HHF2 to make plasmid pJL20 by cotransforming yeast with the CEN-based plasmid pRM102 (24), and the 3HA-Kan PCR product was amplified from pFA6a-3HA-kanMX6 using primers with the following sequences (5′ to 3′): TTTGAAGAGACAAGGTAGAACCTTATATGGTTTCGGTGGTCGGATCCCCGGGTTAATTAA and GATAAAGAAGACAGTCATAAGTGCGGCGACGATAGTCATGGAATTCGAGCTCGTTTAAAC.
Western blot analysis.
Primary antibody incubations were performed overnight in Tris-buffered saline-Tween with 3% dry milk, anti-HA (1:1,000) (Covance), anti-H4 (1:500) (Abcam), and anti-NuP1 (1:500) (Covance). Enhanced chemiluminescence of horseradish peroxidase-conjugated secondary antibodies was detected by film autoradiography. The images shown are in the linear range of detection for the film exposures.
Cell cycle analysis.
For cell cycle analysis, ∼5 × 106 cells for each sample were stained with propidium iodide as described previously (38). Ten thousand cells from each sample were scanned with a Beckman Coulter XL-MCL machine.
Chromatin immunoprecipitation analysis.
Cells (1 × 108) were treated with 1% formaldehyde (final concentration) for 20 min at room temperature. Cross-linking was quenched by addition of glycine to a final concentration of 125 mM. Cells were centrifuged and washed twice in ice-cold Tris-buffered saline. Cells were resuspended in 400 μl lysis buffer (0.1% deoxycholate, 1 mM EDTA, 50 mM HEPES, pH 7.5, 140 mM NaCl, 1% Triton X-100), an equal volume of 0.5-mm glass beads was added, and the cells were vortexed for 10 min at 4°C. Cell lysate was removed from the beads and placed into a new tube. Chromatin was sheared with a Branson 350 Sonifier to an average size of 500 bp. Immunoprecipitations were performed as described previously (18) using a mouse antibody to the HA epitope (Covance) or using an antibody to the C terminus of H3 (Abcam 1791). The linear range of template for multiplex PCR was determined empirically for every primer pair and for every template in every experiment, and PCR-amplified products were quantitatively measured using Labworks (UVP Inc.). PCR products were resolved on a 3% agarose gel and visualized with ethidium bromide. Primer sequences are available upon request. Where quantification is shown, the ratio of immunoprecipitation signal to input signal was plotted on the y axis. All results obtained in the semiquantitative chromatin immunoprecipitation (ChIP) analysis were reproducible in multiple independent experiments.
Mononucleosome purification.
Yeast (JKT0010) transformed with pJL20 was grown overnight in medium lacking uracil and supplemented with 2% raffinose and galactose to an optical density at 600 nm of 1. Nuclei were isolated as described previously (10). Aliquots containing nuclei from 100 mg (wet weight) of the original cells were distributed, and three such aliquots were used for the mononucleosome purification. Three aliquots were combined, washed, and resuspended in 400 μl digestion buffer (15 mM Tris, pH 7.9, 50 mM NaCl, 1.4 mM CaCl2, 0.2 mM EDTA, 0.2 mM EGTA, 5 mM β-mercaptoethanol). Micrococcal nuclease (Sigma) was added to a concentration of 2 U/ml, and samples were incubated for 20 min at 37°C. EDTA was added to a concentration of 10 mM to stop the reaction, and samples were incubated on ice for 30 min. Reaction mixtures were spun for 5 min at 16,000 × g to remove insoluble debris. Samples were layered on top of a 5 to 30% linear sucrose gradient (10 mM Tris, pH 7.5, 1 mM EDTA, 0.5 M NaCl, 0.3 mM phenylmethylsulfonyl fluoride, 5 or 30% sucrose) and spun at 26,000 × g for 16 h in a Beckman SW-41 ultracentrifugal rotor. Fractions (1 ml) were pulled from the top down. Three hundred microliters of each fraction was used for DNA analysis, where samples were RNase treated for 1 h and then phenol-chloroform extracted. The aqueous phase was then ethyl alcohol precipitated and resuspended in 1× Tris-EDTA. Half of each sample was run on a 2% agarose gel and stained with SYBR gold. The remaining volume of each fraction was trichloroacetic acid precipitated, washed with acetone, and then resuspended in 30 μl sodium dodecyl sulfate loading buffer; 10 μl of each fraction was run on 15% polyacrylamide gels, transferred, and subjected to Western blot analysis.
RESULTS
Replication-independent chromatin assembly of H4HA.
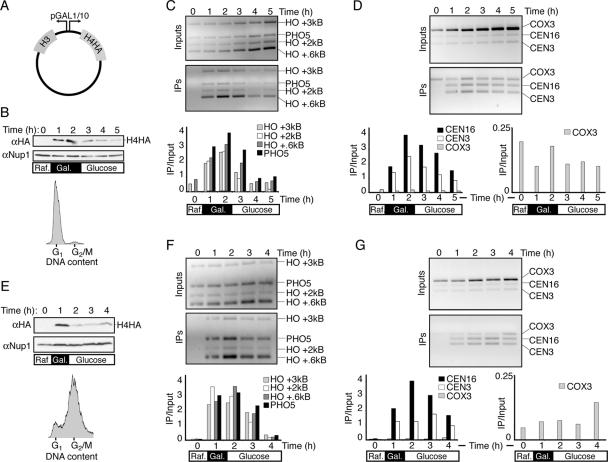
We initially sought to determine whether histones were preferentially incorporated around a double-strand DNA break. To achieve this, we induced a unique double-strand break at the MAT locus by expressing the HO endonuclease under the control of the GAL1 promoter in Saccharomyces cerevisiae. Our yeast additionally harbored a low-copy-number plasmid carrying the HHT2 and HHF2 genes expressing histones H3 and H4 C-terminally tagged with three repeats of the influenza virus hemagglutinin epitope (3HA) (22) under the control of the divergent GAL1/10 promoter (Fig. 1A). As histones were thought to be predominantly assembled into chromatin during DNA replication, we arrested cells in G1 phase via the addition of α-factor to reduce background due to histone incorporation during DNA replication. Upon G1 arrest, galactose was added for 2 h to induce expression of the HO endonuclease and H3/H4HA. Glucose was then added for an additional 3 h to repress the GAL1/10 promoters, as seen by the reduced amount of H4HA in the cells (Fig. 1B). Using ChIP analysis directed against the 3HA epitopes on H4, we sought to determine whether histones were preferentially incorporated around the DNA break site. Surprisingly, and providing the basis for this study, we found that following induction of H4HA and H3, H4HA was incorporated into chromatin in G1-phase cells to similar extents in all of the regions that we examined, including 0.6 kb, 2 kb, and 3 kb from the HO site, the repressed PHO5 ORF, and centromeres 3 and 16 (Fig. 1C and D). It was also apparent that H4HA is removed from DNA at a rate similar to that of its incorporation into chromatin following its transcriptional repression (Fig. 1C and D). As such, the DNA that is immunoprecipitated by the HA antisera is clearly specific to the HA epitope on H4. Importantly, we saw no increase in immunoprecipitation (when normalized to the input signal) of a mitochondrial DNA region (COX3) upon the induction of histones (Fig. 1D), indicating that H4HA incorporation is specific to nuclear DNA. These results indicate that extensive replication-independent histone exchange can occur in budding yeast.
FIG. 1.
Histones expressed outside of S phase are globally incorporated into chromatin. (A) Schematic of pJL20 plasmid used in this study, where copies of H3 and H4 tagged with 3HA epitopes are driven by the divergent GAL1/10 promoters. (B to D) Analysis of H4HA incorporation in G1-arrested cells of strain JKT0010. All samples in this figure were taken from the same experimental time course. (B, top) Western blot analysis of H4HA expression and Nup1 loading control in G1-arrested cells growing in raffinose (Raf.), followed by the addition of galactose (Gal.) for 2 h and then addition of glucose for a further 2 h, as indicated by the schematic below the gel. (Bottom) Flow cytometry analysis of DNA content at T = 0, ensuring that cells were arrested in G1 prior to the start of the experiment. Additional flow cytometry analysis shows that this arrest was maintained throughout the time course (not shown). (C) Gels and quantification of ChIP analysis using primers near the HO recognition sequence or the PHO5 open reading frame as a control. (D) Gels and quantification of anti-HA ChIP analysis using primers that recognize the core of centromeres 3 and 16 and the mitochondrial COX3 open reading frame as a control. Quantification of the COX3 PCR product is additionally shown on a separate graph with a smaller scale. (E to G) Same as B to D, respectively, except that following G1 arrest, cells were released into the cell cycle and rearrested in G2/M using nocodazole. (E, bottom) Flow cytometry analysis of DNA content is shown at T = 0, ensuring that cells were arrested in G2/M prior to the start of the experiment. Additional flow cytometry analysis showed that this arrest was maintained throughout the time course (not shown).
To determine whether replication-independent histone exchange is a G1-phase-specific phenomenon, we examined cells arrested in G2/M phase of the cell cycle. Upon G2/M arrest using nocodazole (Fig. 1E), cells were subjected to the same analyses as described above (Fig. 1B to D). The results were essentially the same for cells arrested in G1 and G2/M (Fig. 1F and G); the tagged H4 was incorporated efficiently at all regions tested in a replication-independent fashion.
H4HA is incorporated into mononucleosomes, and the distribution of incorporation is not affected by its expression level.
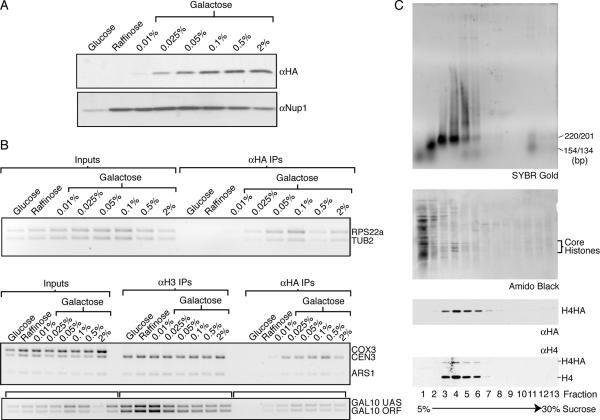
To investigate whether the high level of H4HA expression leads to global replication-independent histone exchange, we lowered the galactose concentrations to reduce H4HA expression (Fig. 2A). We specifically examined incorporation of H4HA into chromatin during G1 phase at regions that are known to have reduced nucleosome density (RPS22a, ARS1, and GAL10). We reasoned that if histones were being forced onto DNA by histone overexpression, then these regions would be more susceptible to forced histone incorporation. The ChIP analyses showed that while altering histone expression changes the absolute level of histone incorporation, it does not grossly affect the relative genomic distribution of replication-independent incorporation (Fig. 2B). Note that the apparent reduction of H4HA incorporation at high galactose concentrations was not observed in independent experiments and most likely reflects an individual experimental variation.
FIG. 2.
Altering the expression of H4HA does not change the relative distribution of its incorporation, and it is incorporated as a nucleosomal component. Strain JKT0010 was grown in medium lacking uracil with various carbon sources to alter the expression of H4HA. (A) Western blot analysis of H4HA using an anti-HA antibody (Covance) and the loading control anti-Nup1 (Covance). (B) A 3% agarose gel stained with ethidium bromide of ChIP analysis using primers that recognize the regions listed on the side. The RPS22a and TUB2 primers amplify a region within the ORFs. UAS, upstream activation sequence. (C) H4HA-containing mononucleosome purification. Top to bottom: SYBR gold-stained 2% agarose gel of fractions recovered from a sucrose gradient, an amido black-stained nitrocellulose membrane of fractions, an anti-HA Western blot showing H4HA, and an anti-H4 Western blot showing both endogenous H4 and H4HA.
To investigate whether H4HA was incorporated into nucleosomes, as opposed to simply coating the DNA in a nonnucleosomal manner, we purified mononucleosomes from yeast expressing H4HA. Mononucleosomes were purified via sucrose gradient sedimentation (Fig. 2C) and migrated in fractions 3 to 6, as indicated by the DNA analysis (Fig. 2C). This corresponds exactly with the fractions containing H4HA protein, as examined by Western blotting with an HA antibody and a C-terminal H4 antibody (Fig. 2C). In accordance with the smaller amount of H4HA than H4 in the nucleosomes (Fig. 2C), comparison of total protein levels in whole-cell extracts revealed that H4HA was less abundant than H4 (data not shown). Taken together, these results indicate that the epitope-tagged histone is incorporated into chromatin as a nucleosomal component.
Deletion of known H3/H4 histone chaperones has no obvious effect on histone incorporation.
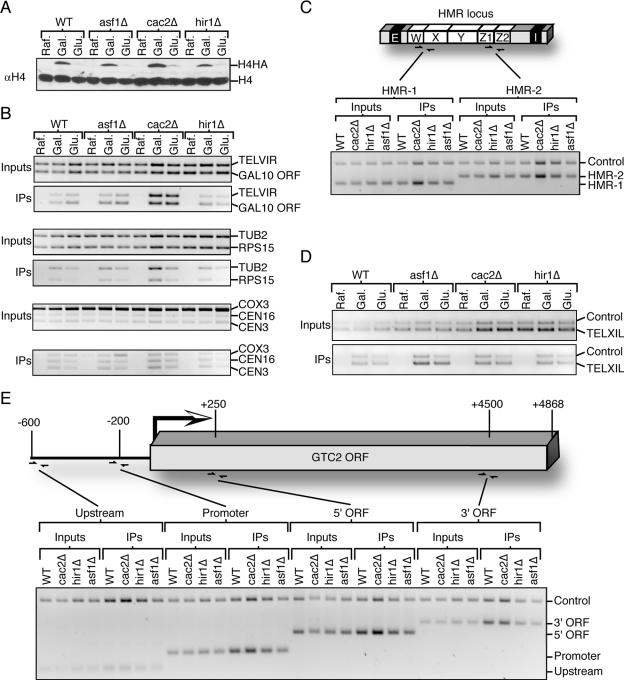
After determining that H4HA was incorporated into nucleosomes throughout the genome in a replication-independent manner, we investigated whether known chaperones of H3/H4 were required for this process. We examined the ability of yeast deleted for ASF1, HIR1, and the CAC2 component of the CAF-1 complex to incorporate H4HA into chromatin in G1-phase-arrested cells. Cells were grown overnight in raffinose-containing media, at which time galactose was added for 1 h to induce transcription of H4HA, and then glucose was added for an additional hour to repress H4HA expression. At each time point, samples for Western blotting (Fig. 3A), ChIP (Fig. 3B to E), and flow cytometry analyses (data not shown) were taken. Remarkably, no general or region-specific differences in H4HA incorporation were seen between the mutant and wild-type yeast at any region examined, including a telomere-proximal region, multiple centromeres, and various transcribed genes (Fig. 3B). Given that increased H4HA expression leads to increased incorporation into chromatin (Fig. 2), the apparent increase in incorporation of H4HA into chromatin in the cac2Δ strain is likely due to the higher expression level of H4HA that occurred in this strain (Fig. 3). We also examined whether histone exchange was less prevalent at transcriptionally silenced regions and whether the histone chaperones influence this histone exchange. Compared to an intergenic (“control”) region, histones were just as efficiently incorporated into the HMR mating type locus (Fig. 3C) and a region proximal to the left telomere of chromosome eleven (Fig. 3D). Inactivation of CAF-1, Hir1, or Asf1 did not noticeably reduce the extent of histone exchange (Fig. 3C and D). These results indicate that the repressive chromatin state of the silent regions did not reduce replication-independent histone exchange and further that the individual disruption of Asf1, CAF-1, and Hir1 had no effect on replication-independent histone exchange in the regions that we examined. Note that the 1 h of glucose repression of H4HA in these ChIP experiments was sufficient to dramatically reduce the total H4HA levels (Fig. 3A). However, this was not enough time to detectably decrease the ChIP signals (Fig. 3B and D), preventing us from drawing conclusions about the ability of H4HA to be removed from the chromatin in these particular experiments.
FIG. 3.
Histones are incorporated in G1 in the absence of the histone chaperones Asf1, CAF-1, and Hir1. (A) Anti-H4 Western blot showing relative expression of H4 and H4HA in strains JKT0010 (WT), JKT0018 (asf1Δ), NWY008 (cac2Δ), and JLY091 (hir1Δ). The “Raf.” samples were taken following G1 arrest in medium containing raffinose, the “Gal.” samples were taken 1 h after addition of galactose, and the “Glu.” samples were taken 1 h after addition of glucose to a final concentration of 2%. Samples from the same time course were used for all of the analyses in this figure. (B) A 2% agarose gel stained with ethidium bromide of anti-HA ChIP analysis using primers that recognize the regions listed on the side. The TELVIR region is internal to the right telomere of chromosome six, while RPS15 and TUB2 are within the ORFs. (C) At the top is shown a schematic of the HMR mating type locus to show the location of HMR primers. Below is shown the gel of the anti-HA ChIP analysis using primers that recognize the regions listed on the side. All samples were taken from G1-arrested cells following 1 h in galactose. The intergenic “Control” primer pair recognizes a gene-free region on chromosome three. (D) Anti-HA ChIP analysis using primers that recognize the regions to the left side of telomere 11 (TELXIL) and the same intergenic control region used in panel C. (E) Schematic of the GTC2 gene to show the location of primers and gel of anti-HA ChIP analysis using primers that recognize the regions listed on the side, where the intergenic control region is the same as that used in panel C. All samples were taken from G1-arrested cells following 1 h in galactose.
Given the recent observations that histones may be removed and subsequently replaced during transcription, we sought to determine whether H4HA would be preferentially incorporated into open reading frames undergoing transcription. When we analyzed H4HA incorporation across the constitutively transcribed GTC2 gene, we found no detectable increase in H4HA exchange into the open reading frame or promoter region compared to an intergenic control region in either the wild-type or mutant histone chaperone strains (Fig. 3E). Inactivation of CAF-1, Hir1, or Asf1 did not noticeably reduce the extent of this replication-independent histone exchange (Fig. 3C and D). Consistent with this result, we did not observe enhanced H4HA incorporation into any of the actively transcribed ORFs that we analyzed, including TUB2, RPS15, RPS22a, and GAL10 (Fig. 2 and 3). As such, our data indicate that replication-independent H4 histone exchange occurs throughout the genome, irrespective of transcriptional silencing or expression, in a manner that is not fully dependent on any one of the Asf1, Hir1, or CAF-1 H3/H4 histone chaperones.
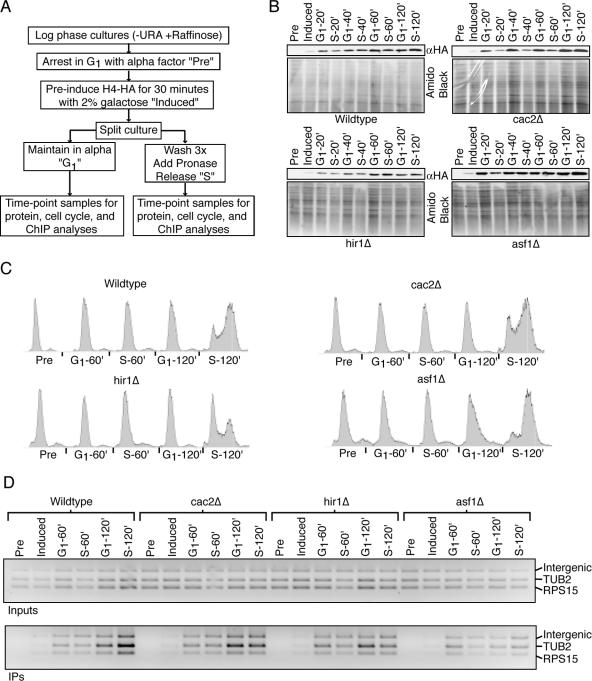
In order to assess the extent of replication-independent histone exchange, we directly compared it to replication-dependent histone exchange. To achieve this, cultures from the wild type or histone chaperone mutants were arrested in G1 phase with α-factor. Upon arrest, galactose was added to preinduce H4HA, followed by splitting of the cultures into “G1” and “S” cultures, where the G1 culture had the α-factor arrest maintained and the S culture was released into the cell cycle (Fig. 4A). Samples were taken over the subsequent 2 h for cell cycle, protein, and ChIP analyses (Fig. 4B to D). In most cases, and especially at the later time points, the anti-HA Western blots showed comparable levels of H4HA being expressed in the G1 and S cultures at equivalent time points. This is important given the dependence of histone incorporation levels on protein expression levels (Fig. 2A and B and 3). For example, the minor difference we do see in H4HA incorporation by ChIP in the wild-type G1-arrested sample at 120 min versus the wild-type S-phase sample at 120 min directly correlates with H4HA expression. Cell cycle analysis revealed that in all cases the S cultures didn't release from G1 arrest until some time between the 60- and 120-min time points. Strikingly, we observed no measurable difference in H4HA incorporation at any genomic region tested when comparing G1-phase and S-phase cultures in either the wild type or the cac2Δ, hir1Δ, or asf1Δ strain (Fig. 4D and data not shown). These results demonstrate that the level of replication-independent histone exchange that can occur in budding yeast is comparable to that of replication-dependent histone exchange. Furthermore, these results suggest that the absence of the individual histone chaperone CAF-1, Hir1, or Asf1 has no drastic effect on the ability of histones to be incorporated during DNA replication in yeast.
FIG. 4.
Histone H4HA is incorporated in cycling or arrested cells, regardless of the presence of the histone chaperone Hir1, CAF-1, or Asf1. (A) Experimental schematic where the sample labels used in panels B to D are described. URA, uracil. (B) The strains described in the legend to Fig. 3 were used for this experiment. Anti-HA Western blots show H4HA expression levels, and an amido black-stained membrane was used to ascertain equal loading. (C) Cell cycle analysis by flow cytometry of propidium iodide-stained yeast. (D) A 2% agarose gel stained with ethidium bromide of anti-HA ChIP analysis using primers that recognize the regions listed on the side, as described in the legend to Fig. 3.
DISCUSSION
This study demonstrates that replication-independent H4 histone exchange can occur efficiently and extensively across the budding yeast genome, regardless of transcriptional activity or chromatin silencing of the DNA region. Furthermore, this work indicates that the individual yeast H3/H4 chaperones Asf1, CAF-1, and Hir1 do not play an essential role in histone incorporation.
We focused on exchange of the H3/H4 histones due to their central position in the nucleosome. Incorporation or removal of H3/H4 from chromatin necessitates the complete disassembly and reassembly of nucleosomes. Although extensive replication-independent exchange of the peripheral H2A/H2B histones is known to occur (11, 15), it was widely believed that histones H3/H4 do not generally undergo replication-independent histone exchange, except during progression of the RNA polymerases (6, 16, 34, 35). Furthermore, it was not clear whether the old histones are replaced following RNA polymerase passage or whether new histones could be incorporated into the chromatin. Our study indicates that replication-independent H3/H4 exchange is extensive in budding yeast and that new histones can be incorporated in trans behind RNA polymerases.
There are many lines of evidence to indicate that overexpression of H3 and H4 does not just force histones inappropriately onto the genome. First, it has previously been shown that the overexpression of histones H3/H4 does not lead to an increase in nucleosome density on the endogenous yeast 2μm plasmid (20). Second, exchange of H4HA “out” of the chromatin occurred at a rate similar to that of exchange “into” the chromatin (Fig. 1). Essentially, since histone H4HA incorporation presumably occurs through histone exchange, the disappearance of immunoprecipitated H4HA-bound DNA that is seen upon repression of H4HA transcription would be due to its replacement by native H4, which is not overabundantly present. Third, greatly reducing the amounts of H4HA expression did not increase the specificity of histone incorporation at transcribed regions versus intergenic regions, for example (Fig. 2). Fourth, we did not observe increased histone H4HA incorporation into known nucleosome-depleted regions of the nuclear genome (RPS22a, ARS1, and transcriptionally active GAL10) (Fig. 2B). Fifth, incorporation of excess H4HA would be accompanied by an increase in total levels of H3 on the genome, and this was not observed (Fig. 2B), although the difference in H3 incorporation might fall below our threshold of detection. Finally, H4HA was seen to be incorporated into mononucleosomes isolated from yeast (Fig. 2C). As such, we are confident that the histone exchange that we saw is physiologically relevant.
Our findings of global replication-independent histone H4 exchange into budding yeast chromatin are consistent with a recent study performed with fission yeast (7). However, there are some key differences between that study and ours. First of all, the fission yeast study expressed only histone H3 (not H4). It is important to express H3 and H4 together in budding yeast, as it has been shown that expression of only one of the histones has a more detrimental effect on the cell than overexpressing pairs of histones (25). Histones H3 and H4 always exist as heterodimers in the cell, making it difficult to understand how H3 expressed on its own would be incorporated into chromatin. The study in fission yeast also observed reduced replication-independent histone H3 incorporation in telomeric regions (7). This was not the case in budding yeast (Fig. 3D), indicating a potential intrinsic difference in the heterochromatin structures in these two yeast species. Indeed, unlike budding yeast, fission yeast has a homologue of heterochromatin protein 1 (Swi6) that may limit histone exchange into these regions. Both the Schizosaccharomyces pombe study and the current study found global histone incorporation that appears to be independent of ongoing transcription. It has recently been estimated that over 10% of all intergenic regions in budding yeast are likely to be transcribed by RNA polymerase II (44). Although it is possible that this intergenic transcription may account for some of the histone H4 exchange that we observed throughout the genome, it is unlikely to account for the histone H4 exchange in all of the intergenic regions we examined.
Our data indicate that replication-independent chromatin assembly can be as extensive as that which occurs during DNA replication. This is surprising, in that it is widely assumed that the majority of histone H3/H4 incorporation happens during DNA replication. For example, density labeling analyses with tissue culture cells indicated that the vast majority of H3/H4 assembly occurs during DNA replication, with a small amount being dependent on transcription and an even smaller amount dependent on neither transcription nor DNA replication (14). It is possible that the extensive and high level of replication-independent incorporation of histones H3/H4 in yeast, compared to the limited histone exchange observed in tissue culture cells, is a reflection of yeast having only the H3.3 variant of histone H3, which readily assembles into chromatin in a replication-independent manner.
This work suggests that the histone chaperones Asf1, CAF-1, and Hir1 do not individually make extensive contributions to chromatin assembly in yeast. This is consistent with the relatively minor growth defects seen in strains lacking the individual histone chaperones (17, 19, 37). Furthermore, there is only about a 10% underassembly of the yeast genome in yeast lacking CAF-1, while yeast lacking Asf1 have about a 10% overassembly of their genome (1). Additionally, the contribution of Asf1 to histone exchange within the ORFs of highly transcribed genes is relatively minor and was detected by quantitative real-time PCR analysis (35). These subtle differences in chromatin assembly and disassembly have not been detected by the current study and may reflect the differences in our experimental approach. The lack of absolute requirement for the individual histone chaperones Asf1, Hir1, and CAF-1 in histone exchange may suggest that there is some redundancy among these factors. In the future, it will be interesting to determine if the combined deletion of multiple H3/H4 chaperones leads to defects in histone exchange that are undetectable in our hands in single-mutant yeast. Consistent with this idea, yeast strains with both CAF-1 and Asf1 inactivated have greatly enhanced growth defects (42). It remains to be determined whether other known or unknown H3/H4 histone chaperones, such as Rtt106 (13), Hif1 (4), or Spt6 (6), also contribute to the histone H3/H4 exchange that we observed in yeast. Alternatively, budding yeast may be able to manage some degree of self-assembly of the chromatin in the absence of these H3/H4 histone chaperones. Nevertheless, there are clear differences between the essential role of the Asf1, Hir1, and CAF-1 histone chaperones for chromatin assembly in higher eukaryotes (12, 28, 29, 32, 33) and their dispensability in budding yeast.
In summary, yeast is capable of extensive histone H3/H4 exchange independent of DNA replication, without regard to genomic position or transcriptional activity of the underlying DNA and with no obvious individual requirement for the histone chaperones Asf1, Hir1, and CAF-1.
Acknowledgments
We thank Melissa Adkins, Christine English, Jason Feser, and Christopher Ramey for critical reading of the manuscript. We are very grateful to Melissa Adkins for advice and discussions, to Michael Grunstein for the pRM102 plasmid, and to the UCCC flow cytometry core facility.
This work was supported by NIH grant GM064475 to J.K.T. J.K.T. is a Leukemia and Lymphoma Scholar.
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Adkins, M. W., and J. K. Tyler. 2004. The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 279:52069-52074. [DOI] [PubMed] [Google Scholar]
- 2.Adkins, M. W., and J. K. Tyler. 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell 21:405-416. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 4.Ai, X., and M. R. Parthun. 2004. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol. Cell 14:195-205. [DOI] [PubMed] [Google Scholar]
- 5.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2004. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol. Cell 14:667-673. [DOI] [PubMed] [Google Scholar]
- 6.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473-1476. [DOI] [PubMed] [Google Scholar]
- 7.Choi, E. S., J. A. Shin, H. S. Kim, and Y. K. Jang. 2005. Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucleic Acids Res. 33:7102-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green, C. M., and G. Almouzni. 2003. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 22:5163-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, E. M., A. J. Antczak, A. O. Bailey, A. A. Franco, K. J. Wu, J. R. Yates III, and P. D. Kaufman. 2005. Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregory, P. D., and W. Horz. 1999. Mapping chromatin structure in yeast. Methods Enzymol. 304:365-376. [DOI] [PubMed] [Google Scholar]
- 11.Hays, S. M., J. Swanson, and E. U. Selker. 2002. Identification and characterization of the genes encoding the core histones and histone variants of Neurospora crassa. Genetics 160:961-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoek, M., and B. Stillman. 2003. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA 100:12183-12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, S., H. Zhou, D. Katzmann, M. Hochstrasser, E. Atanasova, and Z. Zhang. 2005. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc. Natl. Acad. Sci. USA 102:13410-13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson, V. 1990. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry 29:719-731. [DOI] [PubMed] [Google Scholar]
- 15.Jackson, V., and R. Chalkley. 1981. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell 23:121-134. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman, P. D., R. Kobayashi, and B. Stillman. 1997. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11:345-357. [DOI] [PubMed] [Google Scholar]
- 18.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 19.Le, S., C. Davis, J. B. Konopka, and R. Sternglanz. 1997. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13:1029-1042. [DOI] [PubMed] [Google Scholar]
- 20.Ling, X., T. A. Harkness, M. C. Schultz, G. Fisher-Adams, and M. Grunstein. 1996. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 10:686-699. [DOI] [PubMed] [Google Scholar]
- 21.Linger, J., and J. K. Tyler. 2005. The yeast histone chaperone chromatin assembly factor 1 protects against double-strand DNA-damaging agents. Genetics 171:1513-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 23.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 24.Mann, R. K., and M. Grunstein. 1992. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 11:3297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meeks-Wagner, D., and L. H. Hartwell. 1986. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44:43-52. [DOI] [PubMed] [Google Scholar]
- 26.Mello, J. A., and G. Almouzni. 2001. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11:136-141. [DOI] [PubMed] [Google Scholar]
- 27.Mito, Y., J. G. Henikoff, and S. Henikoff. 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 37:1090-1097. [DOI] [PubMed] [Google Scholar]
- 28.Moshkin, Y. M., J. A. Armstrong, R. K. Maeda, J. W. Tamkun, P. Verrijzer, J. A. Kennison, and F. Karch. 2002. Histone chaperone ASF1 cooperates with the Brahma chromatin-remodelling machinery. Genes Dev. 16:2621-2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nabatiyan, A., and T. Krude. 2004. Silencing of chromatin assembly factor 1 in human cells leads to cell death and loss of chromatin assembly during DNA synthesis. Mol. Cell. Biol. 24:2853-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramey, C. J., S. Howar, M. Adkins, J. Linger, J. Spicer, and J. K. Tyler. 2004. Activation of the DNA damage checkpoint in yeast lacking the histone chaperone anti-silencing function 1. Mol. Cell. Biol. 24:10313-10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 32.Roberts, C., H. F. Sutherland, H. Farmer, W. Kimber, S. Halford, A. Carey, J. M. Brickman, A. Wynshaw-Boris, and P. J. Scambler. 2002. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell. Biol. 22:2318-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanematsu, F., Y. Takami, H. K. Barman, T. Fukagawa, T. Ono, K. Shibahara, and T. Nakayama. 2006. Asf1 is required for viability and chromatin assembly during DNA replication in vertebrate cells. J. Biol. Chem. 281:13817-13827. [DOI] [PubMed] [Google Scholar]
- 34.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwabish, M. A., and K. Struhl. 2006. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol. Cell 22:415-422. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz, B. E., and K. Ahmad. 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherwood, P. W., S. V.-M. Tsang, and M. A. Osley. 1993. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:28-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stone, E. M., and L. Pillus. 1996. Activation of an MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J. Cell Biol. 135:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 41.Tyler, J. K. 2002. Chromatin assembly. Eur. J. Biochem. 269:2268-2274. [DOI] [PubMed] [Google Scholar]
- 42.Tyler, J. K., C. R. Adams, S. R. Chen, R. Kobayashi, R. T. Kamakaka, and J. T. Kadonaga. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555-560. [DOI] [PubMed] [Google Scholar]
- 43.Wirbelauer, C., O. Bell, and D. Schubeler. 2005. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 19:1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725-737. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, J., J. Herrera-Diaz, and D. S. Gross. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 25:8985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]